Abstract
Earlier studies have demonstrated both p53-dependent and -independent tumor suppressive functions of B56γ-specific protein phosphatase 2A (B56γ-PP2A). In the absence of p53, B56γ-PP2A can inhibit cell proliferation and cell transformation by an unknown mechanism. In the presence of p53, upon DNA damage, a complex including B56γ-PP2A and p53 is formed which leads to Thr55 dephosphorylation of p53, induction of the p53 transcriptional target p21, and inhibition of cell proliferation. Despite its significance in inhibition of cell proliferation, no B56γ mutations have been linked to human cancer to date. In this study, we first differentiate between the p53-dependent and -independent functions of B56γ-PP2A by identifying a domain of the B56γ protein required for interaction with p53. Within this region we identify a B56γ mutation, F395C, in lung cancer that disrupts the B56γ-p53 interaction. More importantly, we show that F395C is unable to promote p53 Thr55 dephosphorylation, transcriptional activation of p21, and the p53-dependent tumor suppressive function of PP2A. This finding provides a mechanistic basis for the p53-dependent and -independent functions of B56γ-PP2A and establishes a critical link between B56γ-PP2A p53-dependent tumor suppressive function and tumorigenesis.
Keywords: p53, PP2A, B56γ, Thr55 phosphorylation, lung cancer
INTRODUCTION
Protein phosphatase 2A (PP2A) is a family of serine/threonine phosphoprotein phosphatases that has been implicated as a potential tumor suppressor. PP2A consists of a heterotrimeric complex composed of a scaffolding (A) subunit, catalytic (C) subunit, and regulatory (B) subunit. The B subunit is postulated to confer substrate specificity to the PP2A holoenzyme (Janssens and Goris, 2001; Schonthal, 2001; Eichorn et al 2009; Virshup and Shenolikar, 2009). The role of PP2A in control of tumor progression is thought to be governed by a small subset of specific B subunits directing PP2A to dephosphorylate and regulate key tumor suppressors or oncogenes (Eichorn et al., 2009; Virshup and Shenolikar, 2009). Indeed, several members of the B56 family have been described as having a role in directing PP2A potential tumor suppressive activity. It has been shown that B56α-PP2A (PPP2R5A) dephosphorylates the c-myc oncogene at Ser62 leading to its inactivation (Arnold and Sears, 2006), B56δ-PP2A (PPP2R5D) dephosphorylates Cdc25 blocking cell cycle progression (Margolis et al., 2006; Forester et al., 2007), and B56γ-PP2A (PPP2R5C) was shown to be important in SV40 mediated cell transformation (Chen et al., 2004). Interestingly, mutational analyses of PP2A genes have yielded data suggesting PP2A Aα and Aβ mutations are present in some cancers (Chen et al., 2005; Esplin et al., 2006; Ruediger et al., 2001a; Ruediger et al., 2001b; Wang et al., 1998). Importantly, the unifying effect of these mutations is loss of interaction with B56 subunit family members. Although one mutation in B56ε (PPP2R5E) gene of the B56 family has been reported to associate with increased risk of soft tissue sarcoma (Grochola et al, 2009), no mutations in the B56α, B56δ and B56γ tumor suppressors have been linked to human cancer to date.
p53 is a very important highly studied tumor suppressor protein that functions primarily as a transcription factor (Vousden and Prives 2009; Kruse and Gu, 2009). In response to DNA damage, p53 is highly post-translationally modified promoting its activation and nuclear localization, whereby it acts as a transcription factor turning on genes responsible for apoptosis or cell cycle arrest including the CDK inhibitor, p21 (Bode and Dong, 2004). Our laboratory has previously demonstrated that in response to DNA damage, B56γ-PP2A dephosphorylates p53 at Thr55 and promotes its transcriptional activation. In addition, we were able to describe both a p53-dependent and –independent tumor suppressive function of B56γ-PP2A (Li et al., 2007; Shouse et al., 2008). These results, along with recent crystallography studies of the B56γ-PP2A complex (Cho and Xu, 2006; Xu et al., 2006), led us to focus our investigations toward identifying the region of B56γ required for interaction with p53 as an attempt to better understand its p53-dependent and –independent functions.
In the present study we demonstrate that a small region of B56γ (aa391-401) is critical for interaction with the tumor suppressor protein p53. Within this region we identify a cancer derived mutation, F395C that disrupts the B56γ-p53 interaction. Although this mutation is only found from one human cancer sample thus far, we were able to go further to show that F395C lacks the ability to promote p53 Thr55 dephosphorylation and transcriptional activation of the p21 gene. Importantly, we demonstrate that F395C is unable to drive the previously described p53-dependent tumor suppressive functions of PP2A either by inhibiting cell proliferation or decreasing anchorage independent growth of tumor derived cells, while retaining its p53-independent functions. Taken together, these results provide evidence that PP2A-mediated activation of p53 may play an important role in prevention of tumor progression.
RESULTS
Identification of a domain of B56γ required for interaction with p53
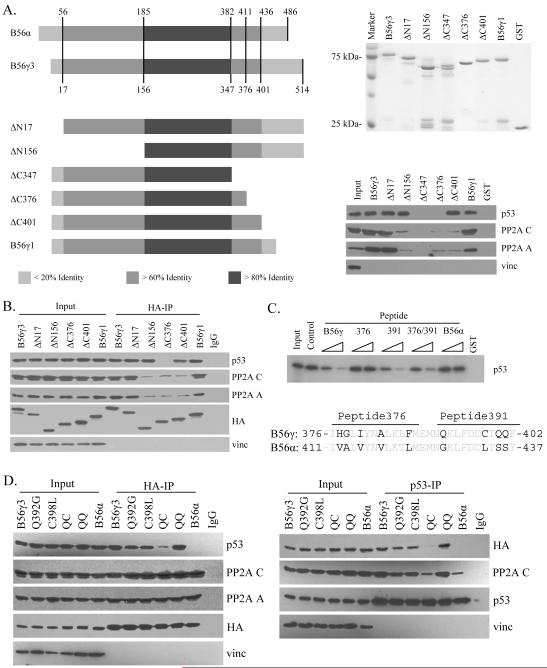
The B56γ gene product directs PP2A phosphatase activity toward p53, promoting p53 Thr55 dephosphorylation, activation, and tumor suppressive function. We sought to better understand the region of the B56γ protein required for interaction with p53. To this end, we compared the amino acid sequences of two different spliced forms of B56γ protein, B56γ1 and B56γ3, able to interact with p53, and the highly related B56α protein, that is unable to interact with p53 (Li et al., 2007). We posited that the more variable regions found toward the C- and N-terminus (Fig. 1A) of B56γ were most likely the areas of potential interaction with p53 due to their variation from the sequence of the non-interacting B56α protein and their position in the potential substrate binding area of the B56γ-PP2A holoenzyme complex as described in crystallographic studies (Cho and Xu, 2006; Xu et al., 2006). To test this, we constructed several B56γ deletion mutants as GST fusion proteins and assayed their ability to interact with the PP2A core and p53 (Fig. 1A). Deletion of the N-terminal 156 amino acids (ΔN156) or the C-terminus after amino acid 401 (ΔC401) disrupted the interaction between B56γ and the AC core of PP2A. Interestingly, although the ΔN156 and ΔC401 mutants were no longer able to interact with the PP2A core, they were still able to bind to p53 to a similar level as wild type B56γ3 and B56γ1. Significantly, the ΔC347 and ΔC376 mutants both lost p53 interaction, suggesting that the deleted region may be required for interaction with p53. To confirm this result, we show that the ΔN156, ΔC376, and ΔC401 mutants also disrupted interaction with the PP2A core in vivo (Fig. 1B). In addition, the ΔC376 mutant lost interaction with p53 in vivo, while the ΔC401 and ΔN156 mutants still maintained interaction (Fig. 1B).
Figure 1.
Mapping of the B56γ domain required for interaction with p53. (A) Left: Diagram of amino acid sequences of B56α, B56γ and various B56γ deletion constructs aligned based on sequence homology. Shades of gray indicate increasing percentage of identical amino acids within that region. Right: Each of the B56γ constructs were expressed in bacteria as GST-fusion proteins, purified, and analyzed by SDS-PAGE (upper). U2OS cell lysates were incubated with the purified GST-fusion proteins indicated, then analyzed by western blot against p53, PP2A A and C, and vinculin (vinc) (lower). (B) U2OS cells were transfected with the HA-tagged B56γ constructs listed, lysed, and subjected to immunoprecipitation with anti-HA antibody. The precipitated proteins were analyzed by western blot against p53, PP2A A and C, HA, and vinculin (vinc). (C) 35S-p53 was incubated with GST protein (GST), GST-B56γ3 protein, or GST-B56γ3 with peptides corresponding to amino acids 376 to 402 (B56γ), 376 to 388 (376), or 391 to 402 of B56γ3 (391), or amino acids 411 to 437 of B56α (B56α). Amino acid sequence of the p53-interaction domain of B56γ aligned with the homologous sequence of B56α is shown below. (D) Lysates of U2OS cells transfected with HA-tagged B56γ3, Q392G, C398L, Q392G/C398L (QC), Q400S/Q401S (QQ) mutant B56γ3, or B56α were either immunoprecipitated with anti-HA antibody, then analyzed by western blot against p53, PP2A A and C, HA, and vinculin (vinc) (left) or with anti-p53 antibody, then analyzed by western blot against HA, PP2A C, p53, and vinculin (vinc) (right).
To exclude the possibility that our result could be explained by a loss of proper conformation due to the deletion, we performed peptide competition experiments utilizing a peptide consisting of the amino acids of B56γ from 376 to 402 (peptide B56γ, Fig. 1C). The assay showed that the B56γ peptide was clearly able to compete out the p53 interaction. An additional smaller peptide containing amino acids 391 to 402 (peptide 391) was also able to block the interaction, while a peptide containing amino acids 376 to 388 (peptide 376) was unable to do so. Combination of both smaller peptides showed similar results to those of peptide 391 only. As a control, a peptide consisting of amino acids from the corresponding region of homology from B56α (peptide B56α) was unable to block the p53 interaction, as expected. These data suggest that the amino acids on B56γ from 391 to 402 constitute a domain required for interaction with p53.
To provide further evidence, we designed and generated four B56γ3 point mutants, Q392G, C398L, the double mutant Q392G/C398L (QC), and the double mutant QQ400/401SS (QQ), where the B56γ amino acids were mutated to the corresponding amino acids present on B56α (Fig. 1C). We assayed for the effects of the mutations on interaction with p53 and the PP2A core in vivo (Fig. 1D). While the interaction was constant between the PP2A core and each of the B subunit constructs expressed, p53 interaction with the QC mutant was almost undetectable and no interaction was detected with the B56α negative control. Interestingly, both Q392 and C398 single mutants retained their ability to interact with p53. To confirm these results, we performed p53 IP followed by immunoblotting with anti-HA antibody and showed that the QC mutant and B56α are indeed unable to interact with p53 (Fig. 1D, right). We previously demonstrated an endogenous interaction between PP2A and p53 that can be strongly enhanced by B56γ3 overexpression (Li et al., 2007). Blotting with the PP2A C subunit showed that the enhanced interaction between p53 and PP2A seen in the presence of overexpressed B56γ3 was lost in the presence of the QC mutant, suggesting that the QC mutant lost its ability to interact with p53. Overall, these results provide additional evidence as to the importance of the region of B56γ between amino acids 376 and 401 for p53 interaction.
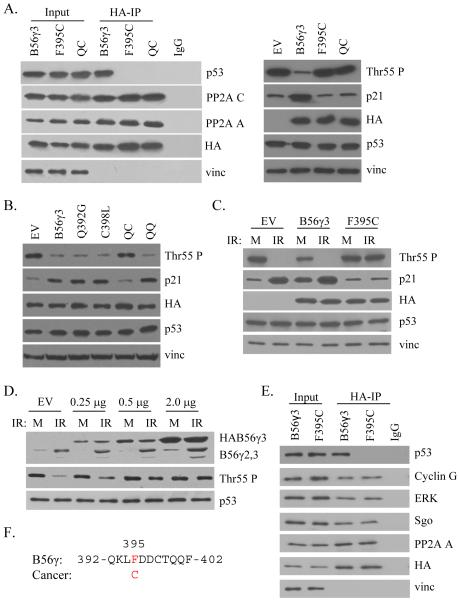
A cancer-derived B56γ3 mutant, F395C, is unable to interact with p53 or promote Thr55 dephosphorylation
To continue to investigate the importance of this region in tumor progression, we analyzed annotated cDNA sequences within our p53 interaction domain present in public databases that were derived from human cancer cell lines and tumor samples and identified a mutation of a phenylalanine residue at amino acid position 395 of B56γ3 that was mutated to cysteine present in a lung tumor sample (Neto et al., 2006). Since this mutation was present within our identified p53 interaction domain (Fig. 2A), we tested whether this mutation could disrupt interaction with p53. Although the F395C mutant was able to interact normally with the PP2A A and C subunits, it specifically lost its ability to interact with p53 (Fig. 2A). The interaction with the QC mutant was shown as a negative control. To provide further evidence for specificity, we tested the ability of F395C to bind to several other PP2A substrates, such as Cyclin G2 (Bennin et al., 2002), ERK (Letourneux et al., 2006), Shugosin (Riedel et al., 2006). As shown in Fig. 2E, F395C, although defect in p53 binding, retains its ability to bind to Cyclin G2, ERK and Shugosin. Together, these results suggest that the F395C mutant was unable to specifically interact with p53 and thus unable to recruit the PP2A core to p53.
Figure 2.
Mutant B56γ3 is unable to promote p53 Thr55 dephosphorylation. (A) Lysates of U2OS cells transfected with HA-tagged B56γ3, F395C, or Q392G/C398L (QC) mutant B56γ3, were either immunoprecipitated with anti-HA antibody, then analyzed by western blot against p53, PP2A A and C, HA, and vinculin (vinc) (left); or analyzed for p53 Thr55 phosphorylation, p21 protein levels, HA, p53, and vinculin (vinc) (right). Amino acid sequence of the p53-interaction domain of B56γ showing the cancer-derived F395C mutation is present on the top. (B) Lysates of U2OS cells transfected with empty vector control (EV), HA-tagged B56γ3, Q392G, C398L, Q392G/C398L (QC), or Q400S/Q401S (QQ) mutant B56γ3 were analyzed for p53 Thr55 phosphorylation, p21 protein levels, HA, p53, and vinculin (vinc). (C) Lysates of U2OS cells transfected with empty vector control (EV), HA-tagged B56γ3, or F395C mutant B56γ3 and either mock treated (M) or treated with ionizing radiation (IR) were analyzed for p53 Thr55 phosphorylation, HA, p53, and vinculin (vinc). (D) Lysates of U2OS cells transfected with varying amounts of HA-tagged F395C mutant B56γ3, were either mock (M) treated or treated with ionizing radiation (IR), then analyzed by western blot against endogenous (endo) and exogenous (exo) B56γ, as well as p53 and Thr55 phosphorylation. (E) Lysates of U2OS cells transfected with HA-tagged B56γ3 or F395C were immunoprecipitated with anti-HA antibody, then analyzed by western blot against p53, Cyclin G, ERK, Sgo, PP2A A, HA, and vinculin (vinc) antibodies.
To test this directly, we assayed the ability of the F395C mutant to promote p53 Thr55 dephosphorylation and p21 induction. As shown in Figure 2A, overexpression of wild-type B56γ3 promoted efficient dephosphorylation of p53 at Thr55, compared to the empty vector control. This dephosphorylation correlated with an induction of the p53 transcriptional target p21, underscoring the functional significance of the dephosphorylation. Overexpression of the F395C mutant, however, was unable to promote p53 Thr55 dephosphorylation or p21 induction, suggesting that this mutant has lost its ability to direct PP2A phosphatase activity toward p53. In addition, the QC mutant was also unable to promote Thr55 dephosphorylation or p21 induction, providing evidence that interaction with p53 is required for the tumor suppressive function of B56γ-PP2A. In addition, as shown in Figure 2B, mutants able to interact with p53, including Q392G, C398L, and the QQ double mutant, promoted p53 Thr55 dephosphorylation and induction of p21 protein to similar levels as wild-type B56γ3. Taken together with the interaction results, we have demonstrated that a cancer derived mutation of B56γ3 may disrupt a physiologically significant function of PP2A in preventing tumor progression through activation of p53.
To investigate the potential for the F395C mutant to block the function of the endogenous B56γ protein, we assayed for p53 Thr55 dephosphorylation and p21 induction after DNA damage in the presence of exogenous wild-type B56γ3, F395C mutant B56γ3 or an empty vector control (Fig. 2C). In the control case, efficient Thr55 dephosphorylation and induction of p21 protein occurs after DNA damage. In the presence of exogenous wild type B56γ3, Thr55 phosphorylation levels are already lower under mock conditions, and further dephosphorylation is observed after IR, while p21 levels are higher in the mock lane and increase further after IR. Importantly, in the presence of the F395C mutant, Thr55 phosphorylation levels do not decrease after DNA damage and induction of the p53 transcriptional target p21 is also blocked. This result raises the question whether the F395C mutant could potentially function as a dominant negative mutant when present at a level similar to the endogenous wild type protein. To test this possibility, we titrated the amount of overexpressed protein and assayed for the effect on Thr55 phosphorylation after DNA damage (Fig. 2D). Decreasing the level of F395C expression to a level similar to endogenous protein (0.25 to 0.5 μg) allowed for some Thr55 dephosphorylation after DNA damage, while higher levels of F395C expression completely blocked this function. This finding suggests that F395C may be able to function as a dominant negative mutant by competing with wild type B56γ for binding to the PP2A core, thereby blocking C subunit-substrate interactions. To further support this view, we assayed for the ability of either wild type or F395C mutant B56γ3 to interact with the endogenous B56γ protein (Fig. S1). No interaction was detected between either exogenous protein and the endogenous B56γ, suggesting that the inhibitory mechanism of F395C overexpression is probably through competitive binding to the AC core of PP2A or some unknown factors.
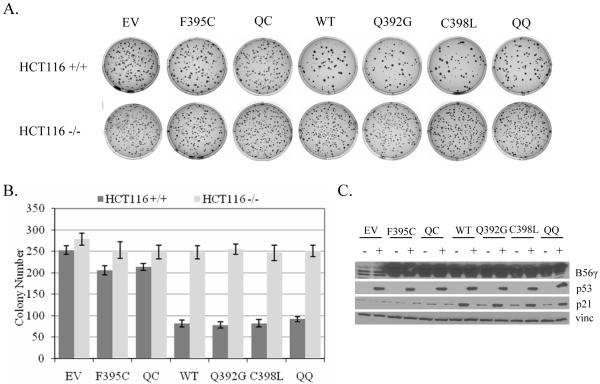
Cancer derived mutant B56γ3 lacks p53-dependent tumor suppressive functions
Although the F395C mutation was found in a tumor sample, we wanted to elucidate whether the mutation could directly contribute to a cancer phenotype. We previously showed that wild type B56γ overexpression decreased anchorage independent growth and cell proliferation rates utilizing a p53-dependent mechanism (Li et al., 2007). Based on our findings that F395C mutant B56γ3 lacks interaction with p53 and ability to promote p53 activation, we first tested whether it could function to block anchorage independent cell growth in a p53-dependent manner. HCT116 cells with either a p53−/− or p53 wild type background were transfected with each of the B56γ3 constructs, including F395C, QC, wild type, Q392G, C398L, and QQ, or an empty vector control and seeded in soft agar. As shown in Figure 3, overexpression of each of the B56γ3 constructs into the HCT116 cells lacking p53 led to a similar decrease in the number of colonies present in the agar from about 280 colonies in the empty vector control to about 250 colonies. In the presence of p53, however, only those B56γ3 constructs able to interact with p53, namely wild type, Q392G, C398L, and QQ, showed a more dramatic decrease in colony number from about 250 colonies in the empty vector control to around 75 colonies. Expression of the F395C mutant led to a decrease in colony number, but only to a similar degree seen in the absence of p53 corresponding to around 210 colonies. Statistical analysis of the data is shown in Figure S2. This finding suggests that F395C mutant B56γ3 is no longer able to function as a p53-dependent tumor suppressor protein. Samples from the transfected cells were analyzed for p21 induction. A strong correlation was demonstrated between the ability of a B56γ3 construct to interact with p53, promote p21 induction, and the ability of that construct to block anchorage independent growth in a p53-dependent manner.
Figure 3.
Overexpression of cancer derived mutant B56γ3 is unable to block anchorage independent growth in a p53-dependent manner. (A) Anchorage-independent growth of HCT116 cells transfected with wild type (WT), F395C, Q392G/C398L (QC), Q392G, C398L, Q400S/Q401S (QQ), or an empty CMV vector control (EV). (B) Colony numbers. The values are the averages +/− standard deviations of the results of three representative experiments. (C) Immunoblots of the transfected HA-B56γ3 and endogenous B56γ3/B56γ2, p53, p21, and vinculin (vinc) proteins. -: HCT116 −/−; +: HCT116 +/+.
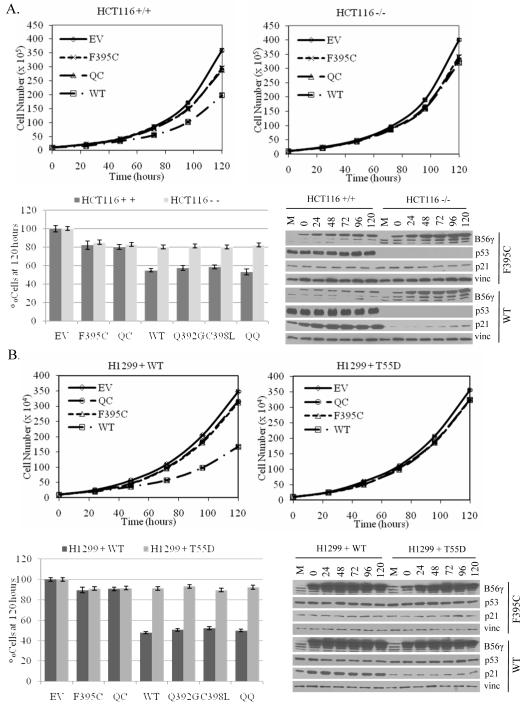
To provide further evidence that F395C mutant B56γ3 lacks p53-dependent tumor suppressive function, we tested whether expression of this mutant protein could inhibit cell proliferation in a p53-dependent manner. As shown in Figure 4A, expression of wild type B56γ3 in the presence of p53 led to a dramatic decrease in cell proliferation corresponding to a 45% reduction in cell number by 120 hours. Interestingly, the F395C mutant was unable to block cell proliferation to a similar degree with only a 20% decrease in cell number. The reduced inhibition of cell proliferation observed in the presence of the F395C mutant can be attributed to its p53-independent function as expression of the mutant in an isogenic p53 null background (HCT116 −/−) led to a decrease that showed no significant difference from wild type B56γ3 (p = 0.08) (Fig. 4A: Fig. S2). In order to provide further data correlating p53 interaction with p53-dependent functions of B56γ3, we expressed the non-interacting QC mutant, as well as the interacting Q392G, C398L, and QQ mutants in the same assay. The non-interacting QC mutant showed similar results to the F395C mutant, while those mutants that retained p53 interaction also retained similar p53-dependent functions comparable to wild type B56γ3 (Fig. 4A). As a control, the lysates of the cell proliferation experiment were analyzed. As expected, increase in p21 protein levels occurred in the case of wild type B56γ3 but not F395C overexpression in HCT116 +/+ but not HCT116 −/− cells, which correlated with the p53-dependent inhibition of cell proliferation (Fig. 4A).
Figure 4.
Overexpression of cancer derived mutant B56γ3 is unable to block cell proliferation in a p53-dependent manner. Representatives of cell proliferation of the HCT116 human colon cancer cell lines (A) or the H1299 human lung cancer cell line (B) transfected with either wild type (WT), F395C, Q392G/C398L (QC), Q392G, C398L, Q400S/Q401S (QQ) mutant B56γ3, or a control CMV empty vector. Numbers of cells present at the 120 hour time point were normalized against the representative empty vector controls and plotted in a bar graph. Error bars show average +/− s.d. from triplicate plates in one representative experiment. Cell lysates were analyzed by immunoblotting of the transfected HA-B56γ3, endogenous B56γ3/B56γ2, p53 and p21 proteins. H1299 + WT: H1299 transfected with wild-type p53; H1299 + T55D: H1299 transfected with T55D. M: empty vector-transfected cell lysate at seeding.
In order to provide evidence that p53 Thr55 phosphorylation status was contributing to the p53-dependent tumor suppressive function of B56γ, similar cell proliferation experiments were performed in H1299 cells that lack functional wild type p53. These cells were transfected with either wild type p53 or a Thr55 to Asp mutant (T55D) that cannot be dephosphorylated at Thr55. As shown in Figure 4B, expression of wild type B56γ3 along with wild type p53 led to a significant inhibition of cell proliferation corresponding to a 50% decrease in cell number at 120 hours. Expression of F395C mutant B56γ3, however, led to only a small inhibition of cell proliferation corresponding to about a 10% decrease in cell number. This small inhibition can be attributed to the Thr55 dephosphorylation-independent function, as a similar inhibition was observed in the presence of T55D mutant p53. Likewise, expression of the non-interacting QC mutant also led to a small inhibition of cell proliferation supporting the notion that p53 interaction and Thr55 dephosphorylation are required for the p53-dependent tumor suppressive function of B56γ3 (Fig. 4B). Statistical analysis of the data is shown in Figure S2. Cell lysates from the experiment were subjected to western blot against to assess p21 protein levels (Fig. 4B). The correlation between p21 induction and decrease in cell proliferation was again documented in the presence of wild type p53 and B56γ3. The lack of p21 induction in the presence of the F395C mutant B56γ3 provides further evidence that this protein has lost its Thr55 dephosphorylation-dependent tumor suppressive function. Taken together our findings demonstrate for the first time that a cancer derived mutant of B56γ3 lacks the physiologically significant p53-dependent tumor suppressive function, providing evidence for the importance of this pathway in protecting cells from a cancerous phenotype.
DISCUSSION
In the present study, we describe a novel molecular mechanism behind a cancer derived loss of function mutation in the PP2A B56γ regulatory subunit gene. This mutation was previously described in a lung tumor sample, although the functional significance of this mutation in tumorigenesis was not previously characterized. The missense mutation changes a phenylalanine residue at amino acid 395 to a cysteine within a region we mapped as being required for B56γ-PP2A-p53 interaction. According to recent crystal structure data, this region maps to an area of high structural variability positioned in such a manner to allow docking of the PP2A core with the substrate (Cho and Xu, 2006; Xu et al., 2006). This provides further support to the notion that this region directly bridges the p53-PP2A interaction. In terms of tumorigenic function, we showed that the mutant protein has lost its ability to mediate the p53-dependent tumor suppressive function of PP2A. Specifically we demonstrated that F395C mutant B56γ3 is no longer able to interact with p53 or inhibit cultured cancer cell proliferation or growth on soft agar in a p53-dependent manner. Unfortunately, we are unable to assess p53 status in this identified cancer sample. However, recent data suggests that as many as 61% of lung tumors have wild type p53 (Petitjean et al., 2007), suggesting the importance of the B56γ3-p53 interaction in regulating wild type p53 tumor suppression function.
Although the F395C mutant protein lacks p53 interaction, it maintains its ability to interact with the PP2A core similar to the wild type protein (Fig. 2A). As such, we were able to demonstrate a competitive effect of F395C expression affecting the activity of the endogenous protein. It appears that even a single copy of this mutation may affect the function of the wild type protein, because when levels of F395C expression are comparable to endogenous levels, there is a decrease in p53 Thr55 dephosphorylation after DNA damage (Fig. 2D). The mechanism for this interference appears to be due to competition between the mutant and wild type proteins for some other molecules required for function, as increasing the amount of F395C protein expressed enhances the effect. The molecule being titrated out may be the PP2A core complex since both F395C and wild type B56γ can bind the PP2A core to similar levels. In addition, it does not appear as though the B56γ protein can dimerize because HA-B56γ3 was unable to interact with endogenous protein (Fig. S1). These findings suggest that the mechanism for F395C interference with wild type function is not dominant negative in the classical sense but due more to competitive binding to other molecules.
Our investigation of the functional consequences of a previously described cancer derived mutation in the B56γ gene provides additional evidence as to the importance of B56γ in tumor suppression. Although the mutation was identified in a lung tumor sample cDNA library, the mutation was not present in the B56γ gene sequence of normal lung tissue libraries present on the NIH database, suggesting that this mutation may have contributed to the phenotype of the tumor sample. In addition, since the mutation was not present in the NCBI SNP database, it is not likely to be a polymorphism. Although there was only one mutation found on the NIH database directly within the mapped p53 interaction domain, several mutations have been identified immediately outside this region that could potentially disrupt the p53-B56γ interaction. We thus tested one of the mutants, A383G, which is immediately adjacent to the mapped p53 interaction domain (aa391-401; Suzuki et al, 2004). Our results suggest that A383G also specifically lost its ability to interact with p53 (Fig. S3B). Furthermore, we showed that A383G is unable to promote p53 Thr55 dephosphorylation (Fig. S3C) and p53-dependent tumor suppressive function of PP2A (Fig. S3D). This finding supports our conclusion that interaction with p53 plays an important role in PP2A-mediated activation of p53 and prevention of tumor progression. Perhaps it is worthwhile to mention that additional mutations have been detected in the B56γ gene. For example, mutations that disrupt binding to the A and C subunits would potentially disrupt both the p53-dependent and independent tumor suppressive functions of this protein and may therefore be more common. Also, the previously described mutations in the A subunit genes that contribute to tumorigenesis may function to disrupt the same pathway as this B56γ mutation, therefore making B56γ mutations in these cancer types less likely. Nevertheless, our study provides compelling biochemical data suggesting the importance of the F395C mutation and potentially other mutants in that expression of the mutant proteins is no longer able to promote PP2A-dependent activation of the tumor suppressor protein p53 and actually disrupts the normal p53 response to DNA damage stress.
Although the cancer-derived F395C mutant protein no longer promotes a p53-dependent tumor suppressive function, it still demonstrates a less dramatic p53-independent function that has been previously described (Li et al., 2007). Interestingly, this mutant protein is able to distinguish between these two and therefore provides compelling evidence as to the ability of the B subunits of PP2A to direct the holoenzyme to specific substrate targets and the potential for the presence of multiple substrate binding domains within a single B subunit. Clearly, this subunit must promote PP2A-mediated dephosphorylation of other target proteins inside the cell that mediate this p53-independent function and further study may provide insight into these protein targets and their role in PP2A-dependent tumor suppression as well as the domain of B56γ responsible for mediating the interaction with the PP2A substrate. Overall, although the regulation of PP2A functions is complex, our study adds significant support to the role of B56γ-PP2A in tumor suppression specifically through activation of the tumor suppressor protein, p53.
MATERIALS AND METHODS
Identification of cancer derived mutation
The AceView program, developed at NCBI, provides a strictly cDNA-supported view of the human transcriptome and the genes by summarizing all quality-filtered human cDNA data from GenBank, dbEST and the RefSeq. Using this program, we looked for B56γ mutations present within our identified p53 interaction domain where the annotated sequencing data was taken from tumors, tumor samples, and cancer cell lines. Our specific mutant was identified from a lung tumor sample and the corresponding gene accession number is BE829999.
Cell culture and plasmids
U2OS, HCT116 or H1299 cells were cultured in McCoy’s 5A, DMEM or RPMI 1640 medium supplemented with 10% fetal calf serum. The B56γ3 deletion mutants, ΔN17, ΔN156, ΔC347, ΔC376, and ΔC401 were generated by PCR. The B56γ3 point mutants, Q392G, C398L, Q392G/C398L, QQ400/401SS, F395C, were generated using the QuikChange Site-Directed Mutagenesis Kit (Stratagene). All plasmids were verified by sequencing.
Western blot and immunoprecipitation
Whole-cell extract was prepared by lysing the cells in a buffer containing 50 mM Tris–HCl (pH 8.0), 120 mM NaCl, 0.5% NP-40, 1 mM DTT, 2 μg/ml aprotinin and 2 μg/ml leupeptin. Cell lysates were subjected to SDS–PAGE, followed by immunoblotting analysis with anti-p53 (DO1, Santa Cruz Biotechnology), anti-p21 (C-19, Santa Cruz), anti-PP2A A subunit (Upstate), anti-PP2A C subunit (1D6, Upstate), anti-PP2A B56γ3 (against full-length B56γ3), anti-ERK (Santa Cruz Biotechnology), anti-Cyclin G (Santa Cruz Biotechnology), anti-SGOL1 (ABNOVA), or anti-vinculin (VIN-11-5, Sigma) antibodies. For Thr55 dephosphorylation, the cell lysate was immunoprecipitated with phospho-specific antibody for Thr55 (Ab202) and immunoblotted with anti-p53 antibody (Li et al., 2007). For GST pull down experiments, GST fusion proteins were expressed in BL21 bacteria and purified using glutathione sepharose beads. U2OS cell lysate or 35S-labeled p53 was then incubated with the GST protein. Precipitated proteins were then analyzed by SDS-PAGE followed by immunoblotting or autoradiography. The peptide competition experiments were performed by addition of different peptides in the reactions at 100- to 1000-fold molar excess. For interaction of endogenous p53 with the transfected B56 proteins, U2OS cells were transfected with various B56 plasmids using FuGene (Roche) and lysed 28 h after transfection. Immunoprecipitation was performed using either anti-p53 polyclonal antibody (FL393, Santa Cruz) or anti-HA 12CA5 monoclonal antibody. The amounts of co-precipitated proteins were determined by immunoblotting.
Cell proliferation and anchorage-independent growth assays
To generate proliferation curves for HCT116 cells, cells were transfected with wild type, QC, F395C, Q392G, C398L, or QQ mutant B56γ3 or a control CMV empty vector using Fugene, seeded in triplicate and counted at 0, 24, 48, 72, 96 and 120 h post seeding. To generate proliferation curves for H1299 cells, cells were cotransfected with wild type, QC, F395C, Q392G, C398L, or QQ mutant B56γ3 or a control CMV empty vector along with either wild type p53 or T55D mutant p53, using Fugene, seeded in triplicate and counted at 0, 24, 48, 72, 96 and 120 h post seeding. For anchorage-independent growth assays of HCT116 cells, cells were transfected with wild type, Q392G, C398L, QC, QQ, or F395C mutant B56γ3 or a control CMV empty vector seeded in triplicate in 0.35% Noble Agar (Fisher) and counted 4 weeks post seeding.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Dr. B. Shen at City of Hope for assistance searching public databases for mutations in the B56γ gene and to P. Podlesny for generating B56γ mutant plasmid constructs. We thank all members of our laboratory for many helpful discussions. This work was supported by NIH grant CA075180 from the National Institute of Cancer.
REFERENCES
- 1.Arnold H, Sears R. Protein phosphatase 2A regulatory subunit B56α associates with c-Myc and negatively regulates c-Myc accumulation. Mol Cell Biol. 2006;26:2832–44. doi: 10.1128/MCB.26.7.2832-2844.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bennin D, Don A, Brake T, McKenzie J, Rosenbaum H, Ortiz L, DePaoli-Roach A, Horne M. Cyclin G2 associates with protein phosphatase 2A catalytic and regulatory B’ subunits in active complexes and induces nuclear aberrations and a G1/S phase cell cycle arrest. J Biol Chem. 2002;277:27449–67. doi: 10.1074/jbc.M111693200. [DOI] [PubMed] [Google Scholar]
- 3.Bode A, Dong Z. Post-translational modification of p53 in tumorigenesis. Nat Rev Cancer. 2004;4:793–805. doi: 10.1038/nrc1455. [DOI] [PubMed] [Google Scholar]
- 4.Chen W, Possemato R, Campbell K, Plattner C, Pallas D, Hahn W. Identification of specific PP2A complexes involved in human cell transformation. Cancer Cell. 2004;5:127–36. doi: 10.1016/s1535-6108(04)00026-1. [DOI] [PubMed] [Google Scholar]
- 5.Chen W, Arroyo J, Timmons J, Possemato R, Hahn W. Cancer-associated PP2A Aα subunits induce functional haploinsufficiency and tumorigenicity. Cancer Res. 2005;85:8183–92. doi: 10.1158/0008-5472.CAN-05-1103. [DOI] [PubMed] [Google Scholar]
- 6.Cho U, Xu W. Crystal structure of a protein phosphatase 2A heterotrimeric holoenzyme. Nature. 2006;445:53–7. doi: 10.1038/nature05351. [DOI] [PubMed] [Google Scholar]
- 7.Eichorn P, Creyghton M, Bernards R. Protein phosphatase 2A regulatory subunits and cancer. Biochim Biophys Acta. 2009;1795:1–15. doi: 10.1016/j.bbcan.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 8.Esplin E, Ramos P, Martinez B, Tomlinson G, Mumby M, Evans G. The Glycine 90 to Aspartate alteration in the Aβ subunit of PP2A (PPP2R1B) associates with breast cancer and causes a deficit in protein function. Genes, Chromosomes and Cancer. 2006;45:182–90. doi: 10.1002/gcc.20284. [DOI] [PubMed] [Google Scholar]
- 9.Forester C, Maddox J, Louis J, Goris J, Virshup D. Control of mitotic exit by PP2A regulation of Cdc25C and Cdk1. Proc Natl Acad Sci USA. 2007;104:19867–72. doi: 10.1073/pnas.0709879104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grochola L, Vazquez A, Bond E, Wurl P, Taubert H, Muller T, et al. Recent natural selection identifies a genetic variant in a regulator subunit of protein phosphatase 2A that associates with altered cancer risk and survival. Clin. Cancer Res. 2009;15:6301–08. doi: 10.1158/1078-0432.CCR-09-0797. [DOI] [PubMed] [Google Scholar]
- 11.Janssens V, Goris J. Protein Phosphatase 2A: a highly regulated family of serine/threonine phosphatases implicated in cell growth and signaling. Biochem J. 2001;353:417–39. doi: 10.1042/0264-6021:3530417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kruse J, Gu W. Modes of p53 regulation. Cell. 2009;137:609–622. doi: 10.1016/j.cell.2009.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Letourneux C, Rocher G, Porteu F. B56-containing PP2A dephosphorylate ERK and their activity is controlled by the early gene IEX-1 and ERK. EMBO J. 2006;25:727–38. doi: 10.1038/sj.emboj.7600980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li H, Cai X, Shouse G, Piluso L, Liu X. A specific PP2A regulatory subunit, B56γ, mediates DNA damage-induced dephosphorylation of p53 at Thr55. EMBO. 2007;26:402–11. doi: 10.1038/sj.emboj.7601519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Margolis S, Perry J, Forester C, Nutt L, Guo M, Jardim M, et al. Role for the PP2A/B56δ Phosphatase in Regulating 14-3-3 Release from Cdc25 to Control Mitosis. Cell. 2006;127:759–63. doi: 10.1016/j.cell.2006.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Neto E, Correa R, Verjovski-Almeida S, Briones M, Nagai M, da Silva W, et al. Shotgun sequencing of the human transcriptome with ORF expressed sequence tags. Proc Natl Acad Sci USA. 2006;97:3491–6. doi: 10.1073/pnas.97.7.3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Petitjean A, Mathe E, Kato S, Ishioka C, Tavtigian S, Hainaut P, et al. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: lessons from recent developments in the IARC TP53 database. Hum Mutat. 2007;6:622–9. doi: 10.1002/humu.20495. [DOI] [PubMed] [Google Scholar]
- 18.Riedel C, Katis V, Katou Y, Mori S, Itoh T, Helmhart W, et al. Protein phosphatase 2A protects centromeric sister chromatid cohesion during meiosis I. Nature. 2006;441:53–61. doi: 10.1038/nature04664. [DOI] [PubMed] [Google Scholar]
- 19.Ruediger R, Pham H, Walter G. Alterations in protein phosphatase 2A subunit interaction in human carcinomas of the lung and colon with mutations in the Aβ subunit gene. Oncogene. 2001;20:1892–99. doi: 10.1038/sj.onc.1204279. [DOI] [PubMed] [Google Scholar]
- 20.Ruediger R, Pham H, Walter G. Disruption of protein phosphatase 2A subunit interaction in human cancers with mutations in the Aα subunit gene. Oncogene. 2001;12:10–15. doi: 10.1038/sj.onc.1204059. [DOI] [PubMed] [Google Scholar]
- 21.Schonthal A. Role of serine/threonine protein phosphatase 2A in cancer. Cancer Letters. 2001;170:1–13. doi: 10.1016/s0304-3835(01)00561-4. [DOI] [PubMed] [Google Scholar]
- 22.Shouse G, Cai X, Liu X. Serine 15 phosphorylation of p53 directs its interaction with B56γ and the tumor suppressor activity of B56γ-specific protein phosphatase 2A. Mol Cell Biol. 2008;28:448–456. doi: 10.1128/MCB.00983-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suzuki Y, Yamashita R, Shirota M, Sakakibara Y, Chiba J, Mizushima-Sugano J, et al. Sequence comparison of human and mouse genes reveals a homologous block structure in the promoter regions. Genome Res. 2004;14:1711–1718. doi: 10.1101/gr.2435604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Virshup D, Shenolikar S. From promiscuity to precision: protein phosphatases get a makeover. Mol. Cell. 2009;33:537–545. doi: 10.1016/j.molcel.2009.02.015. [DOI] [PubMed] [Google Scholar]
- 25.Vousden K, Prives C. Blinded by the light: the growing complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 26.Wang S, Esplin E, Li J, Huang L, Gazdar A, Minna J, Evans G. Alterations of the PPP2R1B gene in human lung and colon cancer. Science. 1998;282:284–7. doi: 10.1126/science.282.5387.284. [DOI] [PubMed] [Google Scholar]
- 27.Xu Y, Xing Y, Chen Y, Chao Y, Lin Z, Fan E, et al. Structure of the protein phosphatase 2A holoenzyme. Cell. 2006;127:1239–51. doi: 10.1016/j.cell.2006.11.033. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.