Abstract
Giardia duodenalis is an intestinal parasite of many vertebrates. The presence of G. duodenalis in the marine environment due to anthropogenic and wildlife activity is well documented, including the contributions from untreated sewage and storm water, agricultural run-off, and droppings from terrestrial animals. Recently, studies have detected this protistan parasite in the feces of marine vertebrates such as whales, dolphins, seals and shore birds. To explore the population biology of G. duodenalis in marine life, we determined the prevalence of G. duodenalis in two species of seal (Halichoerus grypus, Phoca vitulina vitulina and Phoca vitulina richardsi) from the east and west coasts of the USA, sequenced two loci from G. duodenalis-positive samples to assess molecular diversity, and examined G. duodenalis distribution among these seals and other marine vertebrates along the east coast. We found a significant difference in the presence of G. duodenalis between east and west coast seal species. Only the zoonotic lineages of G. duodenalis, Assemblages A and B and a novel lineage, which we designated as Assemblage H, were identified in marine vertebrates. Assemblages A and B are broadly distributed geographically and show a lack of host specificity. Only grey seal (Halichoerus grypus) samples and one gull sample (Larus argentatus) from a northern location of Cape Cod, Massachusetts, USA showed the presence of Assemblage H haplotypes; only one other study of harbor seals from the Puget Sound region of Washington, USA previously recorded the presence of an Assemblage H haplotype. Assemblage H sequences form a monophyletic clade that appears as divergent from the other seven Assemblages of G. duodenalis as these assemblages are from each other. The discovery of a previously uncharacterized lineage of G. duodenalis suggests that this parasite has more genetic diversity and perhaps a larger host range than previously believed.
Keywords: Giardia duodenalis, Novel assemblage, Population biology, Phoca vitulina vitulina, Phoca vitulina richardsi, Halichoerus grypus, Marine vertebrates
1. Introduction
Giardia duodenalis is a parasitic protist that infects the upper intestines of many terrestrial vertebrates (Thompson and Monis, 2004). Until now, seven genetically distinct but morphologically identical lineages, Assemblages A–G, defined this species (Cacciò and Ryan, 2008). Assemblages A and B infect most vertebrates but are the only two assemblages known to infect humans and are therefore considered zoonotic. Assemblages C–G are considered host-specific: Assemblages C and D predominantly occur in dogs, Assemblage E in hoofed livestock, Assemblage F in cats and Assemblage G in rats (Cacciò and Ryan, 2008). Genetic distances equivalent to those that distinguish some protistan genera separate these assemblages (Cacciò and Sprong, 2009). Recent studies indicate that G. duodenalis also occurs in marine vertebrates, which might pose a health threat to both marine and terrestrial life. Although the intestinal and fecal contents of various marine mammals suggest actual infection and not just passive transfer of G. duodenalis by these animals, the effects of infection require further investigation (Olson et al., 1997; Dixon et al., 2008).
Limited molecular information exists about G. duodenalis in seals. While various studies have revealed the presence of G. duodenalis in ringed (Phoca hispida), harp (Phoca groenlandica), grey (Halichoerus grypus), bearded (Erignathus barbatus), and Atlantic and Pacific harbor seals (Phoca vitulina vitulina and Phoca vitulina richardsi; Olson et al., 1997; Measures and Olson, 1999; Fayer et al., 2004; Hughes-Hanks et al., 2005; Dixon et al., 2008; Gaydos et al., 2008; Lasek-Nesselquist et al., 2008), only three pinpointed the G. duodenalis assemblage(s) responsible for infection or the extent of G. duodenalis molecular variation represented within samples (Dixon et al., 2008; Gaydos et al., 2008; Lasek-Nesselquist et al., 2008). Molecular characterization of G. duodenalis in two ringed seals from Quebec (Canada) identified Assemblage B as the source of infection (Dixon et al., 2008). Sequence analysis of Pacific harbor seal fecal samples from the Puget Sound region of Washington (WA, USA), revealed the presence of G. duodenalis Assemblages B, C and D (Gaydos et al., 2008). Additionally, 11 seals harbored a novel 398 bp sequence at the glutamate dehydrogenase (gdh) locus, designated “HS-1” (Gaydos et al., 2008). Sequencing from fecal samples also revealed the presence of Assemblages A and B in a harp seal and Assemblage B in a harbor seal off the coast of Cape Cod, Massachusetts (MA, USA; Lasek-Nesselquist et al., 2008). Collectively, these studies suggest that many phocids are susceptible to harboring G. duodenalis, but we still know very little about which haplotypes occur in seals and the prevalence and distribution of these haplotypes in seal populations.
We analyzed fecal samples from east and west coast (USA) seals to assess the prevalence and molecular diversity of G. duodenalis in seal populations. Additionally we determined whether factors such as geographic location, host species or host genotype influenced the prevalence and distribution of G. duodenalis in seal populations. Sequences from gdh and/or triose phosphate isomerase (tpi) loci characterized G. duodenalis present in east coast Atlantic harbor seals and grey seals surrounding Cape Cod, MA on the east coast of the USA (Fig. 1A) and Pacific harbor seals of central California (CA) on the west coast of the USA (Fig. 1B). A portion of the mitochondrial genome (mtDNA) provided haplotypes for a subset of seals from the east and west coasts. To detect any non-seal species associations with G. duodenalis and to improve our ability to detect geographical associations, we also sequenced G. duodenalis from gulls (Larus argentatus and Larus sp.), a common dolphin (Delphinus delphis) and a mako shark (Isurus paucus), and combined our dataset with that of Lasek-Nesselquist et al. (2008), which included G. duodenalis sequences derived from gulls, seals, dolphins, porpoises and a shark located along the east coast of the USA.
Fig. 1.
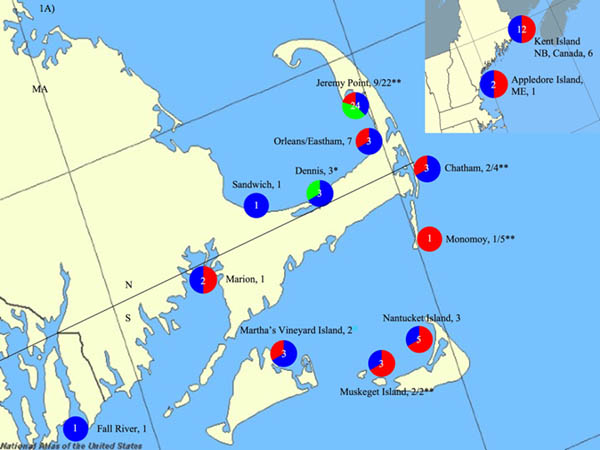
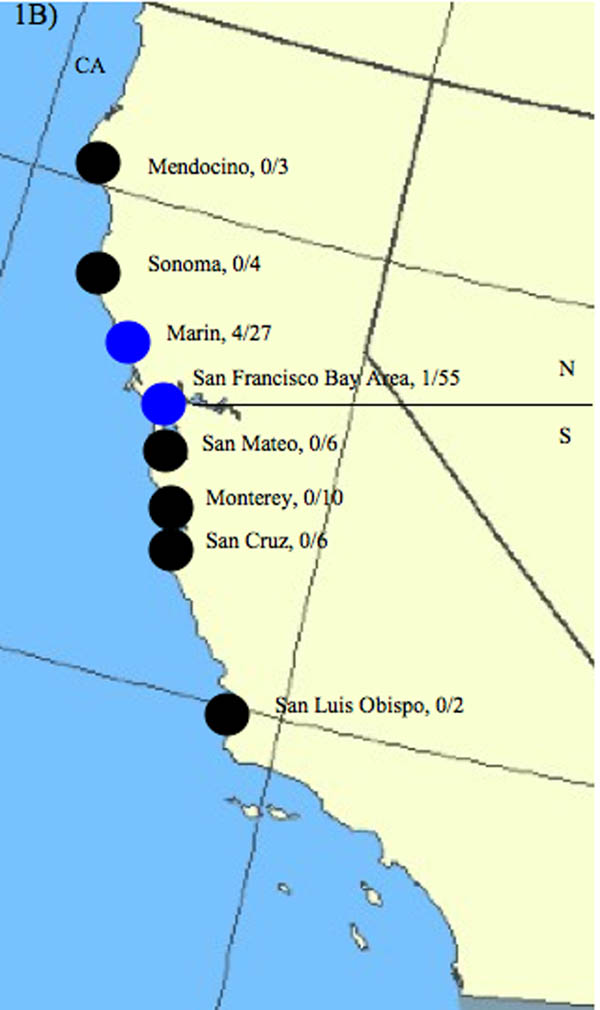
Sampling sites along the east and west coasts of the United States of America (USA). A) East coast sampling sites. East coast samples originated primarily from locations surrounding Cape Cod, Massachusetts (MA), USA but also include areas south of Cape Cod---Marion and Fall River, MA---and north of Cape Cod---Appledore Island, Maine (ME) and Kent Island, New Brunswick (NB), Canada. Pie graphs represent the proportion of times an assemblage was found at each sampling site, with red representing Assemblage A, blue representing Assemblage B and green representing Assemblage H. The white number centered in the pie chart represents the total number of times Assemblages were detected. The number of individual samples taken from each site is indicated after the sampling site. Both sites from this study and that of Lasek-Nesselquist et al. (2008) are indicated in the figure. * Indicates sampling sites from this study only. ** Indicates seal (Atlantic harbor seal and grey seal) sampling sites from this study with the proportion of seals that tested positive listed. N and S indicate the North/South delineation used for dividing sampling sites into regions for analysis of molecular variance (AMOVA). B) West coast Pacific harbor seal sampling sites, which are all located in California, USA. Sites are represented by circles. Black circles indicate sites negative for Giardia duodenalis. Blue circles indicate sampling sites positive for G. duodenalis Assemblage B. The proportion of positive seals at each subdivision is indicated. The San Francisco (SF) Bay Area encompasses Mid, North, and South SF Bay sites. Specifically, the one SF Bay Area positive seal sample derived from Mid SF Bay. The horizontal line indicates the north - south regional division used for AMOVA. Maps were generated by the National Atlas of the USA at www.nationalatlas.gov with the mapmaker application.
2. Materials and methods
2.1. Fecal sample collection and DNA extraction
Fecal samples from Atlantic, harbor and grey seals from the beaches of Cape Cod and Nantucket, MA were collected (Fig. 1A, Table 1) with visual identifications and photographs being taken before approaching the animals. Feces were collected from the sand surface using sterile spoons and 50 ml centrifuge tubes, and transported back to the laboratory on ice. All fecal samples were stored frozen at −80 °C until nucleic acid was extracted. Additional fecal samples were obtained from a herring gull on Appledore Island, Maine (ME, USA) and three gulls in Billings Gate, Dennis, MA using a fecal swab (Fig. 1A, Table 1). The fecal sample from a mako shark derived from an animal caught off Martha’s Vineyard Island in MA (Fig. 1A, Table 1). We re-examined two G. duodenalis-positive samples from Lasek-Nesselquist et al. (2008) - Gull 13 (herring gull) from Kent Island, New Brunswick, Canada and Dolphin 316 (common dolphin) from Orleans, MA (Fig. 1A, Table 1) - to provide positive controls for PCR and to assess whether different primer pairs gave consistent sequence results.
Table 1.
Marine animals sampled including sample size, number of samples positive for assemblage(s) of Giardia duodenalis and G. duodenalis Assemblages present at each sampling site.
| Sample | Sample size | Sampling Site | No. Pos. | Assemblage(s) | Total No. | Total Pos. | Prevalence | No. A |
No. B |
No. H |
No. Mixed |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Phs | 27 | Marin, CA | 4 | B | 112 | 5 | 4.5% | 0 | 5 | 0 | 0 |
| 3 | Mendocino, CA | 0 | B | ||||||||
| 52 | Mid San Fran Bay, CA | 1 | |||||||||
| 10 | Monterey, CA | 0 | |||||||||
| 2 | North San Fran Bay, CA | 0 | |||||||||
| 2 | San Luis Obispo, CA | 0 | |||||||||
| 6 | San Mateo, CA | 0 | |||||||||
| 5 | Santa Cruz, CA | 0 | |||||||||
| 4 | Sonoma, CA | 0 | |||||||||
| 1 | South San Fran Bay, CA | 0 | |||||||||
| Ahs | 7 | Jeremy Pt, Wellfleet, MA | 3 | A, B | 8 | 3 | 37.5% | 3 | 1 | 0 | 1 (A/B) |
| Grey seal | 15 | Jeremy Pt, Wellfleet, MA | 10 | A,B,H | 27 | 17 | 63% | 6 | 5 | 10 | 6 (A/B, B/H, A/H) |
| 5 | Monomoy, MA | 1 | A | ||||||||
| 4 | Chatham, MA | 2 | A,B | ||||||||
| 2 | Muskeget, MA | 2 | A,B | ||||||||
| 1 | undisclosed | 0 | |||||||||
| Gull (sp.?) | 3 | Dennis, MA | 3 | B, H | 4 | 4 | 100% | 1 | 3 | 1 | 2 (A/B, B/H) |
| 1 | Appledore Is., ME | 1 | A, B | ||||||||
| H. gull a | 1 | Kent Island, NB | 1 | A | 1 | 1 | NA | 1 | 0 | ||
| Mako shark | 1 | Martha's Vineyard Is., MA | 1 | A, B | 1 | 1 | 100% | 1 | 1 | 0 | 1 (A/B) |
| Dolphin a | 1 | Wellfleet, MA | 1 | A | 1 | 1 | NA | 1 | 0 | 0 | 0 |
Total No. = total number of samples; Total pos. = total number of G. duodenalis positive samples; No. A, No. B, No. H, No. Mixed, are the total number of samples positive for A, B, H, and the total number of samples with mixed assemblages, respectively.
Pacific harbor seals (Phoca vitulina richardsi) were derived from California (CA) on the west coast of the USA; all other samples were derived from the east coast. Host legend: Phs, Pacific harbor seal; Ahs, Atlantic harbor seal (Phoca vitulina vitulina); Gull (sp.?), herring gull (Larus argentatus) from Appledore Island (Is.) and three gulls of unidentified species (genus Larus) from Dennis; H. gull, Herring gull; Mako shark (Isurus paucus); Localities: San Fran, San Francisco; MA, Massachusetts; ME, Maine; NB, New Brunswick. Mid, North, and South San Francisco Bay encompass the San Francisco Bay Area.
H. gull and Dolphin (Delphinus delphis) derived from a set of samples previously determined to be positive for G. duodenalis so that prevalence cannot be determined (NA). Only gdh detected the presence of Assemblage H while the results for gdh and tpi were combined to calculate the number of samples positive for Assemblages A and B.
Along the west coast, fecal samples were collected from sick and injured harbor seals admitted to the Marine Mammal Center in California during 2007 and 2008. Samples derived primarily from young harbor seals stranding from Mendocino through San Luis Obispo counties (Fig. 1B, Table 1). Samples were collected opportunistically as soon as possible after arrival at the center and before housing them with conspecifics. Other fecal samples came from harbor seals of all ages that were captured and released during a health study in San Francisco Bay and on Clam and Seal Islands on Tomales Bay in Point Reyes National Seashore, CA. Feces were collected into 50 ml tubes using a fecal loop and stored on ice before transport to the Marine Mammal Center, where they were stored at −80 °C. Additional fecal samples were collected from harbor seal haul-out locations within San Francisco Bay, CA for a foraging study (Fig. 1b, Table 1). Feces were stored at −20 °C and sub-sampled for Giardia analysis during the recovery of prey hard parts. Nucleic acids were extracted using the Qiagen stool kit (Valencia, CA, USA) according to kit protocols.
2.2. PCR, cloning and sequencing
We employed a nested PCR approach to amplify gdh and tpi from G. duodenalis-positive samples and designed degenerate primers for both loci in order to amplify DNA from all assemblages. gdh primers amplified a 292–715 bp region of DNA and tpi primers amplified a 490 bp region (Supplementary Table S1). Two previously examined samples served as positive controls for the new gdh primers. We sequenced a portion of the mtDNA for most of the seals in our dataset to examine the influence of seal phylogeography on the presence and haplotypic specificities of G. duodenalis. Seal-specific primers (Supplementary Table S1) amplified nucleotides 7,746-8,736 of the complete seal mitochondrial genome (reference sequence = Phoca vitulina, GenBank accession no. X63726). The mtDNA sequence encompassed the end of the COX1 gene through the genes encoding COX2, the tRNAs Asp, Ser and Lys, to the start of the ATP8 gene.
For G. duodenalis gene amplifications, reactions were set up in 25 ul vols. containing 10 mM Tris-HCl, 50 mM KCl, 2.5 mM MgCl2, 0.1 mM BSA, 0.08 mM dNTP, 0.25 µM primer, 0.11 µl (0.55 units) Taq DNA polymerase (Promega, Madison, WI, USA) and 2 µl of sample DNA or PCR product. Thermocycling conditions were 94 °C for 2 min followed by 30–35 cycles of 95 °C for 30 s, 45–55 °C for 30 s, and 72 °C for 60 s, followed by 72 °C for 7 min. The same PCR and thermal cycling conditions amplified seal mtDNA but nested PCR was not necessary. Products were either separated on agarose gels, gel-extracted and cloned or cloned directly from the PCR with the TOPO TA kit (Invitrogen, Carlsbad, CA, USA). Plasmids were purified from positive clones using standard alkaline-lysis with a Biomek FX liquid handling robot (Beckman Coulter, Fullerton, CA, USA). Clones were sequenced in the forward direction or both directions using universal M13 primers, ABI BigDye 3.1 chemistry, and an Applied Biosystems 3730×l 96 capillary array genetic analyzer (Foster City, CA, USA). For G. duodenalis amplifications, a minimum of 24 clones was sequenced from each PCR. At least 16 clones were sequenced from each seal PCR.
A bioinformatics pipeline using phred, cross match and phrap translated chromatograms into base calls and associated quality scores, removed vectors sequences and assembled forward and reverse reads into full-length sequences for each of the cloned PCR amplicons (Ewing and Green, 1998; Ewing et al., 1998). Sequences were aligned with ClustalW v.1.83 and edited in MacClade (Thompson et al., 1994; Maddison D.R., Maddison W.P., 2001. MacClade 4: Analysis of phylogeny and character evolution. Sinauer Associates, Inc., Sunderland, Massachusetts). We sorted legitimate variation from potential PCR error (including PCR recombination) for reactions that amplified more than one sequence variant as previously described (Lasek-Nesselquist et al., 2008, 2009). We created consensus sequences from clones sequenced in one direction only if two or more clones with high-quality chromatograms confirmed the nucleotide at each position in the gene.
2.3. Characterizing novel G. duodenalis sequences and defining Assemblage H
We used gdh sequences from our study and additional sequences from GenBank (listed in Supplementary Table S2) to examine the evolutionary relationship of the newly discovered Assemblage H to that of the other seven G. duodenalis assemblages (A–G). A Bayesian analysis run in MrBayes v.3.1.2 reconstructed phylogenetic relationships among Assemblages A–H. MrBayes performed 1,000,000 generations with two parallel searches using a GTR + invgamma model and a burn in of 2,500 generations. Also, a maximum likelihood (ML) method was employed in PAUP v.4.0b10 with a GTR+I+G DNA substitution model selected by Akaike information criterion (AIC) in Modeltest v.3.06 (Posada and Crandall, 1998; Swofford, D.L., 2002. PAUP*. Phylogenetic analysis using parsimony (*and other methods). Version 4. Sinauer Associates, Inc., Sunderland, Massachusetts). ML heuristic searches were performed using 100 random taxon-addition replicates with tree bisection and reconnection (TBR) for all ML trees. ML support was determined using 100 bootstrap replicates, each using 10 random taxon-addition replicates with TBR branch swapping. DnaSP v.3.0 calculated the Jukes Cantor corrected genetic distances, and synonymous and non-synonymous substitution rates between Assemblage H and Assemblages A–G (Rozas and Rozas, 1999).
2.4. Genetic differentiation within and between seal populations
Seal mtDNA sequences were used to detect associations between the presence of G. duodenalis and host species haplotype and population structure. ML and Bayesian methods reconstructed evolutionary relationships among mtDNA sequences. PAUP generated the ML tree using a TrN+I DNA substitution model selected by AIC in Modeltest. MrBayes performed 1,000,000 generations with two parallel searches using a GTR + invgamma model and a burn-in of 2,500 generations.
DnaSP estimated nucleotide polymorphism within all three seal species: Pacific harbor seal, Atlantic harbor seal and grey seal. DnaSP also estimated genetic differentiation among all three species and among sampling sites for Pacific harbor and grey seals using Wright’s F statistic (Fst; Wright, 1965; Rozas and Rozas, 1999). Atlantic harbor seal samples derived only from Jeremy Point in Wellfleet and could not be tested for genetic differentiation based on geographical divisions.
Mantel tests, implemented in Isolation By Distance v.3.16 (IBD, http://ibdws.sdsu.edu/) returned significance values for the relationships between genetic differentiation among seal populations and geographic distance (Jensen et al., 2005). The Mantel test measures correlation between two distance matrices (i.e. genetic and geographic) and uses random permutations of one of the matrices to assess significance (Mantel, 1967). For Mantel tests, Pacific harbor seals and grey seals were organized by sampling site and distances between locations were estimated in Google Earth v.5.0. The IBD default parameter settings were used except that IBD generated Fst genetic distance matrices for all pairwise comparisons of mtDNA sequences.
We further examined the population structures of grey and Pacific harbor seals by an analysis of molecular variance (AMOVA) using the ade4 package (Thioulouse et al., 1996) in R v.2.8.1 (R core development team, 2009). AMOVAs estimate the amount of molecular variation within a group of individuals attributable to different hierarchical divisions (Excoffier et al., 1992). For AMOVAs, Pacific harbor seal and grey seal haplotypes were hierarchically organized by North/South regions and sampling sites (Fig. 1). PAUP generated genetic distance matrices for Pacific harbor seal and grey seal haplotypes, which R converted into Euclidean distances. The Rand permutation test in the ade4 package of R determined significance values (Excoffier et al., 1992). Understanding seal population dynamics is important for evaluating how G. duodenalis is transmitted. For example, if seal populations are genetically distinct due to geographic isolation, it is unlikely that shared G. duodenalis profiles will arise from seal interaction between populations. In this scenario, seal haplotype is not a factor in the presence of G. duodenalis but other similarities between their geographical locations, such as a similar terrestrial influence - might contribute to the shared G. duodenalis profiles. Alternatively, if G. duodenalis haplotypes vary among geographically isolated and genetically distinct seal populations the differences could be attributable to host specificities or factors relating to location, such as a different terrestrial impact.
2.5. Molecular variation of G. duodenalis in marine animals
Nucleotide polymorphism for Assemblages A, B and H were estimated in DnaSP v. 3.0. A Pearson’s ℵ2 test with a Yates’ continuity correction or a Fisher’s exact test in R v.2.8.1 tested for a significant correlation between the presence of G. duodenalis in seals and their geographic location (east versus west coast and east coast sampling sites). Using east coast samples, we also tested for significant associations between G. duodenalis haplotype (Assemblage A, B or H) and seal species (Atlantic harbor seal versus grey seal), and G. duodenalis haplotype and sampling site (see Table 1 for sampling sites). Because most G. duodenalis-positive samples showed either gdh or tpi amplification but not both, we combined data from both loci for statistical analyses involving the presence/absence of this parasite.
We analyzed G. duodenalis from seals and other marine vertebrates of this study and from marine vertebrates of Lasek-Nesselquist et al. (2008) to understand the greater context of G. duodenalis within the marine community (Table 1). We used gdh and tpi genealogies to identify geographic and/or species-specific patterns of G. duodenalis distribution and molecular diversity in marine animals (GenBank accession numbers for the sequences used in each tree are listed in Supplementary Table S2). Trees were reconstructed with Bayesian inference and ML methods. MrBayes performed 1,000,000 generations with two parallel searches using a GTR + invgamma model and a burn-in of 2,500 generations. In PAUP, ML methods with a GTR+G or TIM DNA substitution model (selected by the AIC in Modeltest) reconstructed evolutionary relationships among G. duodenalis gdh and tpi sequences, respectively.
2.6. Sequences
All sequences generated in this study were deposited in GenBank, under the accession numbers GUI182371-GUI182396 (tpi sequence set), GUI176069-GUI176101 (gdh sequence set), and GU733611-GU7333706 (mtDNA set).
3. Results
3.1. Identification and molecular characterization of a new G. duodenalis assemblage
We characterized a group of novel G. duodenalis gdh haplotypes from our east coast samples that shared sequence similarity to a novel haplotype identified previously in Pacific harbor seals (Gaydos et al., 2008). Further, we revealed that these haplotypes clustered into a new lineage, which we designated Assemblage H. However, only gdh provided evidence of this new assemblage. Thirty-five seals (eight Atlantic harbor seals and 27 grey seals), four gulls and a mako shark from the east coast, and 112 Pacific harbor seals from the west coast were tested for the presence of G. duodenalis (Table 1). We also re-examined two previously characterized samples – Gull 13 and Dolphin 316 –from Lasek-Nesselquist et al. (2008) using the new gdh primers.
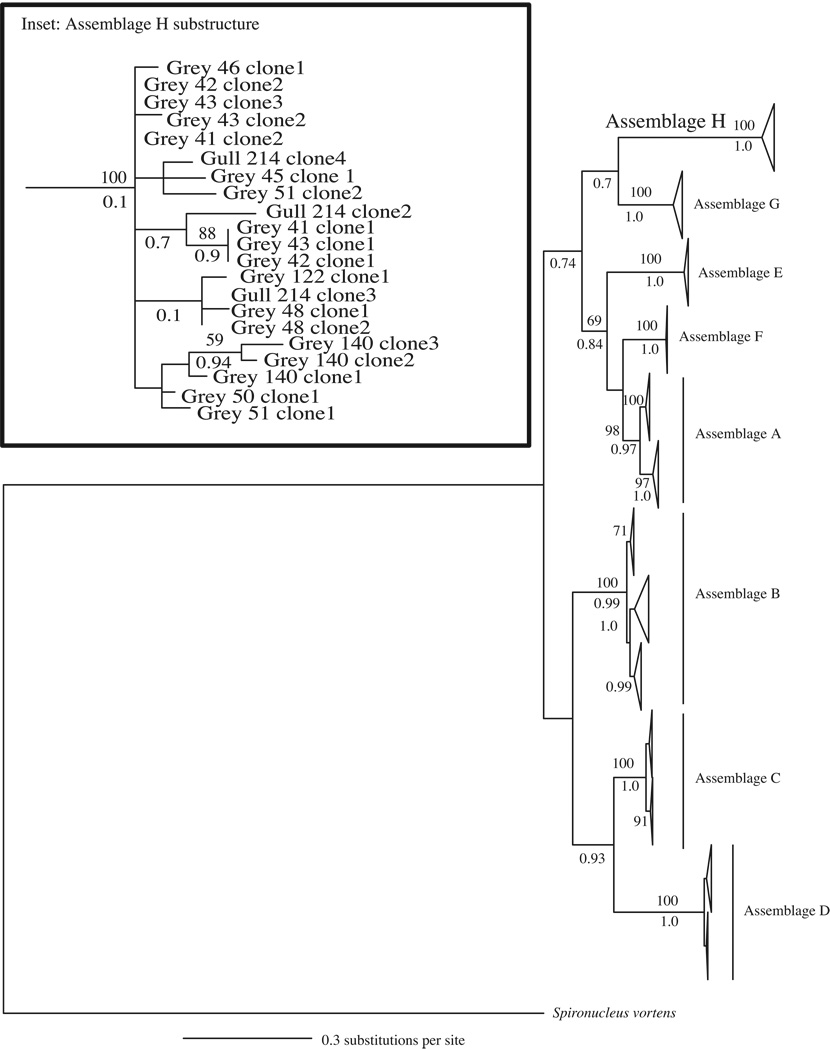
PCR amplification and sequencing of gdh and/or tpi loci from fecal material revealed the presence of G. duodenalis in 20/35 east coast seals, 4/4 gulls, the mako shark and 5/112 Pacific harbor seals (Table 1). As expected, the positive controls – Gull 13 and Dolphin 316 also tested positive for G. duodenalis. BLAST homology searches (Altschul et al., 1990) indicated that most G. duodenalis sequences derived from either human-infecting Assemblages A or B (Table 1). However, 21 G. duodenalis gdh sequences of 295 or 715 bp in length (depending on primers used for amplification) showed only ~80% similarity to either Assemblage A or B. Large genetic distances (14–19%) also separated these gdh sequences from those of Assemblages C–G (Table 2). These novel gdh sequences derived from 10 grey seals and one gull (Table 1) and shared up to 100% sequence homology with a 398 bp G. duodenalis sequence, HS-1, obtained from Pacific harbor seals in the Puget Sound region of WA, USA. Bayesian and ML gene trees revealed that the novel gdh sequences clustered into a well-supported monophyletic clade that was either sister to Assemblage G with moderate support (Bayesian analysis; Fig. 2) or sister to all other assemblages with low bootstrap support (ML analysis; not shown) and equally as divergent from Assemblages A–G as Assemblages A–G were from each other (Fig. 2). Synonymous divergence rates between this lineage and Assemblages A–G are an order of magnitude larger than non-synonymous divergence rates (Table 2). We designated this newly discovered monophyletic group of G. duodenalis Assemblage H.
Table 2.
Genetic distance (K) between Giardia duodenalis Assemblage H and A–G based on 715 bp of the gdh locus.
| Assemblage H | |||
|---|---|---|---|
| K | Ks | Ka | |
| A | 0.144 ± 0.016 | 0.670 | 0.036 |
| B | 0.165 ± 0.015 | 0.529 | 0.032 |
| C | 0.166 ± 0.026 | 0.749 | 0.052 |
| D | 0.187 ± 0.020 | 1.03 | 0.045 |
| E | 0.184 ± 0.039 | 0.980 | 0.047 |
| F | 0.157 ± 0.038 | 0.690 | 0.049 |
| G | 0.140 ± 0.029 | 0.917 | 0.032 |
Ks, synonymous substitution rate; Ka, non-synonymous substitution rate.
Fig. 2.
Glutamate dehydrogenase (gdh) gene tree depicting the relationships among all known Giardia duodenalis assemblages (A–G) and the newly characterized Assemblage H generated using Bayesian analysis. Sequences were 715 bp in length and chosen to maximize phylogenetic resolution while still representing the majority of the Assemblage H variants described. The gene tree is a consensus of 75,000 trees and rooted with Giardia ardeae and Spironucleus vortens. Posterior probabilities > 0.70 are shown below each branch. Bootstrap support > 50 from maximum likelihood (ML) analysis are shown above branches. The area of the triangle corresponds to the amount of variation within a clade. Inset highlights the variation and sub-structuring within Assemblage H and includes all variants (292–715 bp). Grey refers to grey seal.
3.2. Phylogeography of three seal species
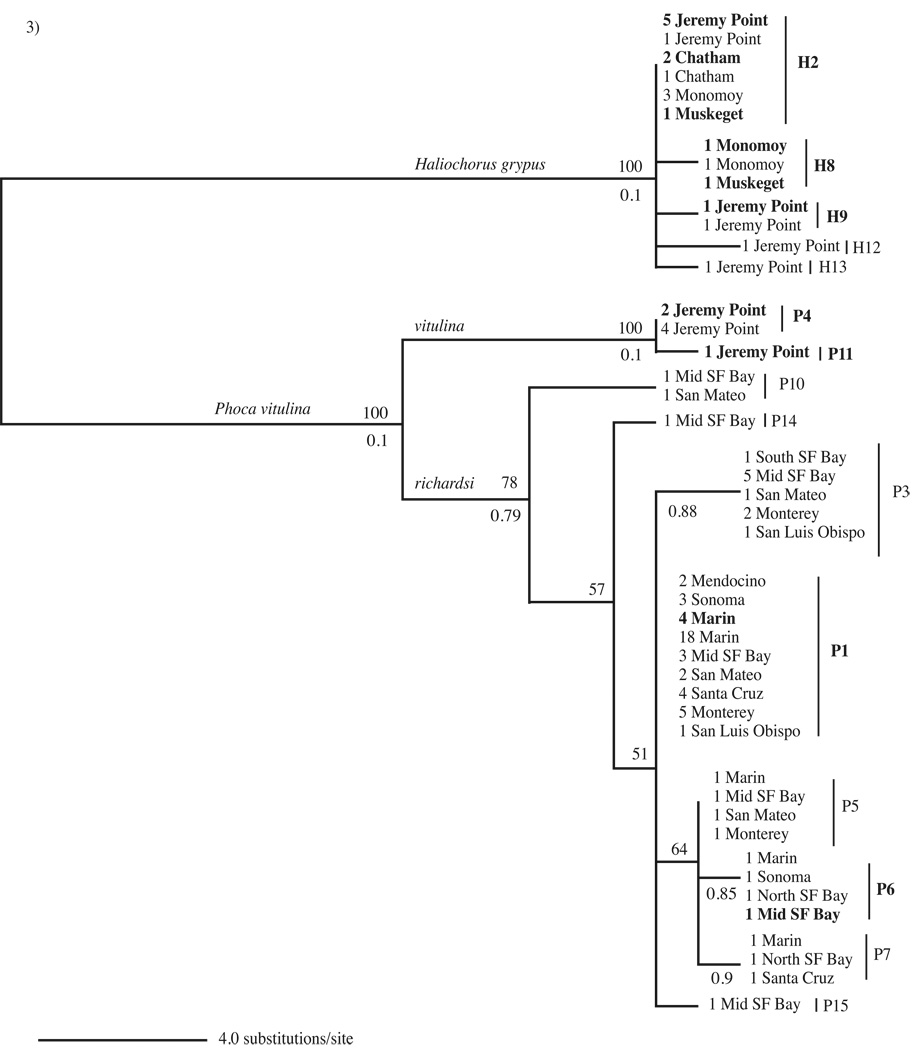
In order to understand whether seal phylogeography affects the prevalence and distribution of G. duodenalis, we sequenced a 963 bp region (after primer removal) of the mtDNA for east and west coast seals (referring only to seals and seal sampling sites of this study). We obtained mtDNA sequences from 7/8 Atlantic harbor seals and 21/27 grey seals from the east coast and 67/112 Pacific harbor seals from the west coast (Fig. 3). East coast samples derived from four sampling locations off the coast of Cape Cod, MA, USA while west coast samples derived from the waters surrounding CA, USA (Table 1, Fig. 1). The seal mtDNA tree reflected the three species divisions: Atlantic harbor seal, Pacific harbor seal and grey seal (Fig. 3). We were unable to determine any correlation between mtDNA haplotype and geographical location for Atlantic harbor seals because these samples comprised only two haplotypes (P4 and P11) from one sampling site: Jeremy Point on Cape Cod, MA (Fig. 3). All grey seal haplotypes fell into a monophyletic group, which lacked sub-structuring. The most common grey seal haplotype (H2) occurred in seals from all four east coast sampling sites and in a grey seal at an undisclosed location off the coast of Cape Cod (Fig. 3). Haplotype H8 only occurred in grey seals from Monomoy and Muskeget in the southern region of Cape Cod while haplotypes H9, H12 and H13 only occurred in grey seals from Jeremy Point in the northern region of Cape Cod, which may reflect population differences or effects of a small sampling size (Fig. 3). Regardless, the fact that grey seals form a monophyletic group and that seals from all sampling sites share a common haplotype (H2) indicate that these seals are not genetically or geographically isolated. Pacific harbor seals also lacked population subdivision as most mtDNA haplotypes were present in the majority of west coast sampling sites and the most common haplotype (P1) occurred ubiquitously (Fig. 3). Although some haplotypes only appeared in one or two locations, this is most likely an effect of sampling size rather than true geographical differences in Pacific harbor seal populations. Indeed, the most frequently sampled sites contained the greatest number of haplotypes, revealing a positive correlation between samples size and haplotype diversity. Low values of π and θ reflected the limited amounts of nucleotide polymorphism sampled within all three seal species (Supplementary Table S3). Fst values confirmed that all three seal species are genetically distinct non-interbreeding entities (Supplementary Table S3) but neither Pacific harbor seals nor grey seals exhibit within-species genetic differentiation based on geographic separation (average Fst value for all sampling site comparisons = −0.041). Mantel tests and AMOVAS also confirmed that these two seal species lacked population subdivisions or sub-structuring (Mantel test: Pacific harbor seal r = −0.248, P = 0.778, grey seal r = −0.109, P = 0.646; AMOVA Supplementary Table S4). Thus, Pacific harbor seals and grey seals each represent undivided populations - sometimes despite large geographic distances - which could contribute to the transmission of G. duodenalis between seals of different locations. Conversely, if there are differences in G. duodenalis haplotypes among these genetically undivided populations it could indicate that the spread of this parasite is tied to factors related to geographical location or it could reflect the effects of seal behavior on parasite transmission.
Fig. 3.
Seal mtDNA maximum likelihood tree. H2, H8, H9, H12 and H13 refer to grey seal (Halichoerus grypus) mtDNA haplotypes. P1, P3-P7, P10, P11, P14 and P15 refer to Atlantic and Pacific harbor seal (Phoca vitulina vitulina and Phoca vitulina richardsi) mtDNA haplotypes. All grey seal and Atlantic harbor seal samples derive from Massachusetts (east coast USA). All Pacific harbor seal samples derive from California (west coast, USA). Mid, North and South SF Bay refer to Mid, North, and South San Francisco Bay, which comprise the San Francisco Bay Area. The tree shows the number of times each haplotype was detected at a sampling site. Boldface indicates Giardia duodenalis positive seal samples. Bootstrap values > 50 are shown above branches and posterior probabilities > 0.70 from Bayesian analysis are shown below branches.
3.3. Giardia duodenalis in east and west coast seals: Prevalence, diversity and distribution
Combining our gdh and tpi datasets, we detected G. duodenalis in seals from all four sampling sites of the east coast, in 3/5 grey seal haplotypes and in both Atlantic harbor seal haplotypes (Fig. 3). In contrast, G. duodenalis occurred in only two adjacent sampling sites along the west coast (Marin County and Mid San Francisco Bay) and in only 2/8 Pacific harbor seal haplotypes (Fig. 3). The presence of G. duodenalis varied significantly by coast (ℵ2 = 43, d.f. = 1, P = 4.7e−11), with 20/35 (55.6%) positive east coast seal samples and only 5/112 (4.5%) positive west coast samples. However, seal species differences along the two coasts could also be responsible for the pattern observed. Atlantic harbor seal and grey seal samples contained G. duodenalis from human-infecting Assemblages A and B (Table 1). Ten grey seal samples from Jeremy Point in Wellfleet also contained the novel G. duodenalis assemblage H (Table 1). Both gdh and tpi primers amplified G. duodenalis from east coast seal samples and mixed assemblages occurred in 7/20 of these seals (Table 1). The presence of Assemblages A and B along the east coast showed no significant association with either the Atlantic harbor seal or grey seal species (ℵ2 = 0.15, d.f. = 1, P = 0.85). The presence of Assemblages A and B also did not depend on sampling site along the east coast (Jeremy Point versus other: Assemblage A, ℵ2 = 0.70, d.f. = 1, P = 0.40 and Assemblage B, ℵ2 = 0.05, d.f. =1, P = 0.70). In contrast, Assemblage H sequences showed a significant association with the Jeremy Point sampling site (ℵ2 = 8.8, d.f. = 3, P = 0.03) as well grey seals (ℵ2 = 3.0, d.f. = 1, P = 0.04). It is difficult to disentangle geographic versus host species influence on the distribution of Assemblage H. While 66.6% (10/15) of Jeremy Point grey seals harbored Assemblage H haplotypes, Assemblage H sequences did not amplify from any of the 11 grey seals from the Chatham, Monomoy or Muskeget sampling sites, stressing the influence of geographical factors (Table 1). However, the only seven Atlantic harbor seals sampled derived from Jeremy Point and none contained Assemblage H haplotypes (Table 1), which is unexpected if there is no host specificity to G. duodenalis distribution.
West coast seal samples contained only Assemblage B haplotypes (Table 1) at the tpi locus; we were not able to amplify gdh from these samples. The lack of Assemblage A or B amplification at the gdh locus from G. duodenalis-positive Pacific harbor seals might indicate that the sequences at this locus are too variable for successful primer annealing. Amplification of only Assemblage B at the tpi locus for Pacific harbor seal samples might reflect coastal differences in the distribution of G. duodenalis haplotypes, different seal species’ susceptibilities to infection or primer bias. The fact that tpi primers preferentially amplified Assemblage B from east and west coast samples suggests a primer amplification bias but this possibility is less convincing given previous success in amplifying both A and B Assemblages from fecal samples (Lasek-Nesselquist et al., 2008) and cultured isolates (data not shown) with these same primers. There was no association between seal haplotype and the presence of G. duodenalis for either Pacific harbor seals (ℵ2 = 3.7, d.f. = 7, P = 0.80) or grey seals (ℵ2 = 0.14, d.f. =1, P = 0.9306). Collectively, east and west coast results suggest that the presence of certain G. duodenalis haplotypes in seals might be dependent on geographical and/or host species factors but not necessarily the haplotype of the seal within a species. However, differences in east and west coast seal behavior or differences in sample collection between coasts might also contribute to the observed higher prevalence of G. duodenalis in east coast seals.
3.4. Diversity and distribution of G. duodenalis in marine vertebrates
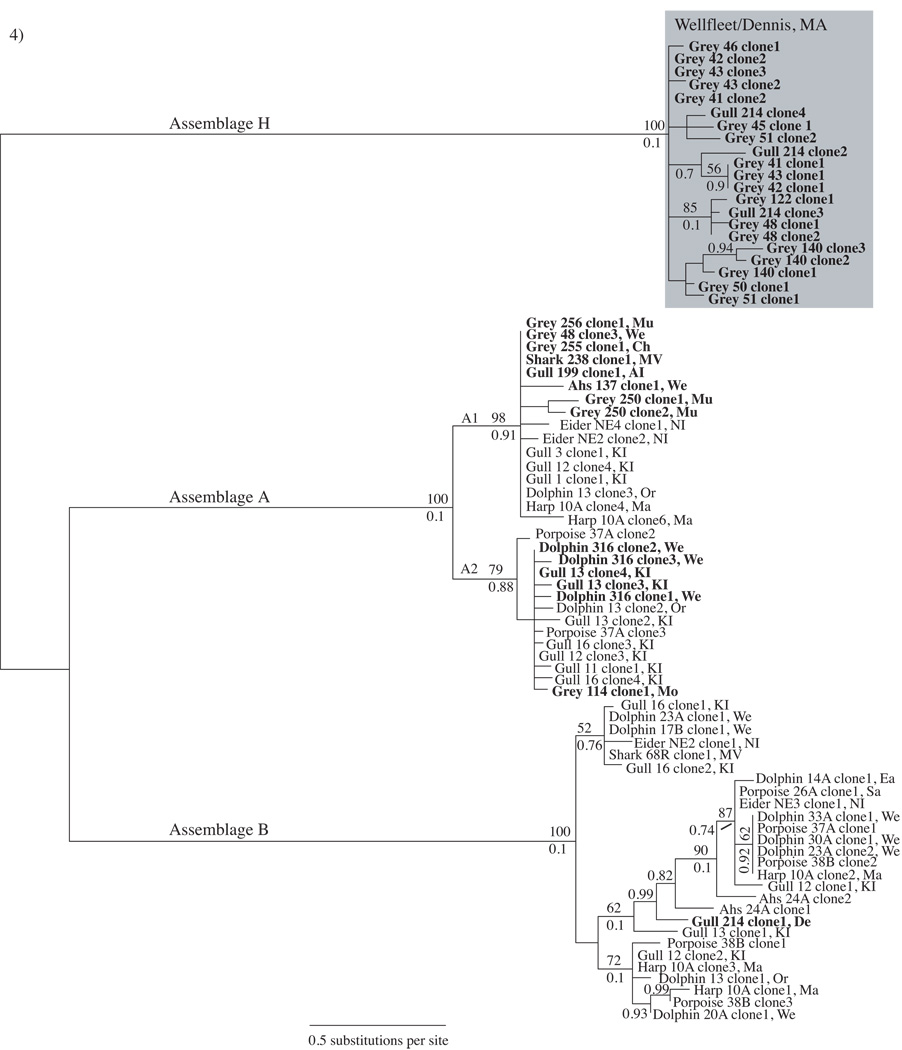
Giardia duodenalis sequences from all east coast vertebrate samples (this study) represent 3/8 Assemblages (A, B and H) and 1–14 haplotypes within each assemblage at the gdh locus (Table 3). Fourteen sequences representing nine Assemblage A haplotypes and 1.17% nucleotide polymorphism were obtained from two gulls, one shark, one dolphin, five grey seals and one Atlantic harbor seal (Table 3). Seventeen sequences from 10 grey seals and one gull represented 14 Assemblage H haplotypes with 0.73% nucleotide polymorphism (Table 3). Only one gull harbored an Assemblage B haplotype. Assemblages A, B and H all showed sub-structuring to varying degrees. Assemblage A haplotypes clustered into two major sub-groups (defined in previous literature as groups A1 and A2) with moderate to strong bootstrap support (Fig. 4). Assemblage B clustered into three moderately supported sub-groups with the majority of sequence variation within one sub-group (Fig. 4). While Assemblage H sequences clustered into five groups, only two show moderate bootstrap support (Fig. 4). Samples frequently contained more than one haplotype and the gdh primers used in this study appeared to preferentially amplify Assemblages A and H (Fig. 4). Samples Dolphin 316 and Gull 13, the positive controls previously analyzed by Lasek-Nesselquist et al. (2008), exemplify this primer bias. In Lasek-Nesselquist et al. (2008), gdh amplification failed using the Dolphin 316 sample and only Assemblage B amplified at the tpi locus while the primers of this study succeeded in amplifying gdh but detected only Assemblage A (Fig. 4). The gdh primers used in Lasek-Nesselquist et al. (2008) and the gdh primers used in this study both amplified Assemblage A from the Gull 13 sample but they amplified different haplotypes (Fig. 4). Additionally, these primers successfully amplified gdh from several Assemblage A and B laboratory cultures (data not shown), suggesting that they react differently when faced with a heterogeneous pool of DNA. Given the amplification biases of the gdh primers, PCR failure for Pacific harbor seal samples at the gdh locus suggests that the sequencing at the tpi locus, which resulted in only Assemblage B haplotypes, accurately represents the assemblage diversity within these samples.
Table 3.
Molecular diversity of Giardia duodenalis from marine mammals.
|
gdh |
tpi |
|||||
|---|---|---|---|---|---|---|
| Assemblage/species | No. seqs | No. haplotypes | π | No. seqs | No. haplotypes | π |
| A | 14 | 9 | 1.17% | 1 | 1 | --- |
| B | 1 | 1 | --- | 26 | 23 | 1.20% |
| H | 21 | 14 | 0.73% | 0 | 0 | --- |
| Atlantic harbor seal | 1 | 1 | --- | 4 | 3 | 0.65% |
| Grey seal | 22 | 12 | 4.50% | 7 | 7 | 7.18% |
| Pacific harbor seal | 0 | 0 | --- | 7 | 6 | 0.82% |
No. seqs.= the total number of Assemblage A, B, and H sequences obtained at the glutamate dehydrogenase (gdh) and triose phosphate isomerase (tpi) loci from all samples and the total number of sequences obtained from three seal species; No. haplotypes = the number of unique haplotypes obtained from our entire dataset for Assemblages A, B, and H and the total number of haplotypes obtained from three seal species; π = percent nucleotide polymorphism for Assemblages A, B, and H for our entire dataset and for three seal species.
Fig. 4.
Giardia duodenalis glutamate dehydrogenase (gdh) Bayesian tree derived from east coast marine animal fecal samples. The gene tree is a consensus of 75,000 sampled trees. Boldface indicates sequences generated from this study. Grey, Ahs, Phs and harp refer to grey seal, Atlantic harbor seal, Pacific harbor seal and harp seal, respectively. Boostrap values > 50 are shown above branches and posterior probabilities > 0.70 are shown below branches unless a boostrap value is not present. The shaded box highlights Assemblage H sequences derived from 10 grey seals from Jeremy Point, Wellfleet, Massachusetts (MA, USA) and one gull from Dennis, MA. Sampling site abbreviations are included with each sample: KI, Kent Island, Canada; We, Wellfleet, MA; De, Dennis, MA; Sa, Sandwich, MA; Or, Orleans, MA; Ch, Chatham, MA; Mo, Monomoy, MA; Mu, Muskeget, MA; MV, Martha’s Vineyard Island, MA; NI, Nantucket Island, MA; AI, Appledore Island, Maine; Ma, Marion, MA. A1 and A2 denote subgroups of Assemblage A.
On the east coast, Assemblages A and B show a broad geographic and host species range but gulls harbor the greatest diversity of G. duodenalis haplotypes (sequences from this study and Lasek-Nesselquist et al., 2008; Fig. 4). Gulls might be particularly susceptible to harboring multiple G. duodenalis haplotypes due to their terrestrial and aquatic lifestyles and their generalist foraging behavior - feeding anywhere from dump sites to the ocean (Julie Ellis, personal communication). Of the three seal species sampled (this study only), grey seals showed the greatest prevalence of G. duodenalis (Table 1) and at the gdh locus, grey seals also showed the greatest haplotypic diversity (Table 3). Identification of Assemblage H occurred only in grey seals and one gull from two nearby sampling sites on Cape Cod, MA: Jeremy Point in Wellfleet and Billings Gate in Dennis (Figs. 1A and 4). Thus, Assemblage H appears to show a geographic association but is not specific to seals.
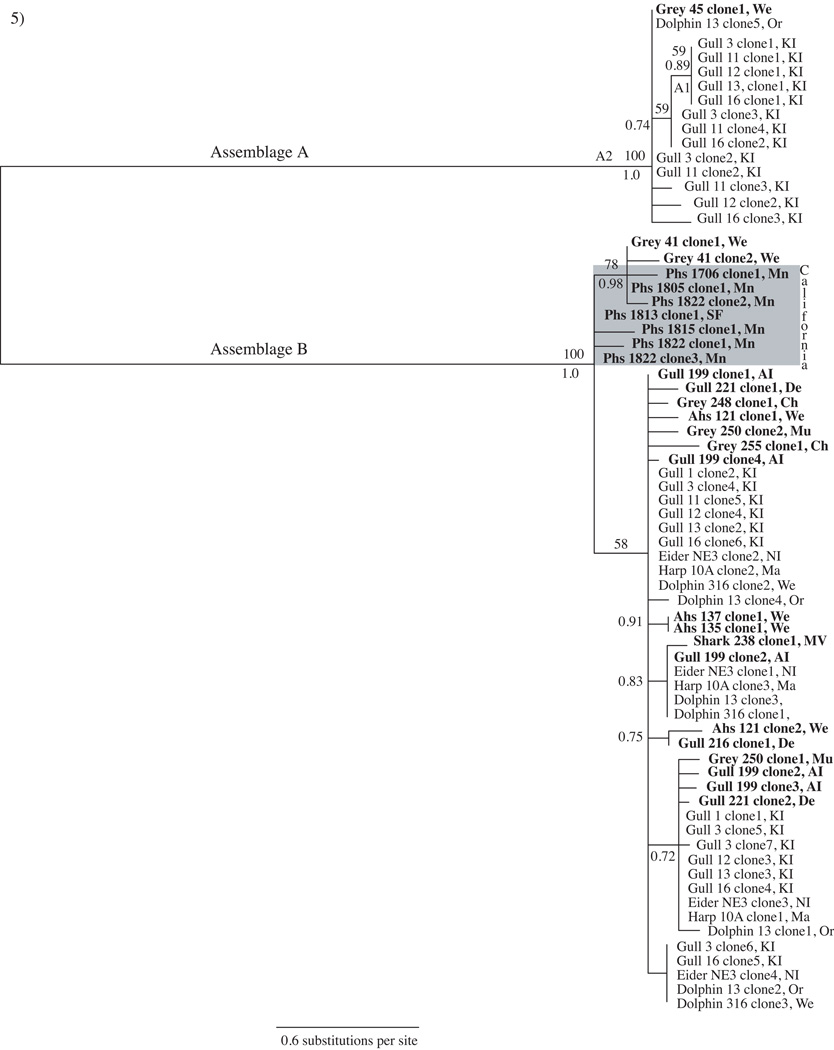
Tpi sequence data from marine vertebrates of this study represent one Assemblage A and 23 Assemblage B haplotypes with a nucleotide diversity of 1.2% (Table 3). Despite previous reports that these tpi primers amplified sequences from non-zoonotic assemblages (Sulaiman et al., 2003; Trout et al., 2004; Lebbad et al., 2009), they failed to amplify a sequence that might correspond to Assemblage H and appeared biased towards Assemblage B amplification (Table 3, Fig. 5). However, previous sequencing results with these primers (Lasek-Nesselquist et al., 2008) do not show an amplification bias, suggesting that our results accurately depict the lack of Assemblage A haplotypes at the tpi locus. As seen with gdh, the tpi genealogy supports a broad host and geographic distribution of G. duodenalis zoonotic haplotypes along the east coast (Fig. 5). Similarly, tpi confirms that gulls harbor the greatest G. duodenalis haplotypic diversity (Fig. 5). Among seal samples, grey seals still maintain the greatest G. duodenalis haplotypic diversity but it is more equally represented among the three seal species at the tpi locus (this study only, Table 3). Results for the west coast are more ambiguous. Only 5/112 Pacific harbor seals tested positive for G. duodenalis and only tpi provided amplification (Fig. 5). These five seals were located at neighboring sampling sites (Marin County and Mid San Francisco Bay) and contained tpi sequences that clustered within two Assemblage B subgroups; primarily to the exclusion of east coast samples (Fig. 5). The differences in the prevalence and genetic diversity of G. duodenalis between east and west coast seal populations could be due to a variety of reasons, including geographical factors, host-specific factors or seal sampling differences and requires further exploration.
Fig. 5.
Giardia duodenalis triose phosphate isomerase (tpi) tree representing a consensus of 75,000 sampled trees from Bayesian analysis. Sequences derive from east and west coast marine animal fecal samples. Boldface indicates sequences generated from this study. The shaded box highlights the cluster of G. duodenalis sequences from California (CA) Pacific harbor seals. Sampling site abbreviations are included with each sample: KI, Kent Island, Canada; We, Wellfleet, Massachusetts (MA); De, Dennis, MA; Sa, Sandwich, MA; Or, Orleans, MA; Ch, Chatham, MA; Mo, Monomoy, MA, Mu, Muskeget, MA; MV, Martha’s Vineyard Island, MA; NI, Nantucket Island, MA; AI, Appledore Island, Maine; Ma, Marion, MA; Mn, Marin, CA; SF, (Mid) San Francisco Bay Area, CA. Posterior probabilities > 0.70 are shown below branches and bootstrap support > 50 from a maximum likelihood analysis are shown above branches. A1 and A2 denote sub-groups of Assemblage A
3.5. Gdh and tpi genealogies combined
Neither gdh nor tpi alone served as a good proxy for determining the presence of G. duodenalis within our fecal samples. In other words, not only did gdh amplification fail in samples where tpi PCR succeeded and vice versa but gdh and tpi also showed conflicting results as to the predominant assemblages present in seals. Of the 28 G. duodenalis-positive seal samples, only seven (25%) provided amplification at both gdh and tpi loci and no sample showed consistent amplification of the same assemblage at both loci (Figs. 4 and 5). Five samples showed a co-occurrence of Assemblages A and B and the other two samples showed a co-occurrence of H and B, and H and A (Table 1). While the gdh tree indicated a strong association between seals and the presence of Assemblage A and H haplotypes, tpi showed that Assemblage B was more prevalent (Figs. 4 and 5). This suggests that either gdh and tpi primers have different amplification biases (even though they were both designed to amplify from all assemblages) or recombination led to an individual with tpi and gdh loci from different ancestries. In the first scenario, when Assemblage A haplotypes are present at gdh and tpi loci, amplification will only occur at the gdh locus and when a sample contains only Assemblage B haplotypes, only tpi will provide amplification. In the second scenario, genetic exchange between individuals of different assemblages must have occurred.
4. Discussion
The high prevalence of Assemblages A and B in east coast marine vertebrates, regardless of species or geographic location, raises health concerns for humans and other animals. The most common Assemblage A and B haplotypes were sequenced from different species and different locations along the east coast, suggesting that these are the most frequently encountered haplotypes in human and/or wildlife populations or that these haplotypes are transmitted rapidly within and between marine vertebrates.
Transmission of G. duodenalis within grey seal, Atlantic harbor seal and Pacific harbor seal populations should be relatively uninhibited as no population was geographically differentiated. Despite lack of apparent population subdivisions, some variation existed in the G. duodenalis assemblages present in seals. This indicates either a geographical influence on the acquisition of this parasite or another contributing factor such as seal behavior. For example, we identified Assemblage H only in grey seals from Jeremy Point, Wellfleet and one gull from a neighboring area. None of the grey seals sampled from more southern locations along the coast of Cape Cod harbored Assemblage H, suggesting that the source was endemic to the Wellfleet area. Perhaps the association between G. duodenalis haplotypes and geography becomes more obscure as vectors efficient at transmitting disease over great distances, such as gulls, are given the opportunity to transport this parasite between different locations. Although Assemblage H was sequenced previously in one study of Pacific harbor seals from Puget Sound, WA, USA (Gaydos et al., 2008) none of our Pacific harbor seal samples from California contained these sequences. This strengthens the idea that the transmission of G. duodenalis haplotypes between seal populations is not necessarily dependent upon host specificities but upon a vector that can accelerate transmission of this parasite across large geographic distances. Whether the source of Assemblage H is seals, gulls or terrestrial remains to be determined. However, the correlation between the discovery of Assemblage H and recent endeavors to molecularly characterize G. duodenalis in marine organisms suggests that Assemblage H might have a marine origin.
While differences in the distribution of Giardia duodenalis assemblages within the three seals species might indicate geographical and behavioral effects, the difference in assemblage distributions among the three seal species might be due to species and/or geographical influences. For instance, even though Atlantic harbor seals and grey seals resided together at Jeremy Point, Wellfleet, none of the Atlantic harbor seal samples contained Assemblage H. The lack of Assemblage A sequences along the west coast and coastal differences in the distribution of Assemblage B haplotypes might indicate a host species- or geographically-related effect. Sampling other marine and terrestrial vertebrates that interact with Pacific harbor seals along the California coastline could elucidate the transmission dynamics of Assemblage B within seal populations and help determine whether Assemblage A is present in this area but not transmitted to Pacific harbor seals.
Coastal location was the most significant factor in determining the presence of G. duodenalis in seals, where less than 5% of west coast seals tested positive compared with over half of the east coast samples. Gaydos et al. (2008) discovered that seals from densely populated haul-out sites in WA, USA were almost five times more likely to have G. duodenalis than those collected from more sparsely populated sites (Gaydos et al., 2008). Perhaps the difference in the prevalence of G. duodenalis between the east and west coast seals of this study is related to haul-out size. Although no specific data on haul-out size was recorded for east coast seals, samples from Jeremy Point where there was high prevalence of G. duodenalis derived from a fairly large haul-out site (>100 seals, personal observation). In contrast, samples obtained from the west coast derived primarily from young seals that had been recently abandoned by their mothers and stranded ashore alone (Colegrove et al., 2005). Subsequently, these young seals might not have had the opportunity to interact with conspecifics at large haul-out sites and become exposed to G. duodenalis.
In general, geographic and host specificities for Assemblage A and B haplotypes are less clear due to inconsistent results between the two loci sequenced. Most samples that provided amplification at one locus did not provide amplification at the second and when amplification occurred within a single sample at both gdh and tpi loci, sequences were always from different assemblages. This indicates either primer amplification biases where Assemblage A and H preferentially amplified at the gdh locus and Assemblage B preferentially amplified at the tpi locus or the presence of G. duodenalis individuals of mixed-assemblage ancestries. In the first scenerio, amplification of only one locus for 75% of G. duodenalis-positive samples would reflect the fact that these samples contain haplotypes from only one assemblage at both loci. Thus, in a sample that contained only Assemblage B at the gdh and tpi loci, only tpi would amplify, which seems likely in the case of Pacific harbor seals. In the second scenerio, genetic exchange between individuals of different assemblages must have occurred. There has been much debate about whether the presence of mixed assemblages in a single sample represents multiple individuals or the haplotype of a single individual (Cacciò and Ryan 2008; Cacciò and Sprong 2009; Monis et al., 2009). Recent evidence indicates that recombination within and between G. duodenalis assemblages occurs (Cooper et al., 2007; Morrison et al., 2007; Teodorovic et al., 2007; Poxleitner et al., 2008; Lasek-Nesselquist et al., 2009); perhaps when gdh and tpi both provide amplification, we are detecting the recombinant haplotype from a single individual. Assemblage-specific primers could be used to determine the identity of the unsequenced locus and a combination of single celled PCR and fluorescence in situ hybridization (FISH) could be used to determine whether the tpi and gdh haplotypes derived from a single G. duodenalis individual, which would have broad implications for the population biology of this parasite. At the least, the inconsistent amplification we observed serves as a cautionary tale for those using PCR to assess parasite molecular diversity and the presence/absence of G. duodenalis assemblages.
The population dynamics of G. duodenalis within marine systems appears to be a result of a tangled web of interacting factors. Teasing apart host-specific and geographic influences requires additional sampling. Giardia duodenalis sequences from gulls represented the majority of the haplotypic diversity sampled, suggesting that gulls are major players in spreading disease between terrestrial and marine environments and within marine environments. A comparative approach involving frequent sampling from conspecific seal and gull populations from two different locations might further elucidate the major contributors to the spread of G. duodenalis with marine systems. Finally, the identification and characterization of a new G. duodenalis assemblage suggest that the diversity of G. duodenalis and the wildlife impacted by this parasite are greater than previously believed.
Supplementary Material
Acknowledgements
This work was funded by grants awarded to Mitchell L. Sogin: NIH Grant # R01 AI058054-02 and NSF Grant # OCE0430724. The authors thank Phillip R. Neil for his advice regarding statistical analyses. We also thank Denise Greig from the Marine Mammal Center, CA for harbor seal feces collected as part of the national Marine Mammal Health and Stranding Response Program, Jim Harvey and Cori Gibble from Moss Landing Marine Laboratories for feces collected from San Francisco Bay harbor seal haulouts under NMFS research permit #555-1870-00, and Michael Moore, Becky Gast, and Andrea Bogomolni for east coast marine vertebrate feces collected under the NMFS research permit #775-1600-10, the NOAA Coastal Ocean Program award # NA05NOS4781247, the NOAA Prescott Program award # NA06NMF4390130 and the COHH award # NIEHS P50ES012742.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Note: Nucleotide sequence data reported in this paper are available in the GenBank database under the accession numbers: GUI182371-GUI182396, GUI176069-GUI176101, and GU733611-GU7333706.
Note: Supplementary material associated with this article.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Cacciò SM, Ryan U. Molecular epidemiology of giardiasis. Mol. Biochem. Parasitol. 2008;160:75–80. doi: 10.1016/j.molbiopara.2008.04.006. [DOI] [PubMed] [Google Scholar]
- Cacciò SM, Sprong H. Giardia duodenalis: genetic recombination and its implications for taxonomy and molecular epidemiology. Exp. Parasitol. 2010;124:107–112. doi: 10.1016/j.exppara.2009.02.007. [DOI] [PubMed] [Google Scholar]
- Colegrove K, Greig DJ, Gulland FMD. Causes of live stranding of Northern Elephant Seals (Mirounga angustirostris) and Pacific Harbor Seals (Phoca vitulina) along the Central California Coast, 1992–2001. Aquatic mammals. 2005;31:1–10. [Google Scholar]
- Cooper M, Adam RD, Worobey M, Sterling CR. Population genetics provides evidence of recombination in Giardia. Curr. Biol. 2007;17:1–5. doi: 10.1016/j.cub.2007.10.020. [DOI] [PubMed] [Google Scholar]
- Deng M, Peterson RP, Cliver DO. First findings of Cryptosporidium and Giardia in California sea lions (Zalophus californianus) J. Parasitol. 2000;86:490–494. doi: 10.1645/0022-3395(2000)086[0490:FFOCAG]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Dixon BR, Parrington LJ, Parenteau M, Leclair D, Santin M, Fayer R. Giardia duodenalis and Cryptosporidium spp. in the intestinal contents of ringed seals (Phoca hispida) and bearded seals (Erignathus barbatus) in Nunavik, Quebec, Canada. J. Parasitol. 2008;94:1161–1163. doi: 10.1645/GE-1485.1. [DOI] [PubMed] [Google Scholar]
- Ewing B, Green P. Basecalling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Smouse PE, Quattro JM. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fayer R, Dubey JP, Lindsay DS. Zoonotic protozoa: from land to sea. Trends Parasitol. 2004;20:531–536. doi: 10.1016/j.pt.2004.08.008. [DOI] [PubMed] [Google Scholar]
- Gaydos JK, Miller WA, Johnson PJ, Zornetzer H, Melli A, Packham A, Jeffries SJ, Lance MM, Conrad PA. Novel and canine genotypes of Giardia duodenalis in harbor seals (Phoca vitulina richardsi) J. Parasitol. 2008;94:1264–1268. doi: 10.1645/GE-1321.1. [DOI] [PubMed] [Google Scholar]
- Hughes-Hanks JM, Rickard LG, Panuska C, Saucier JR, O'Hara TM, Dehn L, Rolland RM. Prevalence of Cryptosporidium spp. and Giardia spp. in five marine mammal species. J. Parasitol. 2005;91:1225–1228. doi: 10.1645/GE-545R.1. [DOI] [PubMed] [Google Scholar]
- Jensen JL, Bohonak AJ, Kelley ST. Isolation by distance, web service. BMC Genet. 2005;6:13. doi: 10.1186/1471-2156-6-13. doi: 10.1186/1471-2156-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebbad M, Mattsson JG, Christensson B, Ljungström B, Backhans A, Andersson JO, Svärd SG. From mouse to moose: multilocus genotyping of Giardia isolates from various animal species. Vet. Parasitol. 2010;168:231–239. doi: 10.1016/j.vetpar.2009.11.003. [DOI] [PubMed] [Google Scholar]
- Lasek-Nesselquist E, Bogomolni AL, Gast RJ, Mark Welch D, Ellis JC, Sogin ML, Moore MJ. Molecular characterization of Giardia duodenalis haplotypes in marine animals: variation and zoonotic potential. Dis. Aquat. Org. 2008;81:39–51. doi: 10.3354/dao01931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasek-Nesselquist E, Mark Welch D, Thompson RCA, Steuart RF, Sogin ML. Genetic exchange within and between assemblages of Giardia duodenalis. J. Eukaryot. Microbiol. 2009;56:504–518. doi: 10.1111/j.1550-7408.2009.00443.x. [DOI] [PubMed] [Google Scholar]
- Mantel N. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967;27:209–220. [PubMed] [Google Scholar]
- Measures LN, Olson M. Giardiasis in pinnipeds from Eastern Canada. J. Wildl. Dis. 1999;35:779–782. doi: 10.7589/0090-3558-35.4.779. [DOI] [PubMed] [Google Scholar]
- Monis PT, Caccio SM, Thompson RCA. Variation in Giardia: towards a taxonomic revision of the genus. Trends Parasitol. 2009;25:93–100. doi: 10.1016/j.pt.2008.11.006. [DOI] [PubMed] [Google Scholar]
- Morrison HG, McArthur AG, Gillin FD, Aley SB, Adam RD, Olsen GJ, Best AA, Cande WZ, Chen F, Cipriano MJ, Davids BJ, Dawson SC, Elmendorf HG, Hehl AB, Holder ME, Huse SM, Kim UU, Lasek-Nesselquist E, Manning G, Nigam A, Nixon JE, Palm D, Passamaneck NE, Prabhu A, Reich CI, Reiner DS, Samuelson J, Svärd SG, Sogin ML. Genomic minimalism in the early diverging intestinal parasite Giardia lamblia. Science. 2007;28:1921–1926. doi: 10.1126/science.1143837. [DOI] [PubMed] [Google Scholar]
- Olson ME, Roach PD, Stabler M, Chan W. Giardiasis in ringed seals from the Western Arctic. J. Wildl. Dis. 1997;33:646–648. doi: 10.7589/0090-3558-33.3.646. [DOI] [PubMed] [Google Scholar]
- Posada D, Crandall KA. MODELTEST: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Poxleitner MK, Carpenter ML, Mancuso JJ, Wang CJ, Dawson SC, Cande WZ. Evidence of karyogamy and exchange of genetic material in the binucleate parasite Giardia duodenalis. Science. 2008;319:1530–1533. doi: 10.1126/science.1153752. [DOI] [PubMed] [Google Scholar]
- Rozas J, Rozas R. DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics. 1999;15:174–175. doi: 10.1093/bioinformatics/15.2.174. [DOI] [PubMed] [Google Scholar]
- Sulaiman IM, Fayer R, Caryn B, Gilman RH, Trout JM, Schantz PM, Das P, Lal AA, Lihua X. Triosephosphate isomerase gene characterization and potential zoonotic transmission of Giardia duodenalis. Emerg. Infect. Dis. 2003;9:1444–1452. doi: 10.3201/eid0911.030084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teodorovic S, Baverman JM, Elmendorf HG. Unusually low levels of genetic variation among Giardia lamblia isolates. Eukaryot. Cell. 2007;6:1421–1430. doi: 10.1128/EC.00138-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thioulouse J, Chessel D, Dolédec S, Olivier JM. ADE-4: a multivariate analysis and graphical display software. Stat. Comput. 1996;7:75–83. [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson RCA, Monis PT. Variation in Giardia: implications for taxonomy and epidemiology. Adv. Parasitol. 2004;58:69–137. doi: 10.1016/S0065-308X(04)58002-8. [DOI] [PubMed] [Google Scholar]
- Trout JM, Santín M, Greiner E, Fayer R. Prevalence of Giardia duodenalis genotypes in pre-weaned dairy calves. Vet. Parasitol. 2004;124:179–186. doi: 10.1016/j.vetpar.2004.07.010. [DOI] [PubMed] [Google Scholar]
- Wright S. The interpretation of population structure by F-statistics with special regards to systems of mating. Evolution. 1965;19:395–420. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.