Abstract
Leukoplakia is the most common premalignant lesion of the oral cavity. Epidermal growth factor receptor (EGFR) abnormalities are associated with oral tumorigenesis and progression. We hypothesized that EGFR expression and gene copy number changes are predictors of the risk of an oral premalignant lesion (OPL) for progressing to oral squamous cell carcinoma (OSCC). A formalin-fixed, paraffin-embedded OPL biopsy specimen was collected from each of 162 patients in a randomized controlled clinical trial. We assessed EGFR expression by immunohistochemistry with two methods: a semi-quantitative analysis (145 evaluable specimens) and an automated quantitative analysis (127 evaluable specimens). EGFR gene copy number was assessed by fluorescence in situ hybridization (FISH) in a subset of 49 OPLs with high EGFR expression defined by the semi-quantitative analysis. We analyzed EGFR abnormalities for associations with OSCC development. High EGFR expression occurred in 103 (71%) of the 145 OPLs and was associated with a nonsignificantly higher risk of OSCC (P = 0.10). Twenty (41%) of 49 OPLs assessed by FISH had an increased EGFR gene copy number (FISH-positive). Patients with FISH-positive lesions had a significantly higher incidence of OSCC than did patients with FISH-negative (a normal copy number) lesions (P = 0.0007). Of note, 10 of 11 OSCCs that developed at the site of the examined OPL were in the FISH-positive group, leaving only one FISH-negative OPL that did so (P < 0.0001). Our data indicate that an increased EGFR gene copy number is common in and associated with OSCC development in patients with OPLs expressing high EGFR, particularly OSCC developing at the site of a high-expression OPL; they also suggest that EGFR inhibitors may prevent oral cancer in patients with OPLs having an increased EGFR gene copy number.
Keywords: epithelial growth factor receptor, oral cancer, oral leukoplakia, immunohistochemistry, fluorescence in situ hybridization, biomarker
Head and neck squamous cell carcinoma (HNSCC) is second only to lung cancer as the most common smoking-related cancer worldwide. Oral squamous cell carcinoma (OSCC) is the most common anatomic site of HNSCC, accounting for approximately 50% of all HNSCC. Despite the tremendous effort to reduce tobacco use, HNSCC remains one of the leading causes of the approximately 443,000 deaths in the U.S. that were attributable to smoking in 2000–2004 (1). The only standard therapeutic option for earlier-stage HNSCC is surgery, but it is a debilitating, substantially morbid procedure that severely impairs quality of life for many patients. Despite recent progress in developing targeted therapies for patients with recurrent and metastatic disease, their prognosis remains poor (2). In light of its continuing burden and evasion of substantial control, HNSCC requires new approaches including prevention.
Leukoplakia and erythroplakia are the most commonly diagnosed oral premalignant lesions (OPLs), with a 17%–24% rate of malignant transformation over a period of up to 30 years (3-6). OPLs also are associated with hyperkeratosis, dysplasia, or in situ carcinoma. OPL histology has little value for marking the risk of oral SCC (OSCC). Although loss of heterozygosity (LOH) profile (7), podoplanin (8), and p63 expression (9) are associated with an increased risk of OSCC, none of these biomarkers is targetable via currently available drugs.
Epidermal growth factor receptor (EGFR) is believed to play an important role in HNSCC development (10-14); EGFR expression and abnormal gene copy number are associated with a poor prognosis of HNSCC patients (15, 16); and the anti-EGFR antibody cetuximab is approved for treating HNSCC (17, 18). An EGFR inhibitor for HNSCC prevention is being tested currently in a randomized, placebo-controlled trial (19). The role of EGFR as a marker of OSCC risk, however, has not been evaluated previously in a large series of OPL patients. Establishing EGFR as a reliable marker of OSCC risk would allow selecting a higher-risk population with a potentially higher likelihood of benefiting from EGFR inhibitors.
We hypothesized that changes in EGFR protein expression and gene copy number marked the risk of developing OSCC and tested the hypothesis in a large population of OPL patients enrolled in a long-term prospective, randomized controlled trial, the first OPL trial that included long-term oral cancer incidence as a prespecified secondary endpoint (20). We demonstrate that EGFR protein expression and gene copy number may be effective markers of the risk of OPLs for progressing to OSCC.
Patients and Methods
Patients and specimens
All of the 162 randomized and eligible patients who were enrolled in a randomized chemoprevention trial at The University of Texas M. D. Anderson Cancer Center (MDACC) were eligible for this study. From 1992 to 2001, the patients had been diagnosed with OPL and randomly assigned to intervention with 13-cis-retinoic acid (13cRA) versus β-carotene (BC) + retinyl palmitate (RP) versus RP alone. Formalin-fixed, paraffin-embedded biopsy specimens were obtained at enrollment or after enrollment but before any event (defined as the diagnosis of OSCC). Clinical-pathologic parameters were obtained from the clinical trial database. The follow-up data were obtained from a combination of chart review and a telephone interview. More detailed clinical information has been previously described in Papadimitrakopoulou et al. (20). The definition of oral cancer development at the same site as an OPL required the cancer and baseline OPL to be on the same side and in the same anatomical structure of the oral cavity. The study was approved by the institutional review board, and written informed consent was obtained from all patients.
EGFR protein expression
Tissue sections (4 μm thick) from formalin-fixed, paraffin-embedded tissue blocks of OPL were mounted on positively charged glass slides. EGFR immunostaining was performed using the avidin-biotin peroxidase complex (ABC) technique, as described previously (8). Briefly, slides were deparaffinized and rehydrated. To retrieve antigenicity, the slides were steamed with 10 mmol/L citrate buffer (pH, 6.0; DakoCytomation, Carpinteria, CA) for 30 minutes. The slides were then incubated in 10% fetal bovine serum for 30 minutes at room temperature, incubated with monoclonal antibody 31G7 (Zymed Laboratories Inc., South San Francisco, CA, USA) diluted 1/100 for 90 min at room temperature, and subjected to signal development processes using the Vectastain Elite ABC kit according to the manufacturer's protocol (Vector Laboratories, Burlingame, CA). The slides were counterstained with Mayer's hematoxylin (DakoCytomation). One lung SCC sample and one OPL sample known to express high levels of EGFR were used as positive (primary antibody added) and negative (no primary antibody) controls. No staining was observed in negative controls.
For the semi-quantitative evaluation of EGFR expression, each slide was scored for membranous expression as follows: 0, no membrane staining; 1, weak membrane staining in >10% of epithelial cells; 2, intermediate membrane staining in >10% of epithelial cells; 3, intense membrane staining in >10% of epithelial cells. Because no or weak EGFR expression has been described in normal oral mucosa (21, 22), we classified scores 0–1 as low EGFR expression; we classified scores 2–3 as high EGFR expression. All scores were based on examining the whole section in each biopsy under a multiheaded microscope by three observers (M.T.B, P.S., and L.M.), who were blinded to clinical patient information.
A second evaluation via an automated analysis system provided an independent, blinded quantitative assessment of EGFR expression. Entire sections were scanned with an Olympus BX61 microscope. Images were acquired and analyzed in the Ariol SL-50 image analyses software. Cytoplasmic and membrane staining were assessed. Composite scores of membrane staining and cytoplasmic staining were obtained by multiplying the number of positive cells by the staining intensity. The sum of the membrane and cytoplasmic scores was used to generate a total composite score. Distribution plots showed that transforming the composite score to its square root divided by 10,000 stabilized variance and brought the data closer to the Gaussian distribution. Therefore, the transformed composite score was used for the final analysis.
EGFR gene copy number
We evaluated EGFR copy number via fluorescence in situ hybridization (FISH), as described previously (23) and in brief here. Tissue sections (4 μm thick) from formalin-fixed, paraffin-embedded tissue blocks were deparaffinized in citrazol washes, digested with proteinase K and incubated in denature solution. Sections were then hybridized using the dual-target, dual-color LSI EGFR SpectrumOrange/CEP 7 SpectrumGreen probe (Vysis Inc., Downers Grove, IL); CEP 7 SpectrumGreen targets the chromosome 7 centromere and serves as a control for copy-number normalization. The analysis was performed on a BX61 brightfield and epifluorescence microscope (Olympus Bx61, Olympus America, Lake Success, NY) equipped with the Quips XL genetic workstation (Applied Imaging, Santa Clara, CA). The EGFR sequence was visualized with a Texas red filter; the chromosome 7 centromere sequence was visualized with a fluorescein isothiocyanate (FITC) filter; and the nuclei were identified with a DAPI filter. Double (FITC and Texas red) and triple band pass filters (DAPI, FITC, and Texas red; Chroma Technology, Brattleboro, VT) were also used. Representative images of each specimen were acquired with a SenSys cooled CCD camera (Photometrics, Tucson, AZ) in monochromatic layers that were subsequently merged by the SmartCapture software (Vysis).
At least 100 non-overlapping interphase nuclei from whole samples were scored by two independent observers (M.T.B. and P.S.) blinded to clinical information. The number of copies of EGFR probes was assessed independently from that of chromosome 7 probes. Copies of probes for EGFR are usually equal to and balanced in number with copies of probes for chromosome 7, except in the case of EGFR amplification, defined by clustered unbalanced gains of EGFR, or a high ratio of EGFR copy number to chromosome 7 copy number. Patients were classified according to the 6 following and previously described FISH patterns of balanced or unbalanced EGFR and chromosome 7 copy numbers (24): a) balanced disomy (in more than 90% of cells); b) low, balanced trisomy (10% to 40% of cells with 3 copies and < 10% of cells with 4 copies); c) high, balanced trisomy (≥ 40% of cells with 3 copies and < 10% of cells with 4 copies; d) low, balanced polysomy (10% to 40 % of cells with 4 copies); e) high, balanced polysomy (≥ 40% of cells with 4 copies); and f) EGFR amplification (clustered unbalanced gain of EGFR). Because of the preinvasive nature of our samples, we expected an extremely low frequency of polysomy (pattern e) and gene amplification (pattern f) compared with what has been reported for HNSCC. Therefore, our definition of FISH positivity (an increased EGFR gene copy number) for OPLs was expanded (beyond HNSCC definitions) to include any one of patterns “b” through “e” (any increase in EGFR gene and chromosome 7 copy number); pattern “a” was considered to reflect a normal EGFR gene copy number.
Statistical methods
The associations between the biomarker expressions, protocol response, and other patient prognostic factors were tested using the Chi-square test or Fisher's exact test for categorical variables and the Wilcoxon rank sum test for continuous variables. Time to event, such as time to death or time to oral cancer development, was calculated from the treatment randomization date to the event date or last follow-up date if no event had been recorded. The Kaplan-Meier method was used to estimate the event-free rate. The median time to event with 95% confidence intervals (CIs) and the event-free rates at years 5 and 10 with 95% CIs by prognostic factors were provided. The log-rank test was used to compare the difference in survival between the prognostic factor groups. Cox proportional hazard models were utilized for multicovariate analysis. Hazard ratios with 95% CIs and P-values were reported. Martingale residual plots were used to visually examine the nature of the relationship between the residuals from a null Cox proportional hazard model (without covariates) and the transformed composite EGFR scores. All tests are two-sided, and P-values less than 0.05 are considered statistically significant.
Results
Patient characteristics
Biopsies from 17 (11%) of the 162 patients enrolled in the chemoprevention trial were excluded from the analyses because of a lack of sufficient tissue in blocks (N = 12) or the absence of evaluable epithelial cells in the H&E section (N = 5). In 18 (12%) of the 145 remaining patients, biopsy specimens were obtained after enrollment because the baseline-biopsy paraffin blocks were unavailable. Median follow-up was 7.5 years, with 35 (24%) of the 145 patients developed oral cancer. Seventeen oral cancers developed at the site of a baseline OPL, and 18 oral cancers developed at a site that was contralateral to and/or different from the site of any baseline OPL. The study population was balanced for gender; the majority of the patients were white, with current or former smoking and alcohol history. Half of the patients received 13cRA, and half received RP or RP plus BC. Two-thirds of the OPLs were classified as hyperplasia, one-third as dysplasia of various degrees.
Semi-quantitative EGFR protein expression in OPLs
Analyzed in 145 specimens, EGFR expression was mostly membranous, predominant in the basal layers of the epithelium, and observed in the vast majority of samples. A total of 42 (29%) OPLs were scored 0 or 1—17 (12%) 0s and 25 (17%) 1s—and thus were considered to have a low EGFR expression. A total of 103 OPLs (71%) were scored 2 or 3—51 (35%) 2s and 52 (36%) 3s—and thus were considered to have a high EGFR expression. In most cases, EGFR expression was homogeneous in the epithelium. Examples of EGFR expression are shown in Fig. 1A–1C.
Fig. 1.
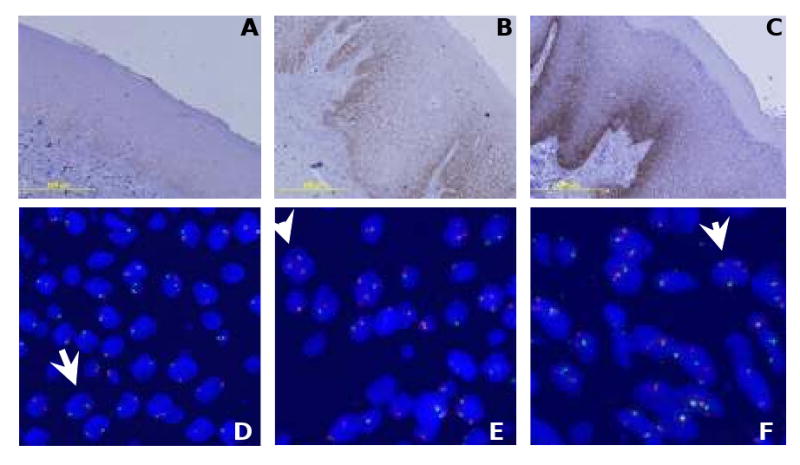
EGFR protein expression and gene copy number in oral leukoplakia (OPL). OPL with EGFR expression scored 1+ (A), 2+ (B), and 3+ (C). Of the 49 patients considered for EGFR gene copy number, disomy (≤ 2 copies in >90% of cells) was observed in 29 (59%) OPLs (D), low trisomy (3 copies in 10 to 40% of cells) in 14 (29%) OPLs (E), high trisomy (≥ 40% of cells with 3 copies and < 10% of cells with 4 copies) in one (2%) case, low polysomy (10% to 40 % of cells with 4 copies) in 4 (8%) OPLs (F), and gene amplification (clustered unbalanced gain of EGFR) in one (2%) case.
Semi-quantitative EGFR expression and clinical-pathologic parameters
High EGFR expression was more frequent in OPLs from females (79.4%) versus males (63.6%, P = 0.03), whites (73.6%) versus non-whites (50%, P = 0.04), and older versus younger patients (high expression in a median age of 59 years versus low expression in a median age of 49 years; P = 0.003). There was no association between EGFR expression and histologic status, smoking history, history of alcoholic consumption, or treatment arm.
EGFR protein expression and oral cancer risk
A trend between OPLs with high EGFR expression (scored 2 or 3) and a higher risk of oral cancer development was observed in the univariate analysis (Table 1 and Fig. 2A), which also showed that oral cancer development had no significant association with sex, race, age, smoking, or alcohol history and had a borderline association with OPL histologic status (P = 0.06). In a multicovariate analysis, neither OPL histology at baseline nor EGFR expression was significantly associated with time to oral cancer (TTOC; data not shown).
Table 1.
The median time (years) to oral cancer development and oral cancer free (OCF) rates at years 5 and 10 for all patients (N=145)
| Variable | Level | N | Event | Median (95% CI) | OCF Rate at 5 Years (95% CI) | OCF Rate at 10 Years (95% CI) | P-value¶ |
|---|---|---|---|---|---|---|---|
| All patients | 145 | 35 | NA (NA, NA) | 0.8 (0.74, 0.87) | 0.72 (0.65, 0.81) | ||
| Sex | Female | 68 | 16 | NA (NA, NA) | 0.78 (0.69, 0.89) | 0.73 (0.62, 0.86) | 0.843 |
| Male | 77 | 19 | NA (NA, NA) | 0.82 (0.74, 0.92) | 0.72 (0.62, 0.84) | ||
| Race | Other | 16 | 3 | NA (NA, NA) | 0.85 (0.67, 1) | 0.75 (0.54, 1) | 0.719 |
| White | 129 | 32 | NA (NA, NA) | 0.8 (0.73, 0.87) | 0.72 (0.64, 0.81) | ||
| Histologic status | Hyperplasia | 97 | 19 | NA (NA, NA) | 0.85 (0.78, 0.92) | 0.78 (0.7, 0.88) | 0.064 |
| Mild dysplasia | 37 | 11 | NA (6.65, NA) | 0.74 (0.61, 0.9) | 0.65 (0.5, 0.85) | ||
| Moderate/severe dysplasia | 11 | 5 | 6.67 (1.85, NA) | 0.61 (0.38, 1) | 0.46 (0.22, 0.97) | ||
| Treatment arm | 13cRA | 73 | 18 | NA (NA, NA) | 0.78 (0.69, 0.89) | 0.72 (0.61, 0.84) | 0.565 |
| BC + RP | 40 | 8 | NA (NA, NA) | 0.84 (0.73, 0.97) | 0.8 (0.68, 0.95) | ||
| RP | 32 | 9 | NA (6.65, NA) | 0.8 (0.67, 0.96) | |||
| Smoking history | Current | 51 | 7 | NA (NA, NA) | 0.87 (0.78, 0.97) | 0.83 (0.72, 0.96) | 0.099 |
| Former | 58 | 20 | NA (6.65, NA) | 0.75 (0.64, 0.87) | 0.62 (0.5, 0.78) | ||
| Never | 36 | 8 | NA (NA, NA) | 0.8 (0.68, 0.94) | 0.76 (0.63, 0.92) | ||
| Alcohol history | Current | 80 | 18 | NA (NA, NA) | 0.84 (0.76, 0.93) | 0.74 (0.64, 0.86) | 0.783 |
| Former | 19 | 5 | NA (6.21, NA) | 0.78 (0.61, 1) | 0.68 (0.48, 0.98) | ||
| Never | 46 | 12 | NA (NA, NA) | 0.75 (0.63, 0.89) | 0.72 (0.6, 0.87) | ||
| EGFR protein expression§ | low | 42 | 6 | NA (NA, NA) | 0.89 (0.8, 1) | 0.82 (0.69, 0.96) | 0.105 |
| high | 103 | 29 | NA (10.7, NA) | 0.77 (0.69, 0.86) | 0.69 (0.6, 0.8) | ||
Abbreviations: CI, confidence interval; NA, not applicable; 13cRA, 13-cis-retinoic acid; BC, β-carotene; RP, retinyl palmitate; EGFR, epidermal growth factor receptor.
P-values are from the log-rank test (univariate analysis)
Semiquantitative evaluation; EGFR low: score 0-1 at immunohistochemistry; EGFR high: score 2-3 at immunohistochemistry
Fig. 2.
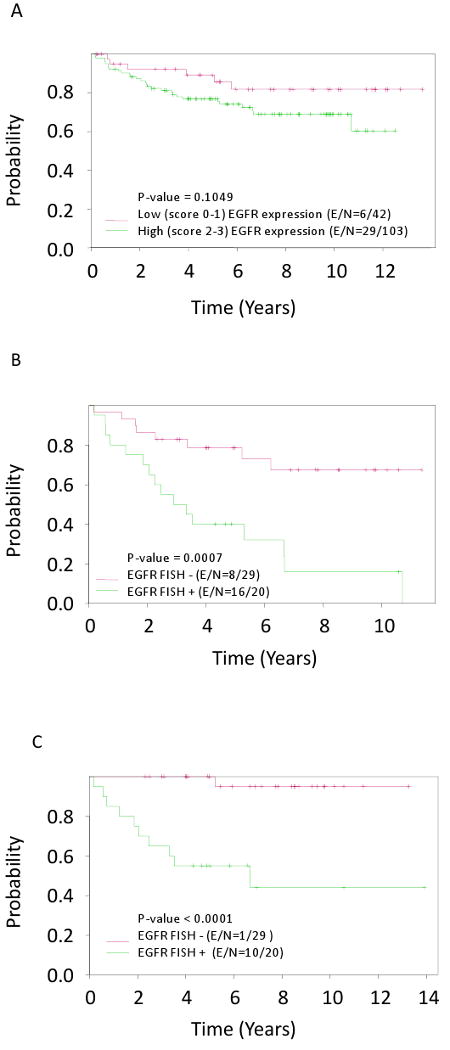
Time to oral cancer in oral leukoplakias (OPL) by EGFR protein expression scored 0-1+ versus 2-3+ (A); Time to oral cancer (B) and time to same site oral cancer (C) in OPL by EGRF gene copy number. E/N: number of events and number of patients.
To further study the association between EGFR expression and oral cancer risk, we quantitatively evaluated EGFR expression using an automated analysis system in 127 samples. Increased total transformed composite EGFR score was significantly associated with oral cancer development, with a hazard ratio of 1.187 (P = 0.012; 95% CI, 1.039–1.356). Increased total transformed composite EGFR score also was significantly associated with oral cancer development in a multicovariate analysis including age, histology at baseline, and treatment arm the (hazard ratio = 1.147; P = 0.036; 95% CI, 1.01-1.30; Table 2). Time to oral cancer was not statistically different between patients with high-EGFR–expression OPLs (defined by median total transformed composite score) and patients with low EGFR-expression OPLs (Fig. 3A). A Martingale residual analysis revealed a linear trend of increasing oral cancer risk beginning with a total transformed composite EGFR score of 7. With a cutoff point at the score 7, time to oral cancer was significantly worse in patients with high EGFR expression (Fig. 3B).
Table 2.
Multicovariate analysis of time to oral cancer on quantitative EGFR expression (E/N = 29/127)*
| Variable | Hazard Ratio | 95% | CI for HR | P-value |
|---|---|---|---|---|
| Age | 1.05 | 1.01 | 1.08 | 0.005 |
| 13cRA vs. BC-RP/RP only | 0.83 | 0.39 | 1.76 | 0.626 |
| Histologic status at baseline: dysplasia versus hyperplasia | 2.99 | 1.42 | 6.29 | 0.004 |
| EGFR transformed total composite score | 1.15 | 1.01 | 1.30 | 0.036 |
Abbreviations: EGFR, epidermal growth factor receptor; E, number of events; N, number of patients; Pr > ChiSq, P-value; HR, hazard ratio; CI, confidence interval; BC, β-carotene; RP: retinyl palmitate; 13cRA, 13-cis-retinoic acid; FISH, fluorescence in situ hybridization.
For this analysis, the Cox proportional models were fitted with other important factors in the models. Composite scores of membrane staining and cytoplasmic staining were obtained by multiplying the number of positive cells by the intensity. The sum of the membrane and cytoplasmic scores was used to generate a total composite EGFR score, which then was transformed by dividing its square root by 10,000. This analysis produced the maximum-likelihood estimates.
Fig. 3.
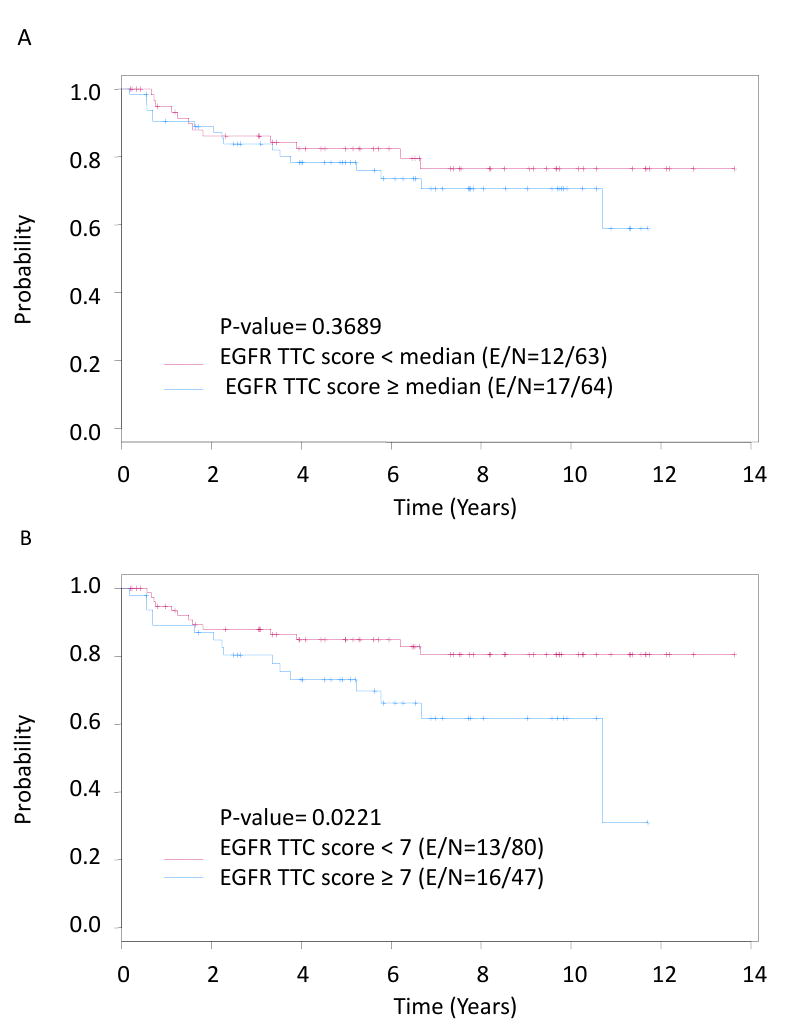
Time to oral cancer in oral leukoplakias (OPL) by EGFR expression evaluated quantitatively by automated analysis: in Fig. 3A, the median of the EGFR total transformed composite (TTC) score was used to dichotomize low versus high EGFR expression; in Fig. 3B, an EGFR TTC score of 7 was used as a cutoff, based on Martingale analysis.
EGFR gene copy number
The border-line association between oral cancer development and high-EGFR-expression OPLs (Fig. 2A and 3) led us to hypothesize that this trend involved the subset of high-expression patients who also had an increased EGFR gene copy number. Because of a scarcity of tissue, we could only evaluate 60 of the 103 high-EGFR–expression OPL patients (semi-quantitative evaluation), including 29 who developed oral cancer and 31 who did not (Table 3). Among these 60 patients, 49 exhibited at least 100 non-overlapping interphase nuclei from the whole sample and so were included in subsequent analyses. Of these 49 patients, 20 OPLs (41%) were FISH-positive, or had a high number of EGFR gene and chromosome 7 copies, distributed as follows: 14 with low trisomy, 1 with high trisomy, 4 with low polysomy, and 1 with gene amplification. The remaining 29 OPLs (59%) were FISH-negative, or had a low copy number (disomy). We did not find any association between FISH positivity and the degree of dysplasia (Supplemental data section, table 4). This comparison, however, is limited by its small sample size (N = 49).
Table 3.
The median time (years) to oral cancer development and oral cancer free (OCF) rates at years 5 and 10 in the subgroup of 49 patients with high EGFR expression and evaluation of EGFR gene copy number
| Variable | Level | N | Event | Median (95% CI) | OCF Rate at 5 Years (95% CI) | OCF Rate at 10 Years (95% CI) | P-value¶ |
|---|---|---|---|---|---|---|---|
| All patients | 49 | 24 | 6.65 (3.52, NA) | 0.62 (0.5, 0.78) | 0.46 (0.32, 0.65) | ||
| Sex | Female | 23 | 11 | 6.67 (2.24, NA) | 0.54 (0.37, 0.8) | 0.47 (0.28, 0.76) | 0.737 |
| Male | 26 | 13 | 6.65 (5.22, NA) | 0.69 (0.53, 0.89) | 0.46 (0.28, 0.74) | ||
| Race | Other | 1 | 0 | NA (NA, NA) | 0.489 | ||
| White | 48 | 24 | 6.65 (3.52, NA) | 0.61 (0.49, 0.77) | 0.45 (0.32, 0.64) | ||
| Histologic status | Hyperplasia | 33 | 14 | 10.7 (5.22, NA) | 0.68 (0.54, 0.87) | 0.53 (0.36, 0.77) | 0.454 |
| Mild dysplasia | 10 | 6 | 5.09 (2.05, NA) | 0.5 (0.27, 0.93) | |||
| Moderate/severe dysplasia | 6 | 4 | 4.45 (1.85, NA) | 0.5 (0.22, 1) | |||
| Treatment arm | 13cRA | 24 | 13 | 6.21 (2.88, NA) | 0.53 (0.36, 0.78) | 0.41 (0.24, 0.69) | 0.452 |
| BC + RP | 18 | 7 | 10.7 (5.3, NA) | 0.72 (0.54, 0.96) | 0.62 (0.4, 0.94) | ||
| RP | 7 | 4 | 5.22 (2.05, NA) | 0.71 (0.45, 1) | |||
| Smoking history | Current | 16 | 5 | NA (6.67, NA) | 0.75 (0.57, 1) | 0.471 | |
| Former | 25 | 15 | 6.21 (2.88, NA) | 0.56 (0.4, 0.79) | 0.39 (0.23, 0.67) | ||
| Never | 8 | 4 | 5.3 (3.36, NA) | 0.6 (0.33, 1) | 0.45 (0.2, 1) | ||
| Alcohol history | Current | 29 | 12 | 10.7 (6.65, NA) | 0.72 (0.58, 0.91) | 0.56 (0.38, 0.81) | 0.272 |
| Former | 8 | 5 | 3.32 (2.27, NA) | 0.47 (0.21, 1) | |||
| Never | 12 | 7 | 3.36 (1.85, NA) | 0.44 (0.21, 0.92) | 0.29 (0.1, 0.87) | ||
| EGFR gene copy number§ | FISH- | 29 | 8 | NA (NA, NA) | 0.79 (0.65, 0.95) | 0.67 (0.5, 0.9) | 0.0007 |
| FISH+ | 20 | 16 | 2.88 (2.05, NA) | 0.4 (0.23, 0.68) | 0.16 (0.05, 0.53) | ||
Abbreviations: CI, confidence interval; NA, not applicable; 13cRA, 13-cis-retinoic acid; BC, β-carotene; RP, retinyl palmitate; EGFR, epidermal growth factor receptor.
P-values are from the log-rank test (univariate analysis); FISH -: disomy; FISH+: low trisomy, high trisomy, low polysomy, or gene amplification.
Table 4.
Multicovariate analysis for time to oral cancer on FISH status for EGFR gene and chromosome 7 copies (E/N = 24/49)*
| Variable | Hazard Ratio | 95% | CI for HR | p-value |
|---|---|---|---|---|
| Age | 1.01 | 0.98 | 1.04 | 0.577 |
| BC+RP/RP-only versus 13cRA | 1.19 | 0.52 | 2.76 | 0.682 |
| Histologic status at baseline: dysplasia versus hyperplasia | 1.02 | 0.42 | 2.46 | 0.965 |
| FISH: positive versus negative | 3.62 | 1.44 | 9.10 | 0.006 |
Abbreviations: E/N, number of events and number of patients; Pr > ChiSq, P-value; HR, hazard ratio; CI, confidence interval; BC, β-carotene; RP: retinyl palmitate; 13cRA, 13-cis-retinoic acid; FISH, fluorescence in situ hybridization.
This analysis produced the maximum-likelihood estimates.
Impact of FISH-positivity on OCF
Patients with a FISH-positive OPL had a significantly higher incidence of OSCC than did those with a FISH-negative OPL (log-rank test, P = 0.0007; Fig. 2B). The difference was even more striking when considering OSCC that developed at the site of a baseline OPL (log-rank test, P<0.0001; Fig. 2C). There was no difference between FISH-positive and FISH-negative OPLs with regard to the incidence of OSCC not at a baseline OPL site (data not shown). The rate of OCF was only 40% for patients with FISH-positive OPLs (95% CI, 0.23–0.68) versus 79% for patients with FISH-negative OPLs (95% CI, 0.65–0.95; P = 0.0007) at 5 years after biopsy. The difference in OCF rate was more pronounced at 10 years after biopsy—only 16% (95% CI, 0.05-0.53) in the FISH-positive group versus 67% (95% CI, 0.5-0.9) in the FISH-negative group (P = 0.0007). In a multicovariate analysis, the only covariate significantly associated with OCF was FISH-positivity, with a hazard ratio of 3.620 (95% CI, 1.439–9.104; Table 4).
Discussion
In the present study, patients with high EGFR-expression OPLs had a statistically significantly decreased OCF if their OPL also carried increased chromosome 7 and EGFR gene copy numbers (16%; 95% CI, 0.05–0.53) versus carrying a normal EGFR gene copy number (67%; 95% CI, 0.5–0.9) at 10 years (P = 0.0007). This finding clearly demonstrates that an increased chromosome 7 and EGFR gene copy number is an early event in oral tumorigenesis that is strongly associated with oral cancer risk. To our knowledge, our study is the first to report EGFR expression and gene copy number in OPLs in a series of longitudinal and prospectively collected samples, which came from the largest, longest-term randomized controlled trial ever conducted in OPL patients (20).
Assessing cancer risk has the potential to help lower cancer incidence and mortality by providing the most appropriate populations for clinical prevention research. Although podoplanin, LOH (7), and p63 (9) have been shown to associate with an OPL's increased risk of OSCC, (8) there are no investigational or clinically approved agents for targeting these abnormalities. EGFR, on the other hand, is a validated cancer treatment target, and an anti-EGFR antibody, cetuximab, has been approved for treating patients with HNSCC and several other types of cancer (17, 18).
A seminal study in NSCLC (25) defined the FISH patterns of chromosome 7 and EGFR gene copy number that we used, but with substantially different definitions of FISH-positive and -negative tumors. The definition for FISH positivity, or a high EGFR copy number, only included high polysomy (≥ 4 gene copies in ≥ 40% cells) or EGFR amplification (unbalanced gene-to-chromosome copy-number ratio of > 2e or ≥ 15 gene copies in ≥ 10% of cells); FISH negativity was defined as low polysomy, high or low trisomy, or disomy. These patterns and definitions of FISH status were applied in previous studies of HNSCC (15, 16). Chung et al. (15) found EGFR gene amplification in 31%, high polysomy in 27%, low polysomy in 17%, trisomy in 17%, and balanced disomy in 8% of 81 cases of HNSCC, resulting in 58% FISH-positive cases of HNSCC. We did not expect to see this high HNSCC frequency of high polysomy or gene amplification in preinvasive lesions. Therefore, we could not use the same definition of FISH positivity in our study, where the majority of OPLs exhibited either a disomy (59%) or trisomy (31%). Therefore, our definition for FISH positivity, or an increased EGFR gene copy number, included trisomy along with polysomy or gene amplification.
Using an independent prospective population, a dual-target and dual-color FISH assay, and recently described FISH patterns, our results are consistent with our earlier findings (26, 27) showing that any increase in copy number of chromosome 7 (called “polysomy” in these early papers) is a major risk factor for oral cancer in OPL patients. These earlier studies built on the then-established association between chromosome “polysomy” and an increased risk of oral cancer (28, 29), evaluating chromosomes 7 and 17 centromeres via chromogenic in situ hybridization. The frequency of chromosome polysomy in the tumor field was shown to increase as the tissue progressed from normal morphology (33% frequency) to hyperplasia (67%) to dysplasia (95%) to SCC (96%). Subsequently, we analyzed OPL biopsies collected in a randomized chemoprevention trial with a median follow-up of 7 years (6). Patients with > 3 chromosome 7 copy numbers in at least 3% of epithelial cells were at an increased risk of oral cancer versus patients with lesser copy numbers (hazard ratio = 1.85, 95% CI, 1.05–3.25; P = 0.03).
Our findings also suggest that an increased EGFR gene copy number in OPLs is a precursor to EGFR gene amplification in HNSCC (as is chromosome 7 increased copy number) and an important oncogenesis-driving effector in oral oncogenesis. Sheu et al. recently conducted functional genomic analyses demonstrating that 7p11.2 was the most frequently amplified region in OSCC, and mapping this region showed a unique amplicon containing SEC61G and EGFR genes (12). The expression level of EGFR but not of SEC61G was up-regulated and tightly correlated with DNA copy number. Furthermore, EGFR downstream effectors such as K-ras, mitogen-activated protein kinase 1, and cyclin D1 also were amplified or mutated, resulting in activation of EGFR signaling in 55% of OSCC patients. Another study validated these findings via array comparative gene hybridization where amplification of 7p12 including EGFR was frequent in HNSCC (30). Taken together, all these findings support our current data indicating that an increased EGFR gene copy number is an early event in oral oncogenesis, consistent with its impact on the oral cancer development of OPL patients. Our and these other data also strongly suggest that EGFR is a major independent driver of oral oncogenesis as it progresses continuously from chromosomal instability to EGFR trisomy to EGFR polysomy and ultimately to EGFR amplification, all resulting in an unbalanced chromosome 7 polysomy.
Previous reports have shown that EGFR expression increases dramatically with progression from dysplastic lesions to HNSCC (22, 31), although EGFR expression as a prognostic factor in HNSCC is controversial (32); it also increases in normal epithelium adjacent to HNSCC compared with normal epithelium of healthy controls, as is consistent with “field cancerization” (31, 33). Analyzing only OPLs in the present study, we found EGFR expression in 88% and high EGFR expression in 71%. Consistent with previous reports (22, 31), EGFR expression did not differ between hyperplasia and dysplasia in the present study. To the best of our knowledge, the impact of EGFR expression on oral cancer development has never been reported in a prospective series of patients (from a randomized controlled trial, in this case). The trend that our semi-quantitative evaluation of EGFR expression showed between high EGFR protein expression and oral cancer development remained in our automated analysis of EGFR expression, which allowed a quantitative, less-subjective evaluation. Our study possibly was underpowered to detect a statistically significant difference in time to oral caner based on this factor alone. EGFR expression is associated with smoking history and is significantly higher in lung SCC (a histologic subtype strongly associated with smoking habits) than in lung adenocarcinoma (34). In our study, we did not observe any association between EGFR expression and smoking history. Tobacco smoke exposure induced an EGFR-centered subnetwork, where EGFR and its ligands were all significantly induced, in a cellular model of oral leukoplakia (35). It is possible that EGFR activation by tobacco smoking preferentially stimulates the expression of EGFR ligands such as amphiregulin rather than EGFR itself (36). Fundamental differences have been reported between human papillomavirus (HPV)-positive and HPV-negative murine models of HNSCC (37) and human oropharyngeal or oral HNSCC (37, 38). In North America, the overall HPV prevalence is 16% in OSCC and 47% in oropharyngeal cancer (39), and HPV-positive oropharyngeal cancer has been associated with a low EGFR expression (40). Therefore, some of the OSCC cases with low EGFR expression in our study possibly were HPV-related, which may have decreased our study power for identifying EGFR-driven OSCC.
It also should be noted that the effect of chemopreventive agents in the present study's clinical trial may be a confounding factor that influenced our results. Although there was no significant difference in oral cancer development among the treatment groups (20), it remains to be determined whether any treatment or treatments in this trial interacted in any way with the status of EGFR expression and copy number in OPLs.
Anti-EGFR antibodies and EGFR tyrosine kinase inhibitors (TKIs) are the most widely used strategies for inhibiting EGFR. For chemoprevention, however, EGFR TKIs, which are oral and convenient, are preferred over the antibodies, which require the inconvenience of intra-venous administration. Increased EGFR protein expression and gene copy number and TK domain activating mutations are the most-studied mechanisms associated with response to EGFR inhibitors (41, 42). EGFR gene copy number abnormalities are well known in HNSCC (15, 16). Because past reports have not shown differences of EGFR gene copy number in laryngeal, pharyngeal, and oral carcinogenesis (15, 16), we speculate that our findings may be relevant to more head and neck sites than just the oral cavity. On the other hand, EGFR mutations have rarely been described in HNSCC tumors or HNSCC cell lines and therefore probably are irrelevant in OPLs (16).
Early reports of a correlation between EGFR expression and response to EGFR TKIs (17, 43, 44) led to controversial subsequent results. Preclinical and clinical HNSCC studies have found an association between EGFR expression or gene copy number and response to EGFR TKIs. Sheu et al. found that only cell lines with EGFR gene amplification or EGFR overexpression were sensitive to an EGFR TKI (12). In 18 HNSCC cell lines, EGFR overexpression correlated with sensitivity to the EGFR TKI gefitinib and EGFR gene amplification occurred in the most sensitive cell lines (13). Other studies found similar results (45). EGFR overexpression and increased gene copy number were associated with a trend toward higher objective clinical response rates in a phase I/II trial of the EGFR TKI erlotinib combined with cisplatin in 37 patients with recurrent or metastatic HNSCC (46). These patient response rates were 36% with strong, 12% with medium, and 0% with weak or absent EGFR expression and 50% with, versus 15% without, high EGFR polysomy or gene amplification.
Tang et al. reported that EGFR levels were elevated in oral premalignant lesions versus in control samples, suggesting that increased EGFR protein expression marks carcinogenesis, in a murine model of oral carcinogenesis induced by the tobacco surrogate 4-nitroquinoline-1 oxide (4NQO; ref.(47); the level of DNA damage in this model is associated with the development of oral premalignant lesions and SCC (21). Sheu et al. showed that the EGFR TKI AG1478 dramatically reduced the incidence of OSCC and high-grade dysplasia in mice that developed oral leukoplakia of the tongue within a model of OSCC induced by 4 weeks of combined arecoline and 4NQO. EGFR gene copy number, however, has not been assessed in 4NQO-based oral mouse models (12). Last, EGFR inhibition has been shown to down-regulate signaling molecules such as cyclin D1 (which is implicated in genetic instability) downstream of the EGFR-signal transducer and activator of transcription 3 (STAT3) pathway (48, 49).
Taken together, these results from in vitro and in vivo models allow us to hypothesize that EGFR gene copy number may be valuable in predicting response to EGFR TKIs in the chemoprevention setting. It is also possible, however, that a change in EGFR copy number may reflect mainly chromosome 7 polysomy or aneuploidy and not a change directly linked to EGFR. If this is the case, increased EGFR gene copy number may be a marker only of oral cancer risk and not of drug sensitivity. Correlative studies in the ongoing phase III Erlotinib Prevention of Oral Cancer (EPOC) trial in OPL patients with a high OSCC risk marked by LOH profile provide a unique opportunity to validate the hypothesis that EGFR gene copy number is a predictive marker of response to EGFR inhibitors (19).
Inhibiting the mammalian target of rapamycin (mTOR) signaling pathway, which is another downstream effector of EGFR, recently prevented the progression of premalignant lesions in an oral-specific chemical carcinogenesis model where Akt-mTOR activation involves both EGFR-dependent and EGFR-independent mechanisms (50, 51). Therefore, it is possible that the effectiveness of EGFR inhibition may be increased by combining it with Akt-mTOR inhibition. Another approach in development is EGFR antisense DNA therapy (EGFR AS), which has been tested in a phase I (dose-escalation) clinical trial involving 17 assessable patients with HNSCC that was injected with EGFR AS once a week for 4 weeks (14). No grades 3–4 or dose-limiting toxicities were reported, and a maximum-tolerated dose was not reached. Five of the 17 patients had an objective response, including 2 complete responses. Disease control (objective response plus stable disease) was associated with baseline EGFR expression. This alternative approach of EGFR targeting may lend itself to oral leukoplakia, which is easily accessible and frequently involves only one or a few lesions.
In conclusion, the present study provides the first demonstration in a prospective longitudinal clinical trial that an increased EGFR gene copy number marks the risk of OSCC development in the substantial subgroup of OPL patients who have EGFR overexpression (71% of our total study population). Follow-on study in larger cohorts will be necessary to validate these findings. Assessments of EGFR protein expression and gene copy number could lead to selecting patients most in need of and most likely to respond to EGFR inhibitors in future oral-cancer chemoprevention trials.
Supplementary Material
Acknowledgments
Grant Support: This work was supported in part by National Cancer Institute grants P01 CA106451, P50 CA97007, and P30 CA16672 and a grant from the Kadorrie Charitable Foundation.
References
- 1.Smoking-attributable mortality, years of potential life lost, and productivity losses--United States, 2000-2004. MMWR Morb Mortal Wkly Rep. 2008;57(45):1226–8. [PubMed] [Google Scholar]
- 2.Gold KA, Lee HY, Kim ES. Targeted therapies in squamous cell carcinoma of the head and neck. Cancer. 2009;115(5):922–35. doi: 10.1002/cncr.24123. [DOI] [PubMed] [Google Scholar]
- 3.Warnakulasuriya S, Johnson NW, van der Waal I. Nomenclature and classification of potentially malignant disorders of the oral mucosa. J Oral Pathol Med. 2007;36(10):575–80. doi: 10.1111/j.1600-0714.2007.00582.x. [DOI] [PubMed] [Google Scholar]
- 4.Papadimitrakopoulou VA, Hong WK, Lee JS, Martin JW, Lee JJ, Batsakis JG, et al. Low-dose isotretinoin versus beta-carotene to prevent oral carcinogenesis: long-term follow-up. J Natl Cancer Inst. 1997;89(3):257–8. doi: 10.1093/jnci/89.3.257. [DOI] [PubMed] [Google Scholar]
- 5.Silverman S, Jr, Gorsky M, Lozada F. Oral leukoplakia and malignant transformation. A follow-up study of 257 patients. Cancer. 1984;53(3):563–8. doi: 10.1002/1097-0142(19840201)53:3<563::aid-cncr2820530332>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 6.Lee JJ, Hong WK, Hittelman WN, Mao L, Lotan R, Shin DM, et al. Predicting cancer development in oral leukoplakia: ten years of translational research. Clin Cancer Res. 2000;6(5):1702–10. [PubMed] [Google Scholar]
- 7.Mao L, Lee JS, Fan YH, Ro JY, Batsakis JG, Lippman S, et al. Frequent microsatellite alterations at chromosomes 9p21 and 3p14 in oral premalignant lesions and their value in cancer risk assessment. Nat Med. 1996;2(6):682–5. doi: 10.1038/nm0696-682. [DOI] [PubMed] [Google Scholar]
- 8.Kawaguchi H, El-Naggar AK, Papadimitrakopoulou V, Ren H, Fan YH, Feng L, et al. Podoplanin: a novel marker for oral cancer risk in patients with oral premalignancy. J Clin Oncol. 2008;26(3):354–60. doi: 10.1200/JCO.2007.13.4072. [DOI] [PubMed] [Google Scholar]
- 9.Saintigny P, El-Naggar AK, Papadimitrakopoulou V, Ren H, Fan YH, Feng L, et al. {Delta}Np63 Overexpression, Alone and in Combination with Other Biomarkers, Predicts the Development of Oral Cancer in Patients with Leukoplakia. Clin Cancer Res. 2009 doi: 10.1158/1078-0432.CCR-09-0498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rubin Grandis J, Melhem MF, Gooding WE, Day R, Holst VA, Wagener MM, et al. Levels of TGF-alpha and EGFR protein in head and neck squamous cell carcinoma and patient survival. J Natl Cancer Inst. 1998;90(11):824–32. doi: 10.1093/jnci/90.11.824. [DOI] [PubMed] [Google Scholar]
- 11.Grandis JR, Tweardy DJ. TGF-alpha and EGFR in head and neck cancer. J Cell Biochem Suppl. 1993;17F:188–91. doi: 10.1002/jcb.240531027. [DOI] [PubMed] [Google Scholar]
- 12.Sheu JJ, Hua CH, Wan L, Lin YJ, Lai MT, Tseng HC, et al. Functional genomic analysis identified epidermal growth factor receptor activation as the most common genetic event in oral squamous cell carcinoma. Cancer Res. 2009;69(6):2568–76. doi: 10.1158/0008-5472.CAN-08-3199. [DOI] [PubMed] [Google Scholar]
- 13.Rogers SJ, Box C, Chambers P, Barbachano Y, Nutting CM, Rhys-Evans P, et al. Determinants of response to epidermal growth factor receptor tyrosine kinase inhibition in squamous cell carcinoma of the head and neck. J Pathol. 2009 doi: 10.1002/path.2515. [DOI] [PubMed] [Google Scholar]
- 14.Lai SY, Koppikar P, Thomas SM, Childs EE, Egloff AM, Seethala RR, et al. Intratumoral epidermal growth factor receptor antisense DNA therapy in head and neck cancer: first human application and potential antitumor mechanisms. J Clin Oncol. 2009;27(8):1235–42. doi: 10.1200/JCO.2008.17.8251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chung CH, Ely K, McGavran L, Varella-Garcia M, Parker J, Parker N, et al. Increased epidermal growth factor receptor gene copy number is associated with poor prognosis in head and neck squamous cell carcinomas. J Clin Oncol. 2006;24(25):4170–6. doi: 10.1200/JCO.2006.07.2587. [DOI] [PubMed] [Google Scholar]
- 16.Temam S, Kawaguchi H, El-Naggar AK, Jelinek J, Tang H, Liu DD, et al. Epidermal growth factor receptor copy number alterations correlate with poor clinical outcome in patients with head and neck squamous cancer. J Clin Oncol. 2007;25(16):2164–70. doi: 10.1200/JCO.2006.06.6605. [DOI] [PubMed] [Google Scholar]
- 17.Burtness B, Goldwasser MA, Flood W, Mattar B, Forastiere AA. Phase III randomized trial of cisplatin plus placebo compared with cisplatin plus cetuximab in metastatic/recurrent head and neck cancer: an Eastern Cooperative Oncology Group study. J Clin Oncol. 2005;23(34):8646–54. doi: 10.1200/JCO.2005.02.4646. [DOI] [PubMed] [Google Scholar]
- 18.Vermorken JB, Mesia R, Rivera F, Remenar E, Kawecki A, Rottey S, et al. Platinum-based chemotherapy plus cetuximab in head and neck cancer. N Engl J Med. 2008;359(11):1116–27. doi: 10.1056/NEJMoa0802656. [DOI] [PubMed] [Google Scholar]
- 19.William WN, Jr, Heymach JV, Kim ES, Lippman SM. Molecular targets for cancer chemoprevention. Nat Rev Drug Discov. 2009;8(3):213–25. doi: 10.1038/nrd2663. [DOI] [PubMed] [Google Scholar]
- 20.Papadimitrakopoulou VA, Lee JJ, William WN, Jr, Martin JW, Thomas M, Kim ES, et al. Randomized trial of 13-cis retinoic acid compared with retinyl palmitate with or without beta-carotene in oral premalignancy. J Clin Oncol. 2009;27(4):599–604. doi: 10.1200/JCO.2008.17.1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ribeiro DA, Favero Salvadori DM, da Silva RN, Ribeiro Darros B, Alencar Marques ME. Genomic instability in non-neoplastic oral mucosa cells can predict risk during 4-nitroquinoline 1-oxide-induced rat tongue carcinogenesis. Oral Oncol. 2004;40(9):910–5. doi: 10.1016/j.oraloncology.2004.04.010. [DOI] [PubMed] [Google Scholar]
- 22.Rubin Grandis J, Tweardy DJ, Melhem MF. Asynchronous modulation of transforming growth factor alpha and epidermal growth factor receptor protein expression in progression of premalignant lesions to head and neck squamous cell carcinoma. Clin Cancer Res. 1998;4(1):13–20. [PubMed] [Google Scholar]
- 23.Hirsch FR, Varella-Garcia M, Bunn PA, Jr, Di Maria MV, Veve R, Bremmes RM, et al. Epidermal growth factor receptor in non-small-cell lung carcinomas: correlation between gene copy number and protein expression and impact on prognosis. J Clin Oncol. 2003;21(20):3798–807. doi: 10.1200/JCO.2003.11.069. [DOI] [PubMed] [Google Scholar]
- 24.Cappuzzo F, Hirsch FR, Rossi E, Bartolini S, Ceresoli GL, Bemis L, et al. Epidermal growth factor receptor gene and protein and gefitinib sensitivity in non-small-cell lung cancer. J Natl Cancer Inst. 2005;97(9):643–55. doi: 10.1093/jnci/dji112. [DOI] [PubMed] [Google Scholar]
- 25.Kayser K. Introducing diagnostic pathology. Diagn Pathol. 2006;1(1):1. doi: 10.1186/1746-1596-1-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hittelman WN, Voravud N, Shin DM, Lee JS, Ro JY, Hong WK. Early genetic changes during upper aerodigestive tract tumorigenesis. J Cell Biochem Suppl. 1993;17F:233–6. doi: 10.1002/jcb.240531034. [DOI] [PubMed] [Google Scholar]
- 27.Voravud N, Shin DM, Ro JY, Lee JS, Hong WK, Hittelman WN. Increased polysomies of chromosomes 7 and 17 during head and neck multistage tumorigenesis. Cancer Res. 1993;53(12):2874–83. [PubMed] [Google Scholar]
- 28.Hittelman WN, Kim HJ, Lee JS, Shin DM, Lippman SM, Kim J, et al. Detection of chromosome instability of tissue fields at risk: in situ hybridization. J Cell Biochem Suppl. 1996;25:57–62. [PubMed] [Google Scholar]
- 29.Pihan GA, Doxsey SJ. The mitotic machinery as a source of genetic instability in cancer. Semin Cancer Biol. 1999;9(4):289–302. doi: 10.1006/scbi.1999.0131. [DOI] [PubMed] [Google Scholar]
- 30.Roman E, Meza-Zepeda LA, Kresse SH, Myklebost O, Vasstrand EN, Ibrahim SO. Chromosomal aberrations in head and neck squamous cell carcinomas in Norwegian and Sudanese populations by array comparative genomic hybridization. Oncol Rep. 2008;20(4):825–43. [PubMed] [Google Scholar]
- 31.Shin DM, Ro JY, Hong WK, Hittelman WN. Dysregulation of epidermal growth factor receptor expression in premalignant lesions during head and neck tumorigenesis. Cancer Res. 1994;54(12):3153–9. [PubMed] [Google Scholar]
- 32.Fischer C, Zlobec I, Stockli E, Probst S, Storck C, Tornillo L, et al. Is immunohistochemical epidermal growth factor receptor expression overestimated as a prognostic factor in head-neck squamous cell carcinoma? A retrospective analysis based on a tissue microarray of 365 carcinomas. Hum Pathol. 2008;39(10):1527–34. doi: 10.1016/j.humpath.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 33.Grandis JR, Tweardy DJ. Elevated levels of transforming growth factor alpha and epidermal growth factor receptor messenger RNA are early markers of carcinogenesis in head and neck cancer. Cancer Res. 1993;53(15):3579–84. [PubMed] [Google Scholar]
- 34.Kang JU, Koo SH, Kwon KC, Park JW, Jung SS. Gain of the EGFR gene located on 7p12 is a frequent and early event in squamous cell carcinoma of the lung. Cancer Genet Cytogenet. 2008;184(1):31–7. doi: 10.1016/j.cancergencyto.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 35.Gumus ZH, Du B, Kacker A, Boyle JO, Bocker JM, Mukherjee P, et al. Effects of tobacco smoke on gene expression and cellular pathways in a cellular model of oral leukoplakia. Cancer Prev Res (Phila Pa) 2008;1(2):100–11. doi: 10.1158/1940-6207.CAPR-08-0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moraitis D, Du B, De Lorenzo MS, Boyle JO, Weksler BB, Cohen EG, et al. Levels of cyclooxygenase-2 are increased in the oral mucosa of smokers: evidence for the role of epidermal growth factor receptor and its ligands. Cancer Res. 2005;65(2):664–70. [PubMed] [Google Scholar]
- 37.Strati K, Pitot HC, Lambert PF. Identification of biomarkers that distinguish human papillomavirus (HPV)-positive versus HPV-negative head and neck cancers in a mouse model. Proc Natl Acad Sci U S A. 2006;103(38):14152–7. doi: 10.1073/pnas.0606698103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pyeon D, Newton MA, Lambert PF, den Boon JA, Sengupta S, Marsit CJ, et al. Fundamental differences in cell cycle deregulation in human papillomavirus-positive and human papillomavirus-negative head/neck and cervical cancers. Cancer Res. 2007;67(10):4605–19. doi: 10.1158/0008-5472.CAN-06-3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kreimer AR, Clifford GM, Boyle P, Franceschi S. Human papillomavirus types in head and neck squamous cell carcinomas worldwide: a systematic review. Cancer Epidemiol Biomarkers Prev. 2005;14(2):467–75. doi: 10.1158/1055-9965.EPI-04-0551. [DOI] [PubMed] [Google Scholar]
- 40.Kumar B, Cordell KG, Lee JS, Worden FP, Prince ME, Tran HH, et al. EGFR, p16, HPV Titer, Bcl-xL and p53, sex, and smoking as indicators of response to therapy and survival in oropharyngeal cancer. J Clin Oncol. 2008;26(19):3128–37. doi: 10.1200/JCO.2007.12.7662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zandi R, Larsen AB, Andersen P, Stockhausen MT, Poulsen HS. Mechanisms for oncogenic activation of the epidermal growth factor receptor. Cell Signal. 2007;19(10):2013–23. doi: 10.1016/j.cellsig.2007.06.023. [DOI] [PubMed] [Google Scholar]
- 42.Uberall I, Kolar Z, Trojanec R, Berkovcova J, Hajduch M. The status and role of ErbB receptors in human cancer. Exp Mol Pathol. 2008;84(2):79–89. doi: 10.1016/j.yexmp.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 43.Dancey JE. Predictive factors for epidermal growth factor receptor inhibitors--the bull's-eye hits the arrow. Cancer Cell. 2004;5(5):411–5. doi: 10.1016/s1535-6108(04)00122-9. [DOI] [PubMed] [Google Scholar]
- 44.Gupta R, Dastane AM, McKenna R, Jr, Marchevsky AM. The predictive value of epidermal growth factor receptor tests in patients with pulmonary adenocarcinoma: review of current “best evidence” with meta-analysis. Hum Pathol. 2009;40(3):356–65. doi: 10.1016/j.humpath.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 45.Erjala K, Sundvall M, Junttila TT, Zhang N, Savisalo M, Mali P, et al. Signaling via ErbB2 and ErbB3 associates with resistance and epidermal growth factor receptor (EGFR) amplification with sensitivity to EGFR inhibitor gefitinib in head and neck squamous cell carcinoma cells. Clin Cancer Res. 2006;12(13):4103–11. doi: 10.1158/1078-0432.CCR-05-2404. [DOI] [PubMed] [Google Scholar]
- 46.Agulnik M, da Cunha Santos G, Hedley D, Nicklee T, Dos Reis PP, Ho J, et al. Predictive and pharmacodynamic biomarker studies in tumor and skin tissue samples of patients with recurrent or metastatic squamous cell carcinoma of the head and neck treated with erlotinib. J Clin Oncol. 2007;25(16):2184–90. doi: 10.1200/JCO.2006.07.6554. [DOI] [PubMed] [Google Scholar]
- 47.Tang XH, Knudsen B, Bemis D, Tickoo S, Gudas LJ. Oral cavity and esophageal carcinogenesis modeled in carcinogen-treated mice. Clin Cancer Res. 2004;10(1 Pt 1):301–13. doi: 10.1158/1078-0432.ccr-0999-3. [DOI] [PubMed] [Google Scholar]
- 48.Boehm AL, Sen M, Seethala R, Gooding WE, Freilino M, Wong SM, et al. Combined targeting of epidermal growth factor receptor, signal transducer and activator of transcription-3, and Bcl-X(L) enhances antitumor effects in squamous cell carcinoma of the head and neck. Mol Pharmacol. 2008;73(6):1632–42. doi: 10.1124/mol.107.044636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nelsen CJ, Kuriyama R, Hirsch B, Negron VC, Lingle WL, Goggin MM, et al. Short term cyclin D1 overexpression induces centrosome amplification, mitotic spindle abnormalities, and aneuploidy. J Biol Chem. 2005;280(1):768–76. doi: 10.1074/jbc.M407105200. [DOI] [PubMed] [Google Scholar]
- 50.Czerninski R, Amornphimoltham P, Patel V, Molinolo AA, Gutkind JS. Targeting mammalian target of rapamycin by rapamycin prevents tumor progression in an oral-specific chemical carcinogenesis model. Cancer Prev Res (Phila Pa) 2009;2(1):27–36. doi: 10.1158/1940-6207.CAPR-08-0147. [DOI] [PubMed] [Google Scholar]
- 51.Molinolo AA, Hewitt SM, Amornphimoltham P, Keelawat S, Rangdaeng S, Meneses Garcia A, et al. Dissecting the Akt/mammalian target of rapamycin signaling network: emerging results from the head and neck cancer tissue array initiative. Clin Cancer Res. 2007;13(17):4964–73. doi: 10.1158/1078-0432.CCR-07-1041. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


