Abstract
The N-terminal Domain (NTD) of Drosophila melanogaster (Dm) Topoisomerase I has been shown to bind to RNA polymerase II, but the domain of RNAPII with which it interacts is not known. Using bacterially-expressed fusion proteins carrying all or half of the NTDs of Dm and human (Hs) Topo I, we demonstrate that the N-terminal half of each NTD binds directly to the hyperphosphorylated C-terminal repeat domain (phosphoCTD) of the largest RNAPII subunit, Rpb1. Thus, the amino terminal segment of metazoan Topo I (1–157 for Dm and 1–114 for Hs) contains a novel phosphoCTD-interacting domain that we designate the Topo I-Rpb1 interacting (TRI) domain. The long-known in vivo association of Topo I with active genes presumably can be attributed, wholly or in part, to the TRI domain-mediated binding of Topo I to the phosphoCTD of transcribing RNAPII.
Keywords: Topoisomerase I, phosphoCTD, RNA polymerase II, transcription, phosphoCTD-interacting domain, Topo I-Rpb1 interacting (TRI) domain
Introduction
Topoisomerase I in metazoan cell nuclei associates with transcriptionally active regions of the genome [1,2], and this attribute maps to the N-terminal domain (NTD) of the protein [3], a non-catalytic segment of variable sequence ranging from about 200 to 400 amino acids in length. The NTD of Drosophila Topo I (amino acids 1 – 315 of the 972-residue protein) is capable of associating with RNA polymerase II (RNAPII) as judged by pull down experiments [3], suggesting a mechanism for targeting Topo I to active genes. Moreover, Topo I was found by proteomics approaches to bind directly to the hyperphosphorylated CTD (PCTD) of Rpb1, the largest subunit of RNAPII [4,5]. Taken together these facts suggest that the NTD can mediate the association of Topo I with elongating RNAPII via direct binding to the PCTD. Herein we confirm this suggestion for fruit fly and human Topo I, and we define the subsegment of the NTD that manifests PCTD binding.
MATERIALS AND METHODS
Plasmids and proteins
Three segments of human Topo I N-terminus, 1–199, 1–114, and 104–199, were constructed by PCR with cDNA as template, and primer pairs CCACTAGTATGAGTGGGGACCACCTCCACA/CCGTCGACCCTCTTCTTTCTTCGGCTTCTT, CACTAGTATGAGTGGGGACCACCTCCACA/CCGTCGACGTGGTGGACTAGAGAAGCCA and GCACTAGTAAGGAGAAGGAAAATGGC/CCGTCGACCCTCTTCTTTCTTCGGCTTCTT were used, respectively. The underlined sequences in the oligo nucleotides correspond to Spe I and Sal I recognition sites, respectively. The fragments were inserted into pET-41a (Novagen) with Spe I and Sal I.
Similarly, the primer pairs GCCAAACTAGTATGAGTGGGGATGTGGCTGC/CGAAGTCGACATCATTGTAGTTCATGGTGC, GCCAAACTAGTATGAGTGGGGATGTGGCTGC/CGAAGTCGACCCGACTGGATGAACTGCT and GCCAAACTAGTCACAAGTCCTCGTCCAGCT/ CGAAGTCGACATCATTGTAGTTCATGGTGC were used for constructing 1–315, 1–157, and 158–315 of Drosophila Topo I, respectively.
All constructs were confirmed by DNA sequencing before they were transformed into the expression strain BL21(DE3) (Novagen). Affinity purification of protein with Ni-NTA (Qiagen) resin was carried out according to the procedure provided by the manufacturer.
Phosphorylation of GST-CTD fusion protein, and Far Western blotting
A βGal-DmCTD fusion protein [6], or a GST-yeast CTD fusion protein [7] was exhaustively phosphorylated, either with or without radioactive ATP, using purified yeast CTDK-I [8]. Far Western blotting (also “reverse” Far Western blotting) was performed as described [8].
RESULTS
We constructed GST fusion proteins carrying part or all of the N-terminal Domain (NTD) from Topo I of Drosophila melanogaster or Homo sapiens (illustrated in Fig. 1A). The fusion proteins were subjected to SDS PAGE and either stained with Coomassie blue or blotted to nitrocellulose and given a chance to renature. The blot was reacted with a 32P-labeled phosphoCTD probe, a βGal-CTD fusion protein exhaustively phosphorylated by yeast CTDK-I (catalytic subunit = Ctk1), and autoradiographed. As shown in Fig. 1B (left, Far Western), the 32P-labeled PCTD probe bound to full length NTD from both Drosophila and human Topo I (lanes 3 & 6, respectively). The Drosophila NTD had apparently suffered some proteolysis during expression and purification (lanes 3), but this afforded a demonstration of binding specificity, as only a subset of the fragments (right, stained gel) bound the PCTD probe (left). Very low background binding was achieved in the experiment, as indicated by the probe’s lack of reactivity with GST or any bacterial proteins (lanes 7).
Fig. 1. N-terminus of Topo I physically interacts with hyperphosphorylated CTD (PCTD).
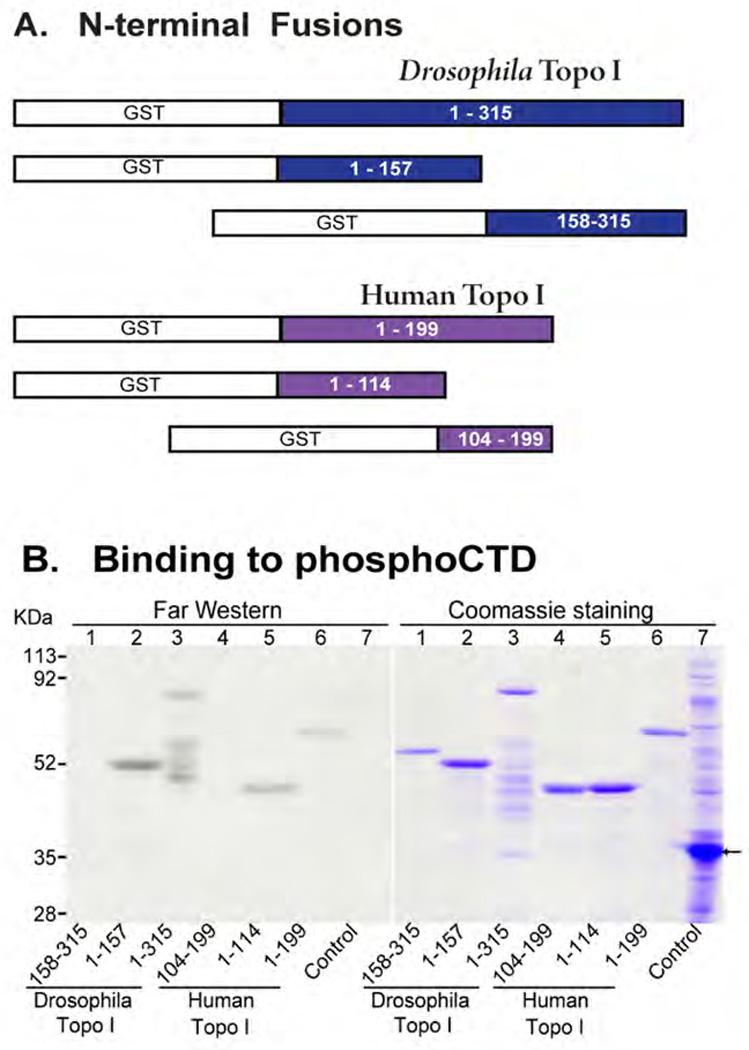
(A) Diagram of GST fusion constructs for Drosophila and human Topo I N-termini. Open (white) portions represent GST tags. Numbers in colored portions indicate amino acid residues. (B) Purified Topo I polypeptides were subjected to SDS-PAGE, transferred to nitrocellulose membrane, and probed (Far Western) with 32P-labeled βGal-CTD fusion protein. A duplicate SDS gel was stained with Coomassie Blue. The Control lane (lane 7) was loaded with lysate of BL21(DE3) harboring pET41a, which was induced with 1 mM IPTG for 5 hr in 30 °C. Arrow indicates a fusion protein encoded by pET41a (GST plus an additional 98 amino acids).
The most N-terminal half of the Drosophila NTD (1 – 157) also bound to the PCTD probe, and the amount of binding (left) relative to the amount of protein (right) was about the same as for full length NTD. Correspondingly, the other half of the Drosophila NTD (158–315) showed no detectable binding to the PCTD probe (lanes 1). In a similar vein, the most N-terminal fragment of human Topo I (lanes 5, 1–114) bound as well as the full length NTD (1–199), whereas the slightly overlapping fragment, 104–199, showed little binding (lanes 4). These results demonstrate that the phosphoCTD-interacting domain (PCID) of Topoisomerase I resides in the N-terminal half of the NTD.
The PCTD binding of the NTD from human Topo I was investigated further, using a “reverse” Far Western approach [8]. A GST-CTD fusion protein, after phosphorylation in the absence of radioactivity, was subjected to SDS PAGE and blotted to nitrocellulose. The blot was then probed with native NTD labeled with 32P. As shown in Fig. 2, the NTD bound to GST-CTD phosphorylated by yeast CTDK-I (duplicates, lanes 1 & 2), but it did not bind to the non-phosphorylated GST-CTD fusion protein (lane 3, NP).
Fig. 2. “Reverse” Far Western blot probed with labeled N-terminus of human Topo I.
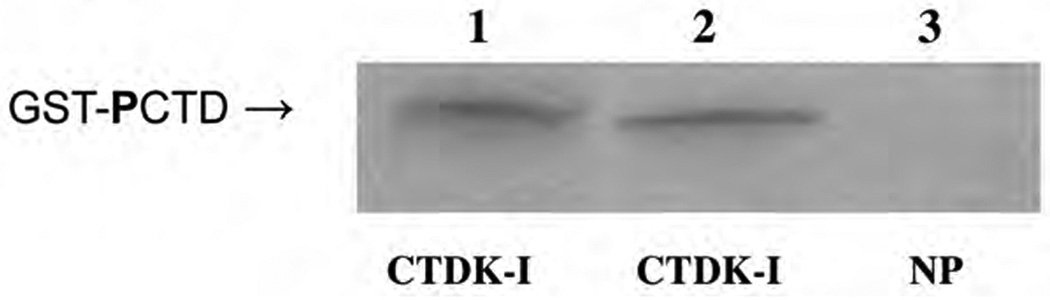
The blot was prepared by SDS PAGE of 1 µg of unlabeled phosphorylated or non-phosphorylated GST-CTD fusion protein per lane, followed by blotting to nitrocellulose [8]. The protein segment comprising amino acids 1–199 of human Topo I was labeled by 32P with casein kinase II and used as a probe. Lanes 1 and 2: GST-CTD fusion protein phosphorylated by CTDK-I; lane 3: non-phosphorylated GST-CTD fusion protein.
DISCUSSION
We show that a phosphoCTD-interacting domain (PCID) resides in the N-terminal segment of Topoisomerase I from Drosophila melanogaster and Homo sapiens. This PCID binds to a CTD exhaustively phosphorylated by yeast CTDK-I; we surmise therefore that it binds to CTD repeats phosphorylated in a Ser2,5P pattern [9] and potentially to repeats phosphorylated in other patterns. The presence of this PCID in the NTD of Topo I provides a molecular explanation for the ability of this domain to target an NTD-βGal fusion protein to transcriptionally active genes in vivo [3]. More importantly, the long-known in vivo association of Topo I with active genes presumably can be attributed, wholly or in part, to the NTD-PCTD interaction. While we do not know the disposition of the Topo I catalytic domain relative to the “downstream” or “upstream” side of RNAPII, we point out that the length and flexibility of the CTD could probably accommodate either orientation [e.g. Fig. 10 in [5].
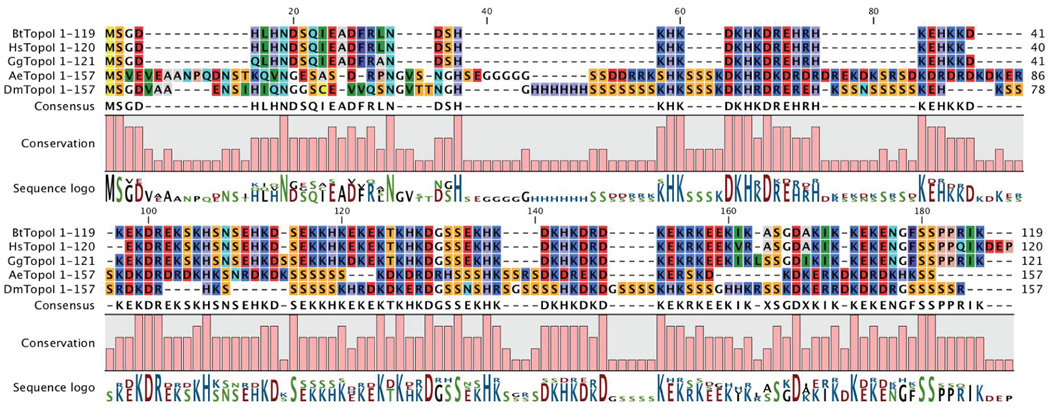
Comparing the amino acid sequences of Topo I N-terminal segments from three vertebrates (Bt, Hs and Gg) and two insects (Ae and Dm) reveals several homologous regions shared by these organisms (Fig. 3). Both insect NTDs contain numerous Serine runs and a few other inserts not found in the vertebrate proteins. The conserved N-terminal regions are very rich in charged residues, and a fair number of completely conserved positions contain Arg, Lys, Asp or Glu. On the other hand, 11 of the 26 completely conserved amino acids (as defined by this alignment, and ignoring the initial Met Ser sequence) are either His, Asn or Ser. The absence of conserved aromatic and hydrophobic residues distinguishes the Topo I PCID from previously-described CID, WW, FF and SRI domains of other proteins that associate with elongating RNAPII [see e.g. [10]. Further experiments will be required to describe more fully the attributes of the Topo I PCID at the molecular level; in the meantime we propose to call this novel PCTD-binding entity the Topo I–Rpb1 interacting (“TRI”) domain.
Fig. 3. N-terminal segments of Topoisomerases I aligned.
The indicated segments of Topo I from Bos taurus (Bt), Homo sapiens (Hs), Gallus gallus (Gg), Aedes aegypti (Ae), and Drosophila melanogaster (Dm) were aligned using the Alignment program within CLC Combined Workbench, v 3.0.3 www.clcbio.com, using a Gap open cost of 6, a Gap extension cost of 0.2, and an End gap cost of Free.
ACKNOWLEDGEMENTS
This work was supported by grants from the National Institutes of Health to ALG (GM040505) and to TSH (GM029006).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Gilmour DS, Pflugfelder G, Wang JC, Lis JT. Topoisomerase I interacts with transcribed regions in Drosophila cells. Cell. 1986;44:401–407. doi: 10.1016/0092-8674(86)90461-7. [DOI] [PubMed] [Google Scholar]
- 2.Gilmour DS, Elgin SC. Association of topoisomerase I with transcriptionally active loci in Drosophila. Nci Monogr. 1987;1987:17–21. [PubMed] [Google Scholar]
- 3.Shaiu WL, Hsieh TS. Targeting to transcriptionally active loci by the hydrophilic N-terminal domain of Drosophila DNA topoisomerase I. Mol Cell Biol. 1998;18:4358–4367. doi: 10.1128/mcb.18.7.4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carty SM, Greenleaf AL. Hyperphosphorylated C-terminal repeat domain-associating proteins in the nuclear proteome link transcription to DNA/chromatin modification and RNA processing. Mol Cell Proteomics. 2002;1:598–610. doi: 10.1074/mcp.m200029-mcp200. [DOI] [PubMed] [Google Scholar]
- 5.Phatnani HP, Jones JC, Greenleaf AL. Expanding the Functional Repertoire of CTD Kinase I and RNA Polymerase II: Novel PhosphoCTD-Associating Proteins in the Yeast Proteome. BIOCHEMISTRY. 2004;43:15702–15719. doi: 10.1021/bi048364h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee JM, Greenleaf AL. A protein kinase that phosphorylates the C-terminal repeat domain of the largest subunit of RNA polymerase II. Proc Natl Acad Sci U S A. 1989;86:3624–3628. doi: 10.1073/pnas.86.10.3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morris DP, Lee JM, Sterner DE, Brickey WJ, Greenleaf AL. Assaying CTD Kinases In Vitro and Phosphorylation-Modulated Properties of RNA Polymerase II In Vivo, METHODS. A companion to Methods in Enzymology. 1997;12:264–275. doi: 10.1006/meth.1997.0478. [DOI] [PubMed] [Google Scholar]
- 8.Phatnani HP, Greenleaf AL. Identifying PhosphoCTD-Associating Proteins. In: Schoenberg DR, editor. Methods in Molecular Biology: mRNA Processing and Metabolism. Totowa, NJ: Humana Press; 2004. pp. 17–28. [DOI] [PubMed] [Google Scholar]
- 9.Jones JC, Phatnani HP, Haystead TA, MacDonald JA, Alam SM, Greenleaf AL. C-terminal Repeat Domain Kinase I Phosphorylates Ser2 and Ser5 of RNA Polymerase II C-terminal Domain Repeats. J Biol Chem. 2004;279:24957–24964. doi: 10.1074/jbc.M402218200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Phatnani HP, Greenleaf AL. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 2006;20:2922–2936. doi: 10.1101/gad.1477006. [DOI] [PubMed] [Google Scholar]