Summary of Recent Advances
The SAGA complex provides a paradigm for multi-subunit histone modifying complexes. Although first characterized as a histone acetyltransferase, due to the Gcn5 subunit, SAGA is now known to contain a second activity, a histone deubiqutinase, as well as subunits important for interactions with transcriptional activators and the general transcription machinery. The functions of SAGA in transcriptional activation are well-established in S. cerevisiae. Recent studies in S. pombe, Drosophila, and mammalian systems reveal that SAGA also has important roles in transcript elongation, the regulation of protein stability, and telomere maintenance. These functions are essential for normal embryo development in flies and mice, and mutations or altered expression of SAGA subunits correlate with neurological disease and aggressive cancers in humans.
Introduction
Histone modifications, such as acetylation and ubiquitination, play a key role in facilitating a number of cellular events, including gene regulation. Acetylation of histones is largely associated with relaxing chromatin structure to support the entry of transcriptional machinery to genomic loci for activation, while histone ubiquitination has been linked to both gene activation and repression [1, 2]. The histone modifying enzymes that catalyze these post-translational modifications are often integrated into large multi-subunit complexes to facilitate their enzymatic activity and substrate specificity. SAGA (Spt-Ada-Gcn5 acetyltransferase) is a 2MDa multi-protein chromatin modifying complex that is conserved between yeast and humans and harbors two known enzymatic modules that mediate the acetylation and deubiquitination of histones as well as non-histone substrates [3-5]. However, like many multi-subunit complexes, the functions of several components in the SAGA complex are still unknown. In addition, it is unclear exactly how the members of SAGA interact, coordinate and regulate the acetyltransferase and deubiquitinase activites of the complex, and whether the acetyltransferase and deubiquitinase (Dub) modules function in a cooperative or antagonistic manner to regulate cellular processes, such as gene regulation. Here we will discuss the most recent advances made toward deciphering how the catalytic activities of the SAGA complex function in development and disease.
The SAGA complex
The Tetrahymena thermophila protein, p55, was the first identified transcription related histone acetyltransferase (HAT) enzyme and is the ortholog of the yeast (y) transcriptional co-activator protein, Gcn5 (general control nonderepressible 5) [1]. Recombinant yGcn5 acetylates non-nucleosomal histone 3 (H3) lysine residue 14 (K14) [6, 7]. However, the incorporation of yGcn5 into native multi-subunit complexes, such as ySAGA, expands its specificity to additional lysines in nucleosomal histones in vitro, including H3 K9, 14, 18, 23 and to a lesser degree histone H2B [7, 8].
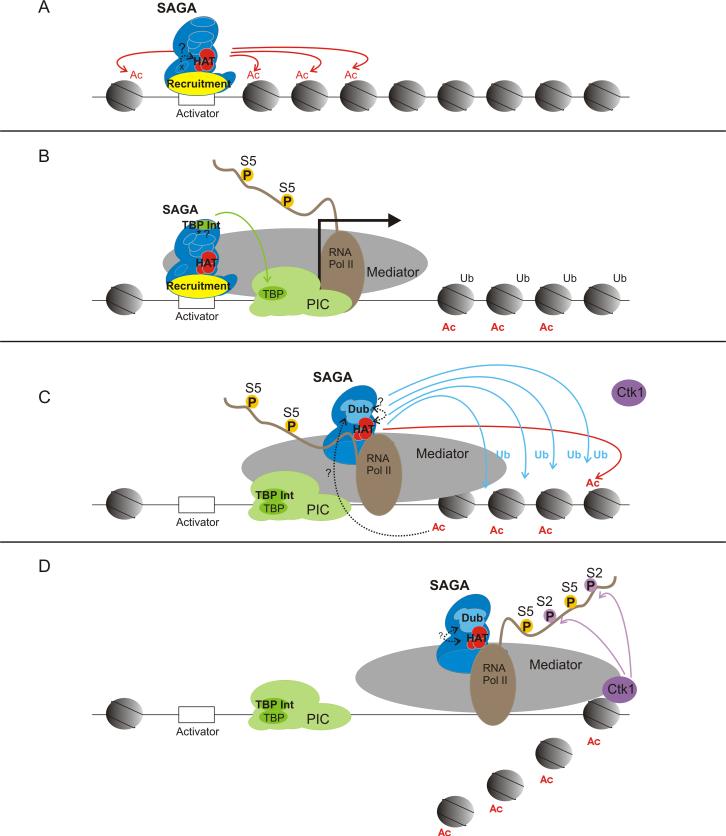
The ySAGA complex is modular in structure and has distinct functional units (Table 1) including a recruitment module (Tra1) [9, 10]; acetylation module (Gcn5, Ada2, Ada3) [11-14]; TBP interaction unit (Spt3, Spt8) [13, 15, 16]; Dub module (Ubp8, Sus1, Sgf11 and Sgf73) [17-21]; and architecture unit (Spt7, Spt20, Ada1, TAF5, -6, -9 and -12) [8, 13, 22, 23]. ySAGA is recruited to gene loci by the interaction of Tra1 with specific transcriptional activators [10], and the bromodomain of yGcn5 binds acetylated H3 and H4 N-terminal tails which potentiates cooperative nucleosome acetylation of histone H3 [24, 25] (Figure 1A). The HAT module composed of yGcn5, yAda2 and yAda3 catalyzes this acetylation, opening up the chromatin landscape for binding of additional transcription factors and the pre-initiation complex (PIC) [14]. The ySpt3 subunit of ySAGA also recruits TBP to facilitate PIC formation and transcriptional activation (Figure 1B) [16]. In addition to promoting gene activation, a modified ySAGA complex, lacking ySpt8, accompanies Pol II during elongation and functions to acetylate and subsequently evict nucleosomes from gene coding regions (Figure 1 C, D) [26]. The Dub module of ySAGA, including Ubp8, Sus1 and Sgf11, also facilitates elongation through deubiquitination of H2B K123ub1 (Figure 1C), which allows for the recruitment of the Ctk1 kinase and subsequent Ser2 phosophorylation of the Pol II C-terminal domain (Figure 1D) [27]. ySAGA contributes to other aspects of gene expression as well. For example, SAGA-mediated histone acetylation functions in co-transcriptional spliceosome assembly and the recruitment of U2 snRNP to intron branchpoints [28], and SAGA-associated Sgf73 and Sus1 proteins link histone deubiquitination to the mRNA export machinery [29].
Table 1.
Components of the SAGA multi-subunit complexes in yeast, Drosophila and human. The protein structures of the SAGA orthologs are shown to scale. The legend denotes the domain names. The protein domains were determined with SMART (http://smart.embl-heidelberg.de/).

|
Figure 1.
The enzymatic activities of SAGA facilitate transcription. Different modules of SAGA are required for various stages of gene activation. A. The recruitment module localizes the SAGA complex to target genes, via an activator protein, allowing the HAT module to acetylate neighboring histones. B. The TBP interaction module strengthens SAGA interactions with the transcriptional machinery and phosphorylation (P) of the Pol II CTD at Ser5 allows transcription to proceed. C. Upon initiation, SAGA is recruited to the Pol II machinery and assists in elongation, promoting further acetylation of histones within the coding region and the Dub module deubiquitinates histones H2B. D. Deubiquitinated histones allow the interaction of Ctk1 with H2B, which phosphorylates Ser2 of the Pol II CTD to extend transcriptional elongation. Some of the questions remaining in this process include: A. Can other SAGA components (X) also contribute to HAT activity? B. Do PTMs (*) also regulate aspects of SAGA module interactions and activities? C. Does histone acetylation have any effect on SAGA Dub activity? C, D. Can the HAT module control Dub activity and vice versa?
The components, modules and functions of the ySAGA are highly conserved across many species and act as a model for understanding the Drosophila (d) and human (h) SAGA complexes. The remainder of our discussion will focus on the enzymatic modules of these metazoan SAGA complexes and how their regulation is intimately linked to both development and disease.
The SAGA histone acetyltransferase module
Although the general compositions of dSAGA and hSAGA complexes are very similar to the ySAGA complex, some differences have been observed. The GCN5 transcript is alternatively spliced in mouse and human cells, generating a short (GCN5S) isoform similar to yGcn5, and more predominantly expressed long (GCN5L) protein isoform that is incorporated into hSAGA and is highly analogous to the GCN5-related family member, PCAF [30, 31]. This long GCN5 isoform is also conserved in Drosophila Gcn5 (dGcn5), suggesting an evolutionary gain of function in ancestral metazoans [32]. GCN5L (hereafter referred to as GCN5) contains both a HAT domain and a bromodomain similar to yGcn5, but also has an extended N-terminal domain which confers the ability to acetylate mononucleosomal H3 as well as free H3 in vitro [30, 33]. Metazoans also have two proteins similar to yAda2, ADA2A and ADA2B, which are found in distinct GCN5-containing complexes [33-36]. In particular, ADA2B is present in hSAGA and serves to bridge the interactions between GCN5 and ADA3 to facilitate acetylation of chromatin substrates [33]. Interestingly, both ADA2B and ADA3 may regulate distinct aspects of chromatin acetylation, as ADA2B-GCN5 dimeric complexes enhance the ability of GCN5 to acetylate mononucleosomes, whereas acetylation of polynucleosomes is strictly mediated by ADA2B-GCN5-ADA3 heterotrimeric complexes in vitro [33]. dAda2b and dAda3 also likely regulate some aspect of chromatin acetylation in vivo, as deletion of either dAda2b or dAda3 leads to decreased H3 K9, 14 acetylation levels on polytene chromosomes [37, 38]. Furthermore, loss of Wda (a yTAF5 homolog) in Drosophila also reduces H3 K9 acetylation levels without compromising the integrity of the dSAGA complex, suggesting a role for Wda in chromatin acetylation [39]. Post-translational modifications identified on these and other subunits [40], as well as the acetylation of hADA3 by GCN5 [33], imply that PTMs may also play an important but unexplored role in regulating the functions of SAGA, including HAT activity.
As in yeast, hSAGA is recruited to specific target genes by activators, such as c-Myc, p53 and E2F, to stimulate the acetylation of histones and the activation of transcription [41-45].
While gene expression profiling for GCN5 has yet to be reported in mammalian cells, experiments in both budding (S. cerevisiae) and fission (S. pombe) yeast support the idea that GCN5 is a gene-specific co-activator, as yGcn5 only appears to regulate 4% and 1.1% of genes, respectively [46, 47]. However, numerous genome–wide mapping studies of histone acetylation patterns demonstrate that yGcn5 globally acetylates H3 K9, 14 and that Gcn5 binds within the promoter and 5’ coding regions of most highly transcribed genes in S. cerevisiae [48-52]. Additionally, genome-wide analysis of yGcn5 in S. pombe indicates that Gcn5 may have a general role in elongation, given that yGcn5 is found within the coding region of actively transcribed genes, and correlates with high H3 K14 acetylation levels [53]. Interestingly, hSAGA contains at least three subunits that have the ability to bind histone tails, including SGF29 which harbors a chromodomain, hSTAF65γ that displays a BTP domain (bromodomain transcription factors and PHD containing), and hADA2B that has a SANT domain (DNA binding domain in the SWI-SNF and ADA complexes, the transcriptional co-repressor N-CoR and TFIIIB). A role for these subunits in SAGA recruitment has not been reported; however, global Gcn5 mediated acetylation may be modulated by such domains. Collectively, these studies indicate that hSAGA functions both in gene specific and in global transcription events and that it can act at multiple steps in the transcription cycle.
The SAGA Deubiquitination module
Although first discovered and characterized as a HAT complex, SAGA was later discovered to harbor a second catalytic module centered on the Ubp8 (yeast) or USP22 (metazoan) deubiquitinases. As with GCN5, the Dub subunit may work most efficiently in the context of a module within SAGA that contains additional components [29]. In yeast, Sus1, Sgf11, and Ubp8 form a structural entity within the ySAGA holoenzyme, and the association of this Dub module with the rest of the complex is mediated by Sgf73 [29]. Sgf11 does not have a substantial role in maintaining the overall composition or integrity of SAGA but appears to mediate either recruitment or stable association of Ubp8 in ySAGA [20, 54]. Sus1 is present in two functionally different protein complexes in both yeast and in flies, the SAGA complex where it regulates its deubiquitination activity through interactions with Ubp8 and Sgf11 and the TREX/AMEX complex, where it plays a role in mRNA export [55-57]. Such dual functions of E(y) 2 (the Drosophila ortholog of Sus1) have implicated dSAGA in the positioning of specific gene loci (like hsp70) to the nuclear periphery, achieving a tightly regulated link between transcription and mRNA export under stress conditions [56]. hUSP22 deubiquitinates H2Bub, like the yeast ortholog, but can also deubiquitinate H2A in vitro , implicating its function in the regulation of Polycomb-mediated H2A ubiquitination [58, 59].
Polyglutamine (polyQ) expansions in the N-terminal region of ATXN7 (the mammalian ortholog of Sgf73) are associated with spinocerebellar ataxia type 7 (SCA7), which is characterized both by motor coordination deficiencies (ataxia) and retinal defects. Mouse models of SCA7 indicate that both the wild type and mutant forms of ATXN7 integrate into the SAGA complex and do not affect SAGA composition. However the effects of polyQ ATXN7 on SAGA HAT activity or on SAGA-mediated gene activation are not clear. In one set of experiments, the acetyltransferase activity of the complex did not appear to be affected by transgenic polyQ ATXN7, even though chromatin immunoprecipitation experiments indicated aberrant SAGA recruitment and hyperacetylation of H3 K9, 14 on rod cell-specific gene promoters in the retinas of the transgenic mice. Surprisingly these hyperacetylation events correlated with decreased mRNA levels of the analyzed genes [60]. In contrast, two other studies which used either cultured cells or yeast expressing the polyQ forms of ATXN7/Sgf73 came to very different conclusions on the effects of the mutant protein on SAGA functions [61, 62]. Reduced levels of acetylated H3 were observed on promoter-enhancer regions of photoreceptor genes, likely caused by impaired acetyltransferase activity of the complex in the presence of the polyQ expanded form of ATXN7 [62]. Neither of these studies addressed the effects of the polyQ allele on the Dub module. Clearly, much more work is needed to understand how the polyQ expansion affects SAGA recruitment and enzymatic activities.
The role of SAGA in development
Given that SAGA is conserved across many species and plays an essential role in regulating gene expression, it is perhaps not surprising that particular subunits of the SAGA HAT module control developmental processes. In Drosophila, loss of dGcn5 is lethal due to a lack of metamorphosis[63]. Furthermore, dAda2b and dAda3, which regulate H3 K9, 14 acetyaltion are also essential for Drosophila viability. In a similar manner, Gcn5 is required for normal development in mice. Gcn5 null (Gcn5-/-) mice are embryonic lethal at 10.5 days post coitum (d.p.c.) and do not acquire normal mesoderm due to enhanced apoptosis in these tissues [64]. However, catalytically inactive Gcn5 mice survive slightly longer (16.5 d.p.c.) and display both neural tube closure defects and exencephaly, implying that Gcn5 HAT activity is required for proper development and that SAGA also has HAT-independent functions [65]. Interestingly, PCAF null mice develop normally, but double homozygous null Gcn5 and PCAF embryos die at 7.5 d.p.c. indicating a functional redundancy for these two HATs during embryonic development [64, 66]. In cell culture, Gcn5 -/- mouse embryonic stem cells remain pluripotent and are capable of differentiation into all three germ layers [67]. The role of GCN5 in ES cells has yet to be defined, but is of considerable interest. H3 K56ac is linked to pluripotency of human embryonic stem cells and GCN5 acetylates H3 K56 on free histone in vitro, suggesting that GCN5 may play a role in maintaining stemness [68, 69].
In addition to the HAT module regulating larval and embryonic development, the SAGA Dub module has been identified to control different aspects of neural development. This is observed by mutations in the Drosophila nonstop (USP22 ortholog) and dSgf11 proteins leading to defects in axonal targeting photoreceptors [70, 71]. Furthermore, retinal defects are also observed in the SCA7 neurodegenerative disease as discussed above, but the roles of USP22 or other Dub module subunits in development have not yet been defined.
The role of SAGA in cancer
The role of hSAGA in cancer is a good paradigm of its multi-functionality. For example, TRRAP and GCN5 regulate the oncogenic activity of the c-Myc oncoprotein and the p53 tumor suppressor [41]. Additional studies underscore that the whole SAGA complex may function in tumorigenesis. STAF65γ (hSPT7) has a role in the transcription of several Myc-dependent genes [43]. Furthermore, SAGA recruitment may require SGF29, a poorly studied subunit of the complex, whose deregulated expression is implicated in malignant transformation [72]. In parallel, GCN5/PCAF acetylates non–histone proteins involved in transcriptional activation or repression, such as c-Myc [73].
The deubiquitination activity of the hSAGA complex has also been implicated in cancer, as USP22 has been grouped by transcriptional profiling into an 11-gene “signature” of poor prognosis in multiple types of cancer [59, 74]. This signature also includes the PRC1 polycomb repressor complex subunit, Bmi1, a known regulator of self renewal in normal and malignant stem cells, suggesting USP22 may be a marker for cancer stem cells. Specifically, USP22 may be required for cell-cycle progression and could function with GCN5 to control certain cell cycle genes. GCN5 has been implicated in the transcriptional regulation of cyclin A, cyclin D3, PCNA and cdc25b, as well as CDK1 and cyclin B1 after DNA damage [75]. Furthermore, GCN5 acetylates the non-histone protein CDC6 to promote S-phase progression of the cell cycle. GCN5 and USP22 also protect telomeres from DNA damage response through the stabilization of a shelterin component called TRF1, and interestingly, this regulation is not transcriptional but involves USP22-mediated deubiquitination of TRF1 [76]. In addition, the Gcn5 related protein, PCAF, appears to regulate ubiquitination of Hdm2 through an intrinsic ubiquitination activity [77] and chk2 turnover [78], adding another level of regulation to an already complicated network.
Conclusions
SAGA is comprised of distinct modules that intimately collaborate to maintain structural integrity, genomic recruitment and interactions with the basal transcription machinery to localize the HAT and Dub activities of the complex to gene loci. Elegantly, SAGA mediates histone acetylation of gene promoters to enhance transcriptional activation and facilitates elongation by deubiquitinating histones downstream of Pol II and acetylating histones within the coding region to promote histone eviction (Figure 1). Functional HAT and Dub modules of the SAGA complex are central to normal mouse and Drosophila development. In addition, SAGA functions in cancer, as USP22 is part of an 11 gene “signature” associated with poor cancer prognosis and SAGA contributes to the activation of c-Myc responsive genes. However, many questions remain unanswered regarding the intricate regulation of this co-activator complex. Do SAGA components outside of the HAT and Dub modules either stimulate or suppress these enzymatic activities (Figure 1A)? What role do posttranslational modifications of the SAGA components play in complex regulation (Figure 1B)? Is SAGA-mediated histone acetylation required for Dub activity downstream of Pol II (Figure 1C), and do members of the HAT and Dub modules cross regulate each other (Figure 1C, D)? In addition to these questions, much remains to be elucidated about the biological function of SAGA. Little is known about the role of USP22 in mammalian development or the function of GCN5 and/or USP22 in embryonic stem cells. Furthermore, it is unclear how tightly the functions of GCN5 and USP22 are linked or whether the HAT and/or Dub module functions in SCA7 disease. Addressing these questions will undoubtedly lead to a clearer understanding for the roles of SAGA in development and disease.
Acknowledgements
We thank Boyko Atanassov, Yi Chun Chen, Andria Schibler and Marenda Wilson-Pham for useful comments and discussions on the manuscript. Calley L. Hirsch is funded as an Odyssey Fellow supported by the Odyssey Program and the Houston Endowment Inc, Award for Scientific Achievement at The Univeristy of Texas M.D. Anderson Cancer Center. Parts of this work were supported by a grant from the NIH to SYRD, GM067718.
Glossary
- Ada
adaptor
- ATXN
ataxin
- BrD
bromodomain
- BTP
bromodomain transcription factors and PHD domain containing
- CDK1
cyclin dependent kinase 1
- Chk2
checkpoint kinase 2
- ChrD
chromodomain
- Ctk1
carboxy-terminal domain kinase 1
- DEXDc
DEAD-like helicase
- dE(y)2
Drosophila enhancer of yellow 2
- Dub
deubiquitinase
- EnY2
human ortholog of enhancer of yellow 2
- FAT
FRAP/ATM/TRRAP domain
- Gcn5
general control nonderepressible 5
- HAT
histone acetyltransferase
- Hdm2
human ortholog of mouse double minute 2 (mdm2)
- LisH
Lis homology
- nonstop
Drosophila analog of Ubp8
- PCAF
p300/CBP-associated factor
- PCNA
proliferating cell nuclear antigen
- PI3Kc
phosphoinositide 3-kinase, catalytic domain
- PIC
pre-initiation complex
- Pol II
RNA polymerase II
- PTM
post-translational modification
- SAGA
Spt-Ada-Gcn5 acetyltranferase complex
- SANT
Swi3/Ada2/N-CoR/TFIIIB domain
- SCA7
spinocerebellar ataxia 7
- Sgf
SAGA-associated factor
- Spt
suppressor of Ty element
- STAF
specificity protein transcription factor - associated factor
- Sus1
S1 gene upstream of ySa1
- SWIRM
Swi3/Rsc8/Moira domain
- TAF
TATA binding protein-associated factor
- TBP
TATA binding protein
- TFIID
transcription factor II D
- Tra1
yeast ortholog of human TRRAP
- TREX/AMEX
transcription and export/mRNA and export
- TRF1
telomeric-repeat binding factor 1
- TRRAP
transformation/transcription domain-associated protein
- Ubp8
ubiquitin specific processing protease
- UBP-ZF
ubiquitin protease – zinc finger
- UCH
ubiquitin carboxyl-terminal hydrolase
- USP22
ubiquitin specific peptidase 22
- WD40
40 amino acid protein domain often terminating in Trp-Asp
- Wda
will decrease acetylation
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Evangelia Koutelou, Department of Biochemistry and Molecular Biology, University of Texas M.D. Anderson Cancer Center, Houston, TX 77030, USA.
Calley L. Hirsch, Program in Genes and Development, University of Texas M.D. Anderson Cancer Center, Houston, TX 77030, USA.
Sharon Y.R. Dent, Center for Cancer Epigenetics, University of Texas M.D. Anderson Cancer Center, Houston, TX 77030, USA.
References
- 1.Brownell JE, Zhou J, Ranalli T, Kobayashi R, Edmondson DG, Roth SY, Allis CD. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- 2.Weake VM, Workman JL. Histone ubiquitination: triggering gene activity. Molecular cell. 2008;29:653–663. doi: 10.1016/j.molcel.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 3.Nagy Z, Tora L. Distinct GCN5/PCAF-containing complexes function as co-activators and are involved in transcription factor and global histone acetylation. Oncogene. 2007;26:5341–5357. doi: 10.1038/sj.onc.1210604. [DOI] [PubMed] [Google Scholar]
- 4.Baker SP, Grant PA. The SAGA continues: expanding the cellular role of a transcriptional co-activator complex. Oncogene. 2007;26:5329–5340. doi: 10.1038/sj.onc.1210603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rodriguez-Navarro S. Insights into SAGA function during gene expression. EMBO Reports. 2009;10:843–850. doi: 10.1038/embor.2009.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuo MH, Brownell JE, Sobel RE, Ranalli TA, Cook RG, Edmondson DG, Roth SY, Allis CD. Transcription-linked acetylation by Gcn5p of histones H3 and H4 at specific lysines. Nature. 1996;383:269–272. doi: 10.1038/383269a0. [DOI] [PubMed] [Google Scholar]
- 7.Grant PA, Eberharter A, John S, Cook RG, Turner BM, Workman JL. Expanded lysine acetylation specificity of Gcn5 in native complexes. J Biol Chem. 1999;274:5895–5900. doi: 10.1074/jbc.274.9.5895. [DOI] [PubMed] [Google Scholar]
- 8.Grant PA, Duggan L, Cote J, Roberts SM, Brownell JE, Candau R, Ohba R, Owen-Hughes T, Allis CD, Winston F, et al. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- 9.Grant PA, Schieltz D, Pray-Grant MG, Yates JR, 3rd, Workman JL. The ATM-related cofactor Tra1 is a component of the purified SAGA complex. Molecular cell. 1998;2:863–867. doi: 10.1016/s1097-2765(00)80300-7. [DOI] [PubMed] [Google Scholar]
- 10.Brown CE, Howe L, Sousa K, Alley SC, Carrozza MJ, Tan S, Workman JL. Recruitment of HAT complexes by direct activator interactions with the ATM-related Tra1 subunit. Science. 2001;292:2333–2337. doi: 10.1126/science.1060214. [DOI] [PubMed] [Google Scholar]
- 11.Marcus GA, Silverman N, Berger SL, Horiuchi J, Guarente L. Functional similarity and physical association between GCN5 and ADA2: putative transcriptional adaptors. Embo J. 1994;13:4807–4815. doi: 10.1002/j.1460-2075.1994.tb06806.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horiuchi J, Silverman N, Marcus GA, Guarente L. ADA3, a putative transcriptional adaptor, consists of two separable domains and interacts with ADA2 and GCN5 in a trimeric complex. Mol Cell Biol. 1995;15:1203–1209. doi: 10.1128/mcb.15.3.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sterner DE, Grant PA, Roberts SM, Duggan LJ, Belotserkovskaya R, Pacella LA, Winston F, Workman JL, Berger SL. Functional organization of the yeast SAGA complex: distinct components involved in structural integrity, nucleosome acetylation, and TATA-binding protein interaction. Mol Cell Biol. 1999;19:86–98. doi: 10.1128/mcb.19.1.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Balasubramanian R, Pray-Grant MG, Selleck W, Grant PA, Tan S. Role of the Ada2 and Ada3 transcriptional coactivators in histone acetylation. J Biol Chem. 2002;277:7989–7995. doi: 10.1074/jbc.M110849200. [DOI] [PubMed] [Google Scholar]
- 15.Bhaumik SR, Raha T, Aiello DP, Green MR. In vivo target of a transcriptional activator revealed by fluorescence resonance energy transfer. Genes Dev. 2004;18:333–343. doi: 10.1101/gad.1148404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mohibullah N, Hahn S. Site-specific cross-linking of TBP in vivo and in vitro reveals a direct functional interaction with the SAGA subunit Spt3. Genes Dev. 2008;22:2994–3006. doi: 10.1101/gad.1724408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henry KW, Wyce A, Lo WS, Duggan LJ, Emre NC, Kao CF, Pillus L, Shilatifard A, Osley MA, Berger SL. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev. 2003;17:2648–2663. doi: 10.1101/gad.1144003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Daniel JA, Torok MS, Sun ZW, Schieltz D, Allis CD, Yates JR, 3rd, Grant PA. Deubiquitination of histone H2B by a yeast acetyltransferase complex regulates transcription. J Biol Chem. 2004;279:1867–1871. doi: 10.1074/jbc.C300494200. [DOI] [PubMed] [Google Scholar]
- 19.Rodriguez-Navarro S, Fischer T, Luo MJ, Antunez O, Brettschneider S, Lechner J, Perez-Ortin JE, Reed R, Hurt E. Sus1, a functional component of the SAGA histone acetylase complex and the nuclear pore-associated mRNA export machinery. Cell. 2004;116:75–86. doi: 10.1016/s0092-8674(03)01025-0. [DOI] [PubMed] [Google Scholar]
- 20.Lee KK, Florens L, Swanson SK, Washburn MP, Workman JL. The deubiquitylation activity of Ubp8 is dependent upon Sgf11 and its association with the SAGA complex. Mol Cell Biol. 2005;25:1173–1182. doi: 10.1128/MCB.25.3.1173-1182.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shukla A, Bajwa P, Bhaumik SR. SAGA-associated Sgf73p facilitates formation of the preinitiation complex assembly at the promoters either in a HAT-dependent or independent manner in vivo. Nucleic Acids Res. 2006;34:6225–6232. doi: 10.1093/nar/gkl844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grant PA, Schieltz D, Pray-Grant MG, Steger DJ, Reese JC, Yates JR, 3rd, Workman JL. A subset of TAF(II)s are integral components of the SAGA complex required for nucleosome acetylation and transcriptional stimulation. Cell. 1998;94:45–53. doi: 10.1016/s0092-8674(00)81220-9. [DOI] [PubMed] [Google Scholar]
- 23.Wu PY, Ruhlmann C, Winston F, Schultz P. Molecular architecture of the S. cerevisiae SAGA complex. Molecular cell. 2004;15:199–208. doi: 10.1016/j.molcel.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 24.Owen DJ, Ornaghi P, Yang JC, Lowe N, Evans PR, Ballario P, Neuhaus D, Filetici P, Travers AA. The structural basis for the recognition of acetylated histone H4 by the bromodomain of histone acetyltransferase gcn5p. Embo J. 2000;19:6141–6149. doi: 10.1093/emboj/19.22.6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25•.Li S, Shogren-Knaak MA. The Gcn5 bromodomain of the SAGA complex facilitates cooperative and cross-tail acetylation of nucleosomes. J Biol Chem. 2009;284:9411–9417. doi: 10.1074/jbc.M809617200. [The authors perform kinetic analysis of SAGA-mediated acetylation, identifying that the Gcn5 bromodomain is necessary to promote nucleosome acetylation activity.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Govind CK, Zhang F, Qiu H, Hofmeyer K, Hinnebusch AG. Gcn5 promotes acetylation, eviction, and methylation of nucleosomes in transcribed coding regions. Molecular cell. 2007;25:31–42. doi: 10.1016/j.molcel.2006.11.020. [DOI] [PubMed] [Google Scholar]
- 27••.Wyce A, Xiao T, Whelan KA, Kosman C, Walter W, Eick D, Hughes TR, Krogan NJ, Strahl BD, Berger SL. H2B ubiquitylation acts as a barrier to Ctk1 nucleosomal recruitment prior to removal by Ubp8 within a SAGA-related complex. Molecular cell. 2007;27:275–288. doi: 10.1016/j.molcel.2007.01.035. [Ubp8 is shown to have an important role in transcriptional elongation. After the initiation of transcription, Ubp8 is responsible for H2B ubiquitin removal within the open reading frame of the GAL1 gene. Ubp8-mediated deubiquitination allows Ctk1 to interact with histones H2A/H2B and in turn phosphorylate the CTD of Pol II to promote elongation.] [DOI] [PubMed] [Google Scholar]
- 28•.Gunderson FQ, Johnson TL. Acetylation by the transcriptional coactivator Gcn5 plays a novel role in co-transcriptional spliceosome assembly. PLoS Genet. 2009;5:e1000682. doi: 10.1371/journal.pgen.1000682. [An elegant genetic analysis in yeast that provides evidence for an additional role of the SAGA complex in pre-messenger RNA splicing. Gcn5 HAT activity is required for the recruitment of specific components of the splicing machinery and contributes to the coordination of splicing with transcription.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29••.Kohler A, Schneider M, Cabal GG, Nehrbass U, Hurt E. Yeast Ataxin-7 links histone deubiquitination with gene gating and mRNA export. Nat Cell Biol. 2008;10:707–715. doi: 10.1038/ncb1733. [This study elucidates the role of Sgf73 (the yeast ortholog of mammalian Ataxin-7) in the integration of the Dub module in SAGA complex. Detailed biochemical experiments clearly show the cooperativity between different components of the Dub module and domain analysis of Sgf73 dissects how it mediates the recruitment of the TREX-2 mRNA export factors to SAGA.] [DOI] [PubMed] [Google Scholar]
- 30.Xu W, Edmondson DG, Roth SY. Mammalian GCN5 and P/CAF acetyltransferases have homologous amino-terminal domains important for recognition of nucleosomal substrates. Mol Cell Biol. 1998;18:5659–5669. doi: 10.1128/mcb.18.10.5659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martinez E, Palhan VB, Tjernberg A, Lymar ES, Gamper AM, Kundu TK, Chait BT, Roeder RG. Human STAGA complex is a chromatin-acetylating transcription coactivator that interacts with pre-mRNA splicing and DNA damage-binding factors in vivo. Mol Cell Biol. 2001;21:6782–6795. doi: 10.1128/MCB.21.20.6782-6795.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith ER, Belote JM, Schiltz RL, Yang XJ, Moore PA, Berger SL, Nakatani Y, Allis CD. Cloning of Drosophila GCN5: conserved features among metazoan GCN5 family members. Nucleic Acids Res. 1998;26:2948–2954. doi: 10.1093/nar/26.12.2948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33••.Gamper AM, Kim J, Roeder RG. The STAGA subunit ADA2b is an important regulator of human GCN5 catalysis. Mol Cell Biol. 2009;29:266–280. doi: 10.1128/MCB.00315-08. [This paper shows that human ADA2b, but not ADA2a, is incorporated into hSAGA. Furthermore, ADA2b and ADA3 form a heterotrimeric complex with GCN5, which efficiently acetylates nucleosomes in vitro, but ADA3-ADA2a-Gcn5 complexes do not facilitate nucleosome acetylation. In agreement, only ADA2b is found to associate with SAGA specific p53-target genes following DNA damage.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guelman S, Suganuma T, Florens L, Swanson SK, Kiesecker CL, Kusch T, Anderson S, Yates JR, 3rd, Washburn MP, Abmayr SM, et al. Host cell factor and an uncharacterized SANT domain protein are stable components of ATAC, a novel dAda2A/dGcn5-containing histone acetyltransferase complex in Drosophila. Mol Cell Biol. 2006;26:871–882. doi: 10.1128/MCB.26.3.871-882.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang YL, Faiola F, Xu M, Pan S, Martinez E. Human ATAC Is a GCN5/PCAF-containing acetylase complex with a novel NC2-like histone fold module that interacts with the TATA-binding protein. J Biol Chem. 2008;283:33808–33815. doi: 10.1074/jbc.M806936200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guelman S, Kozuka K, Mao Y, Pham V, Solloway MJ, Wang J, Wu J, Lill JR, Zha J. The double-histone-acetyltransferase complex ATAC is essential for mammalian development. Mol Cell Biol. 2009;29:1176–1188. doi: 10.1128/MCB.01599-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qi D, Larsson J, Mannervik M. Drosophila Ada2b is required for viability and normal histone H3 acetylation. Mol Cell Biol. 2004;24:8080–8089. doi: 10.1128/MCB.24.18.8080-8089.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38•.Grau B, Popescu C, Torroja L, Ortuno-Sahagun D, Boros I, Ferrus A. Transcriptional adaptor ADA3 of Drosophila melanogaster is required for histone modification, position effect variegation, and transcription. Mol Cell Biol. 2008;28:376–385. doi: 10.1128/MCB.01307-07. [These authors highlight the importance of dAda3 in HAT function and Drosophila development. dAda3 is required for late larval stages and loss of dAda3 leads to decreased H3 and H4 acetylation levels at specific lysine residues. In addition, Ada3 is regulates the expression of specific genes.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guelman S, Suganuma T, Florens L, Weake V, Swanson SK, Washburn MP, Abmayr SM, Workman JL. The essential gene wda encodes a WD40 repeat subunit of Drosophila SAGA required for histone H3 acetylation. Mol Cell Biol. 2006;26:7178–7189. doi: 10.1128/MCB.00130-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40••.Mischerikow N, Spedale G, Altelaar AF, Timmers HT, Pijnappel WW, Heck AJ. In-depth profiling of post-translational modifications on the related transcription factor complexes TFIID and SAGA. J Proteome Res. 2009;8:5020–5030. doi: 10.1021/pr900449e. [This paper provides the first report of post-translational modifications specific to components of ySAGA. In particular, unique sites of protein acetylation and phosphorylation in ySAGA and TFIID were compared. Within the ySAGA complex, ySpt7 and ySgf73 were identified to be significantly acetylated, while ySpt7, yAda3, yTaf10, yTaf12 and ySgf73 were found to be phosphorylated.] [DOI] [PubMed] [Google Scholar]
- 41.McMahon SB, Wood MA, Cole MD. The essential cofactor TRRAP recruits the histone acetyltransferase hGCN5 to c-Myc. Mol Cell Biol. 2000;20:556–562. doi: 10.1128/mcb.20.2.556-562.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gamper AM, Roeder RG. Multivalent binding of p53 to the STAGA complex mediates coactivator recruitment after UV damage. Mol Cell Biol. 2008;28:2517–2527. doi: 10.1128/MCB.01461-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43••.Liu X, Vorontchikhina M, Wang YL, Faiola F, Martinez E. STAGA recruits Mediator to the MYC oncoprotein to stimulate transcription and cell proliferation. Mol Cell Biol. 2008;28:108–121. doi: 10.1128/MCB.01402-07. [The authors show that hSAGA participates in Mediator recruitment during transcriptional activation. Myc functions to recruit the hSAGA components TRRAP and GCN5 to the TERT target gene for histone acetylation, in a manner independent of STAF65γ. However, STAF65γ is required for Myc recruitment of SPT3 and TAF9 to the TERT promoter. As well, both STAF65γ and Myc are needed to recruit the Mediator components MED1, MED16 and MED17 to the TERT gene. This data highlights the modularity of the hSAGA complex.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu X, Tesfai J, Evrard YA, Dent SY, Martinez E. c-Myc transformation domain recruits the human STAGA complex and requires TRRAP and GCN5 acetylase activity for transcription activation. J Biol Chem. 2003;278:20405–20412. doi: 10.1074/jbc.M211795200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lang SE, McMahon SB, Cole MD, Hearing P. E2F transcriptional activation requires TRRAP and GCN5 cofactors. J Biol Chem. 2001;276:32627–32634. doi: 10.1074/jbc.M102067200. [DOI] [PubMed] [Google Scholar]
- 46.Lee TI, Causton HC, Holstege FC, Shen WC, Hannett N, Jennings EG, Winston F, Green MR, Young RA. Redundant roles for the TFIID and SAGA complexes in global transcription. Nature. 2000;405:701–704. doi: 10.1038/35015104. [DOI] [PubMed] [Google Scholar]
- 47.Helmlinger D, Marguerat S, Villen J, Gygi SP, Bahler J, Winston F. The S. pombe SAGA complex controls the switch from proliferation to sexual differentiation through the opposing roles of its subunits Gcn5 and Spt8. Genes Dev. 2008;22:3184–3195. doi: 10.1101/gad.1719908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rosaleny LE, Ruiz-Garcia AB, Garcia-Martinez J, Perez-Ortin JE, Tordera V. The Sas3p and Gcn5p histone acetyltransferases are recruited to similar genes. Genome Biol. 2007;8:R119. doi: 10.1186/gb-2007-8-6-r119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Durant M, Pugh BF. Genome-wide relationships between TAF1 and histone acetyltransferases in Saccharomyces cerevisiae. Mol Cell Biol. 2006;26:2791–2802. doi: 10.1128/MCB.26.7.2791-2802.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, Bell GW, Walker K, Rolfe PA, Herbolsheimer E, et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 51.Robert F, Pokholok DK, Hannett NM, Rinaldi NJ, Chandy M, Rolfe A, Workman JL, Gifford DK, Young RA. Global position and recruitment of HATs and HDACs in the yeast genome. Molecular cell. 2004;16:199–209. doi: 10.1016/j.molcel.2004.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roh TY, Ngau WC, Cui K, Landsman D, Zhao K. High-resolution genome-wide mapping of histone modifications. Nat Biotechnol. 2004;22:1013–1016. doi: 10.1038/nbt990. [DOI] [PubMed] [Google Scholar]
- 53.Johnsson A, Durand-Dubief M, Xue-Franzen Y, Ronnerblad M, Ekwall K, Wright A. HAT-HDAC interplay modulates global histone H3K14 acetylation in gene-coding regions during stress. EMBO Rep. 2009;10:1009–1014. doi: 10.1038/embor.2009.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ingvarsdottir K, Krogan NJ, Emre NC, Wyce A, Thompson NJ, Emili A, Hughes TR, Greenblatt JF, Berger SL. H2B ubiquitin protease Ubp8 and Sgf11 constitute a discrete functional module within the Saccharomyces cerevisiae SAGA complex. Mol Cell Biol. 2005;25:1162–1172. doi: 10.1128/MCB.25.3.1162-1172.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pascual-Garcia P, Govind CK, Queralt E, Cuenca-Bono B, Llopis A, Chavez S, Hinnebusch AG, Rodriguez-Navarro S. Sus1 is recruited to coding regions and functions during transcription elongation in association with SAGA and TREX2. Genes Dev. 2008;22:2811–2822. doi: 10.1101/gad.483308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kurshakova MM, Krasnov AN, Kopytova DV, Shidlovskii YV, Nikolenko JV, Nabirochkina EN, Spehner D, Schultz P, Tora L, Georgieva SG. SAGA and a novel Drosophila export complex anchor efficient transcription and mRNA export to NPC. Embo J. 2007;26:4956–4965. doi: 10.1038/sj.emboj.7601901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kohler A, Pascual-Garcia P, Llopis A, Zapater M, Posas F, Hurt E, Rodriguez-Navarro S. The mRNA export factor Sus1 is involved in Spt/Ada/Gcn5 acetyltransferase-mediated H2B deubiquitinylation through its interaction with Ubp8 and Sgf11. Mol Biol Cell. 2006;17:4228–4236. doi: 10.1091/mbc.E06-02-0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58•.Zhao Y, Lang G, Ito S, Bonnet J, Metzger E, Sawatsubashi S, Suzuki E, Le Guezennec X, Stunnenberg HG, Krasnov A, et al. A TFTC/STAGA module mediates histone H2A and H2B deubiquitination, coactivates nuclear receptors, and counteracts heterochromatin silencing. Molecular cell. 2008;29:92–101. doi: 10.1016/j.molcel.2007.12.011. [The authors found that USP22 and its Drosophila homolog nonstop use deubiquitinating activity on ubiquitinated H2A and H2B and this event facilitates the transactivation by the androgen receptor in human cells. They also identify the subunits of the mammalian Dub module and they observe that this deubiquitination activity enhances the positional effect variegation in Drosophila.] [DOI] [PubMed] [Google Scholar]
- 59•.Zhang XY, Varthi M, Sykes SM, Phillips C, Warzecha C, Zhu W, Wyce A, Thorne AW, Berger SL, McMahon SB. The putative cancer stem cell marker USP22 is a subunit of the human SAGA complex required for activated transcription and cell-cycle progression. Molecular cell. 2008;29:102–111. doi: 10.1016/j.molcel.2007.12.015. [USP22 deubiquitinating activity is important for the transactivation of both Myc and p53 target genes, a function that explains USP22 identification as a member of the “11-gene-signature” of aggressive cancer types.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Helmlinger D, Hardy S, Abou-Sleymane G, Eberlin A, Bowman AB, Gansmuller A, Picaud S, Zoghbi HY, Trottier Y, Tora L, et al. Glutamine-expanded ataxin-7 alters TFTC/STAGA recruitment and chromatin structure leading to photoreceptor dysfunction. PLoS Biol. 2006;4:e67. doi: 10.1371/journal.pbio.0040067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McMahon SJ, Pray-Grant MG, Schieltz D, Yates JR, 3rd, Grant PA. Polyglutamine-expanded spinocerebellar ataxia-7 protein disrupts normal SAGA and SLIK histone acetyltransferase activity. Proc Natl Acad Sci U S A. 2005;102:8478–8482. doi: 10.1073/pnas.0503493102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Palhan VB, Chen S, Peng GH, Tjernberg A, Gamper AM, Fan Y, Chait BT, La Spada AR, Roeder RG. Polyglutamine-expanded ataxin-7 inhibits STAGA histone acetyltransferase activity to produce retinal degeneration. Proc Natl Acad Sci U S A. 2005;102:8472–8477. doi: 10.1073/pnas.0503505102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Carre C, Szymczak D, Pidoux J, Antoniewski C. The histone H3 acetylase dGcn5 is a key player in Drosophila melanogaster metamorphosis. Mol Cell Biol. 2005;25:8228–8238. doi: 10.1128/MCB.25.18.8228-8238.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xu W, Edmondson DG, Evrard YA, Wakamiya M, Behringer RR, Roth SY. Loss of Gcn5l2 leads to increased apoptosis and mesodermal defects during mouse development. Nat Genet. 2000;26:229–232. doi: 10.1038/79973. [DOI] [PubMed] [Google Scholar]
- 65.Bu P, Evrard YA, Lozano G, Dent SY. Loss of Gcn5 acetyltransferase activity leads to neural tube closure defects and exencephaly in mouse embryos. Mol Cell Biol. 2007;27:3405–3416. doi: 10.1128/MCB.00066-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yamauchi T, Yamauchi J, Kuwata T, Tamura T, Yamashita T, Bae N, Westphal H, Ozato K, Nakatani Y. Distinct but overlapping roles of histone acetylase PCAF and of the closely related PCAF-B/GCN5 in mouse embryogenesis. Proc Natl Acad Sci U S A. 2000;97:11303–11306. doi: 10.1073/pnas.97.21.11303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lin W, Srajer G, Evrard YA, Phan HM, Furuta Y, Dent SY. Developmental potential of Gcn5(-/-) embryonic stem cells in vivo and in vitro. Dev Dyn. 2007;236:1547–1557. doi: 10.1002/dvdy.21160. [DOI] [PubMed] [Google Scholar]
- 68.Tjeertes JV, Miller KM, Jackson SP. Screen for DNA-damage-responsive histone modifications identifies H3K9Ac and H3K56Ac in human cells. Embo J. 2009;28:1878–1889. doi: 10.1038/emboj.2009.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xie W, Song C, Young NL, Sperling AS, Xu F, Sridharan R, Conway AE, Garcia BA, Plath K, Clark AT, et al. Histone h3 lysine 56 acetylation is linked to the core transcriptional network in human embryonic stem cells. Molecular cell. 2009;33:417–427. doi: 10.1016/j.molcel.2009.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70•.Weake VM, Lee KK, Guelman S, Lin CH, Seidel C, Abmayr SM, Workman JL. SAGA-mediated H2B deubiquitination controls the development of neuronal connectivity in the Drosophila visual system. Embo J. 2008;27:394–405. doi: 10.1038/sj.emboj.7601966. [In this study the Drosophila orthologs of the Dub module of SAGA nonstop and dSgf11 are analysed, providing strong evidence for the evolutionary conservation of SAGA complex among species and implicating the Drosophila SAGA complex in the regulation of accurate axon guidance in the optic lobe of the fly.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Poeck B, Fischer S, Gunning D, Zipursky SL, Salecker I. Glial cells mediate target layer selection of retinal axons in the developing visual system of Drosophila. Neuron. 2001;29:99–113. doi: 10.1016/s0896-6273(01)00183-0. [DOI] [PubMed] [Google Scholar]
- 72.Kurabe N, Katagiri K, Komiya Y, Ito R, Sugiyama A, Kawasaki Y, Tashiro F. Deregulated expression of a novel component of TFTC/STAGA histone acetyltransferase complexes, rat SGF29, in hepatocellular carcinoma: possible implication for the oncogenic potential of c-Myc. Oncogene. 2007;26:5626–5634. doi: 10.1038/sj.onc.1210349. [DOI] [PubMed] [Google Scholar]
- 73.Patel JH, Du Y, Ard PG, Phillips C, Carella B, Chen CJ, Rakowski C, Chatterjee C, Lieberman PM, Lane WS, et al. The c-MYC oncoprotein is a substrate of the acetyltransferases hGCN5/PCAF and TIP60. Mol Cell Biol. 2004;24:10826–10834. doi: 10.1128/MCB.24.24.10826-10834.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Glinsky GV, Berezovska O, Glinskii AB. Microarray analysis identifies a death-from-cancer signature predicting therapy failure in patients with multiple types of cancer. J Clin Invest. 2005;115:1503–1521. doi: 10.1172/JCI23412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shimada M, Niida H, Zineldeen DH, Tagami H, Tanaka M, Saito H, Nakanishi M. Chk1 is a histone H3 threonine 11 kinase that regulates DNA damage-induced transcriptional repression. Cell. 2008;132:221–232. doi: 10.1016/j.cell.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 76••.Atanassov BS, Evrard YA, Multani AS, Zhang Z, Tora L, Devys D, Chang S, Dent SY. Gcn5 and SAGA regulate shelterin protein turnover and telomere maintenance. Molecular cell. 2009;35:352–364. doi: 10.1016/j.molcel.2009.06.015. [The authors show that Gcn5 is implicated in telomeres maintenance by regulating components of the shelterin complex post-transcriptionally. They provide the first evidence for SAGA-dependent regulation of ubiquitination and protein turnover of TRF1, expanding the specificity and the targeting of SAGA complex Dub activity to non-histone substrates.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Linares LK, Kiernan R, Triboulet R, Chable-Bessia C, Latreille D, Cuvier O, Lacroix M, Le Cam L, Coux O, Benkirane M. Intrinsic ubiquitination activity of PCAF controls the stability of the oncoprotein Hdm2. Nat Cell Biol. 2007;9:331–338. doi: 10.1038/ncb1545. [DOI] [PubMed] [Google Scholar]
- 78.Kass EM, Poyurovsky MV, Zhu Y, Prives C. Mdm2 and PCAF increase Chk2 ubiquitination and degradation independently of their intrinsic E3 ligase activities. Cell Cycle. 2009;8:430–437. doi: 10.4161/cc.8.3.7624. [DOI] [PubMed] [Google Scholar]