Abstract
Positive elongation factor b (P-TEFb) is a cellular protein kinase that is required for RNA polymerase II (RNAP II) transcriptional elongation of protein coding genes. P-TEFb is a set of different molecular complexes, each containing CDK9 as the catalytic subunit. There are two isoforms of the CDK9 protein -- the major 42KDa CDK9 isoform and the minor 55 KDa isoform that is translated from an in-frame mRNA that arises from an upstream transcriptional start site. We found that shRNA depletion of the 55K CDK9 protein in HeLa cells induces apoptosis and double-strand DNA breaks (DSBs). The levels of apoptosis and DSBs induced by the depletion were reduced by expression of a 55K CDK9 protein variant resistant to the shRNA, indicating that these phenotypes are the consequence of depletion of the 55K protein and not off-target effects. We also found that the 55K CDK9 protein, but not the 42K CDK9 protein, specifically associates with Ku70, a protein involved in DSB repair. Our findings suggest that the 55K CDK9 protein may function in repair of DNA through an association with Ku70.
Keywords: CDK9, P-TEFb, Ku70, RNAP II elongation, DNA repair
Introduction
P-TEFb is a cellular protein kinase that is required for RNAP II transcriptional elongation of many, if not most, protein coding genes [reviewed in (1;2)]. P-TEFb stimulates elongation through the phosphorylation of the carboxyl terminal domain of the largest subunit of RNAP II, as well as phosphorylation of DSIF and NELF, two multi-subunit protein factors that associate with RNAP II and limit elongation. P-TEFb is actually a set of distinct molecular complexes, each containing CDK9 as the catalytic subunit and different cyclin regulatory subunits -- Cyclin T1, Cyclin T2a, Cyclin T2b, or Cyclin K. Cyclin T2a and T2b are spliced variants from the same gene, while Cyclins T1 and K are encoded in distinct genes. A large portion of P-TEFb exists as a catalytically inactive snRNP complex with 7SK snRNA and HEXIM1/2, MePCE (BCDIN3) and PIP7S (LARP7) proteins [reviewed in (3)]. P-TEFb that is not in the 7SK snRNP is bound to a bromodomain protein termed Brd4 that can bind to acetylated histones that mark active genes (4;5).
Additional complexity of P-TEFb was demonstrated when two isoforms of the CDK9 protein were identified (6). The major CDK9 isoform is a 42 kDa protein (42K), while a minor isoform is a 55 kDa protein (55K) that is translated from an in-frame mRNA that arises from an upstream transcriptional start site. The 55K protein is identical to the 42K protein except for an additional 117 amino acid residues at its amino terminus.
The expression patterns of the 42K and 55K CDK9 proteins differ. Both the 42K and 55K proteins are expressed at basal levels in quiescent primary CD4+ T cells, but only the 42K protein is up-regulated by T cell activation (7;8). The 55K CDK9 protein is generally not expressed in primary monocytes and it is induced during macrophage differentiation, while the 42K protein is expressed at a relatively high level in monocytes and is only weakly induced during differentiation (7). The relative abundance of the 42K and 55K proteins also varies between different murine tissues (9). Additionally, the sub-nuclear localization of the 42K and 55K CDK9 proteins differs, as a transiently expressed 42K protein is localized diffusely in the nucleoplasm of HeLa cells, while the 55K protein accumulates in the nucleolus (7). The 55K protein is therefore likely to be the CDK9 isoform that was identified in human nucleoli in a proteomics analysis (10). The substrate specificities of the 42K and 55K CDK9 proteins appear to be similar, as in vitro kinase reactions with purified CDK9 proteins showed identical phosphorylation patterns with 144 peptide substrates (7).
To investigate the function of the 55K CDK9 isoform, we used a shRNA vector to deplete the 55K protein in HeLa cells. We observed that the 55K protein is essential for cell viability, as apoptosis is induced following its depletion. Furthermore, we found that Ku70, a protein involved in double-stranded DNA break (DSB) repair, specifically associates with the 55K but not 42K CDK9 protein. Depletion of the 55K protein induced DSBs as indicated by phosphorylation of the histone H2AX (termed γ-H2AX). The levels of apoptosis and DSBs induced by the shRNA vector were reduced by expression of a 55K CDK9 protein variant that is resistant to the shRNA vector, indicating that these phenotypes are the consequence of depletion of the 55K protein rather than that of an off-target effect of the shRNA vector. Our results demonstrate that the 55K CDK9 protein associates with Ku70 and this CDK9 isoform may play a role in repair of DSBs.
Materials and Methods
Cells and reagents
HeLa cells and 293T cells were maintained in DMEM with 10% fetal bovine serum (FBS) and antibiotics. Cyclin T1, Cyclin T2a, CDK7 and CDK9 antisera were purchased from Santa Cruz Biotechnology; γ-H2AX antiserum was from Millipore; β-actin and Flag antisera were from Sigma; PARP antiserum was from Cell Signaling; anti-Flag M2 affinity gel was from Sigma; Ku70 antisera was from Abcam; mouse anti-rabbit light-chain antiserum was from Jackson Immunoresearch. Polybrene and puromycin were purchased from Sigma.
Production of shRNA viruses and flow cytometry
The Dharmacon siDesign program was used to design the siRNA target sequence in the unique region of the CDK9 55K mRNA. The 55K target sequence was: GGC CTC TCG GGA ACT ACA A. The control siRNA target sequence used in this study was: GCT ATA GCT GTT CTA GTT C; this control sequence is a mismatch to human Cyclin T1 RNA (11). DNA oligonucleotides encoding shRNA sequences were purchased from Invitrogen, and inserted under the control of the U6 promoter in the FG12 lentiviral vector (12). The shRNA sequences are expressed in this vector from the U6 promoter; the vector also expresses eGFP as a marker protein but does not express any lentivirus viral gene products. Lentiviral stocks were generated by standard plasmid co-transfections in 293T cells, and viral stocks were collected from culture supernatant at 72hr post-transfection and passed through 0.45um filters. Aliquots of lentiviral stocks were stored at –80°C. HeLa cells were infected with shRNA lentiviruses in the presence of polybrene, and cells were collected at the indicated time points for GFP detection, PI staining, and preparation of cell extracts for immunoblots. For GFP detection, cells were removed from culture dishes with trypsin, washed with 2% FBS PBS, followed by 1% paraformaldhyde fixation. GFP positive cells were analyzed by flow cytometry using a Beckman-Coulter XL-MCL Cytometer. Propidium Iodide staining was performed with the cellular DNA Flow Cytometric Analysis Kit (Roche Molecular Biochemicals) and DNA content was analyzed by flow cytometry.
Immunoblots
Immunoblots were performed as described previously (13). Briefly, cells were collected, washed with PBS, and lysed with EBCD buffer (50 mM Tris-HCl pH 8.0, 120mM NaCl, 0.5% NP-40, 5 mM dithiothreitol) containing a protease inhibitor cocktail (Sigma). Total protein concentrations were determined by the Bio-Rad protein assay, and 20 μg of total protein was analyzed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis followed by transfer to nitrocellulose membranes. After binding with the primary antiserum and reactions with secondary HRP-conjugated antibodies, blots were washed extensively with TBST (50mM Tris.Cl pH8, 200mM NaCl and 0.2% Tween 20), and the membranes were developed by enhanced chemiluminescence methods (Pierce). Dilutions of antibodies used in immunoblots were: CDK9 (1:5,000), Cyclin T1 (1:2,000), Cyclin T2 (1:1,000), CDK7 (1:1,000), γ-H2AX (1:1,000), PARP (1:1,000), β-actin (1:5,000), Ku70 (1:5,000).
Construction of HeLa cell pools expressing 55K proteins
The CDK9 55K cDNA containing a Flag epitope tag was inserted into pBabe-puro, an MLV retroviral vector containing a puromyocin-resistance gene (14). A variant of the 55K CDK9 cDNA was constructed by PCR mutagenesis that contained synonymous mutations that render the expressed mRNA resistant to the shRNA vector; the sequence differences between this variant and the wild type CDK9 mRNA are shown in Figure 1A. The CDK9-MLV vector identities were confirmed by DNA sequencing. Stocks of MLV viruses were generated by plasmid co-transfection in 293T cells. The MLV viral stocks were used to transduce HeLa cells, and transduced cell pools were selected by resistance to puromyocin.
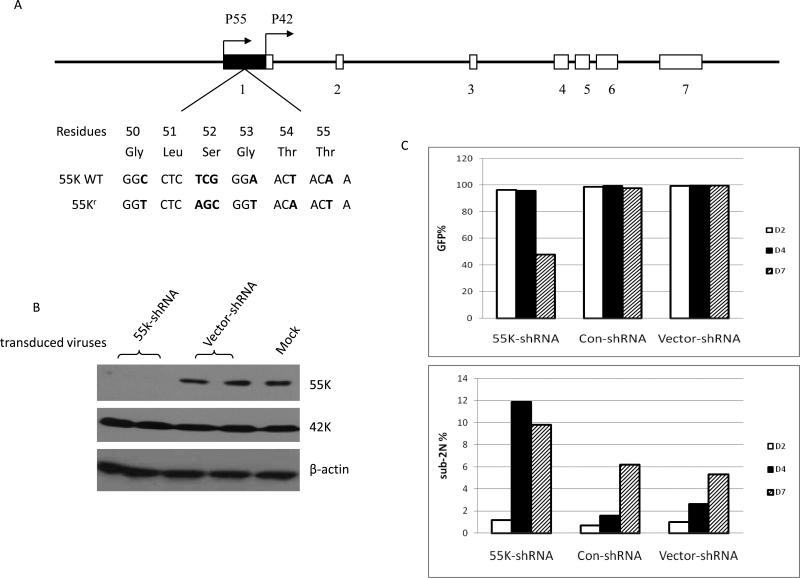
Figure 1. Specific shRNA depletion of 55K CDK9 protein.
A. The genomic structure of the human CDK9 gene and transcription start sites of mRNAs encoding the 55K and 42K proteins are shown (24). The sequence of the wild type 55K mRNA and the sequence of the mRNA encoding the shRNA-resistant 55K protein, termed 55Kr, are shown. B. At four days post-infection, HeLa cell extracts were prepared from cultures infected in duplicate with 55K-shRNA lentivirus, parental Vector-shRNA lentivirus; a single culture was mock-infected. Expression levels of the indicated proteins were examined in immunoblots. C. Expression of the eGFP reporter protein in the cultures infected with the indicated shRNA viruses was examined by flow cytometry at two, four, and seven days post-infection (top panel). Portions of cell cultures were also stained with propidium iodide; the percentage of cells that had a sub-2N DNA content as determined by flow cytometry are shown (bottom panel).
Mass Spectrometry
Mass s spectrometry analysis to identify the association of Ku70 with the 55K CDK9 protein was carried out in the laboratory of Dr. Jun Qin, Baylor College of Medicine.
Results
ShRNA depletion of 55K CDK9 protein induces apoptosis
Although the RNA sequences unique to the 55K mRNA are 83% GC rich, we identified a region in this sequence that appeared suitable as a target for siRNA depletion of the 55K CDK9 protein (Fig. 1A). We therefore constructed a lentiviral shRNA vector, termed 55K-shRNA, which targets these sequences. To examine whether this vector was effective in depleting the 55K protein, duplicate HeLa cell cultures were infected with the 55K-shRNA or (parental) Vector-shRNA lentivirus; a single culture was also mock-infected. Cell extracts were prepared at four days post-infection and expression levels of 55K and 42K CDK9 proteins and β-actin were determined in an immunoblot (Fig. 1B). The 55K protein was effectively depleted by the 55K-shRNA lentivirus, while the 42K protein was unaffected, indicating that this lentiviral vector can be used to deplete the 55K protein. We also performed RT-PCR assays to verify that the 55K-shRNA vector reduced the steady-state level of CDK9 RNA (see Supplemental Data).
The 55K-shRNA lentiviral vector expresses an enhanced green fluorescent protein (GFP) as a marker for infection. To examine effects of depletion of the 55K protein on cellular growth, HeLa cell cultures were infected with the 55K-shRNA, the Vector-shRNA, or a control shRNA lentivirus termed Con-shRNA. The Con-shRNA expresses a shRNA that is a mismatch to human Cyclin T1 mRNA and it has no measurable effect on cellular growth and little detectable effect on the transcriptional profile of cells (11;15). GFP expression was monitored at two, four and seven days post-infection with these lentiviral vectors (Fig. 1C). Cultures infected with the 55K-shRNA, Con-shRNA, or Vector-shRNA lentiviruses were greater than 95% GFP-positive at two and four days post-infection. At seven days post-infection, however, GFP expression was observed in only about 50% of the culture infected with the 55K-shRNA, while cultures infected with the Vector-shRNA or Con-shRNA maintained GFP expression in greater than 98% of the cells. These data suggest that cells in which the 55K protein has been depleted may be lost from the culture or a growing significantly slower. We performed an immunoblot analysis of cell extracts prepared during the course of this experiment and observed that the 55K shRNA lentivirus caused a small reduction in expression of the 55K protein at two days post-infection and an effective depletion at four and seven days post-infection, similar to the depletion shown in Figure 1B (data not shown).
During this time course, we also stained portions of infected cultures with propidium iodide to determine the rate of apoptosis as measured by sub-2N DNA content (Fig. 1C). At two days post-infection, cultures infected with either the 55K-shRNA, Con-shRNA, or Vector-shRNA lentivirus contained less than 2% apoptotic cells. At four days post-infection, the percentages of apoptotic cells in Con-shRNA and Vector-shRNA infected cells were less than 3%. In contrast, cells infected with the 55K-shRNA lentivirus contained 12% apoptotic cells at four days post-infection. At seven days post-infection, the percentage of apoptotic cells in the Con-shRNA and Vector-shRNA infected cells were 5-6%; this increase in the level of apoptosis in the control cultures is likely due to increased cell density as the cultures were not diluted after day four post-infection. However, at day seven post-infection the percentage of apoptotic cells in the culture infected with the 55K-shRNA vector was approximately 10%, almost two-fold higher than the control cultures. These data suggest that loss of GFP expression in the culture infected with the 55K-shRNA lentivirus is the result of apoptosis following depletion of the 55K protein.
Identification of Ku70 as a 55K CDK9-associated protein
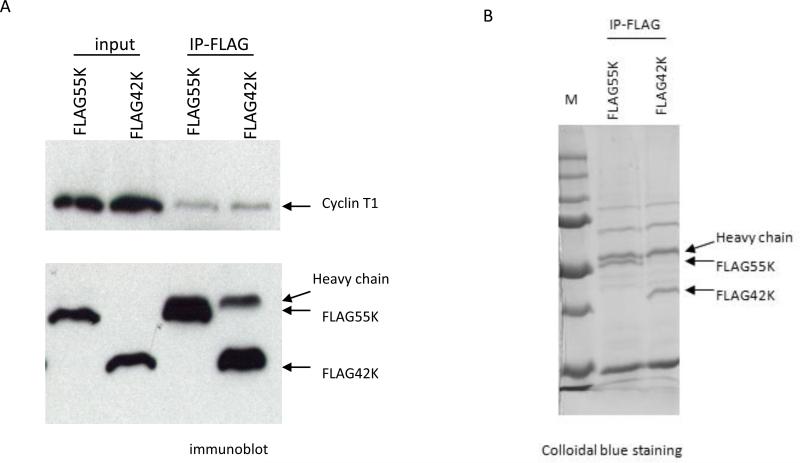
Because our shRNA depletion experiments suggested that the 55K CDK9 protein is essential for cell viability, we wished to identify 55K-associated proteins. We constructed expression vectors for Flag-tagged 42K and 55K CDK9 proteins for a proteomic analysis of proteins that associate with the 55K but not 42K protein. The 42K and 55K proteins could be efficiently immunoprecipitated with a Flag antibody from cells transfected with the expression vectors as determined by an immunoblot and Colloidal blue staining of immunoprecipitates (Fig. 2). We carried out a mass spectrometry analysis of proteins that co-immunoprecipitate with the 55K or 42K CDK9 protein. This analysis identified Ku70 as a protein that was present in 55K but not 42K immunoprecipitations. Ku70 in association with Ku80 functions as a single-stranded DNA-dependent ATP-dependent helicase and is involved in repair of double-strand DNA breaks [reviewed in (16)].
Figure 2. Immunoprecipitations of Flag-55K and Flag-42K CDK9 proteins for proteomic analysis.
A. Plasmid expression vectors for Flag-tagged 55K and 42K CDK9 proteins were transfected into HeLa cells and immunoprecipitations with the Flag antibody were performed at Day 2 post-transfection. An immunoblot was performed to measure the expression levels of the Flag-tagged proteins and the relative amount of Cyclin T1 that co-immunoprecipitated with each protein. B. The plasmid vectors were transfected into HeLa cells and immunoprecipitations with the Flag antibody were performed. A portion of the immunoprecipitate was analyzed on a SDS-polyacrylamide gel which was stained with Colloidal Blue. The remaining portion was subjected to a proteomics analysis to identify proteins present in the precipitations.
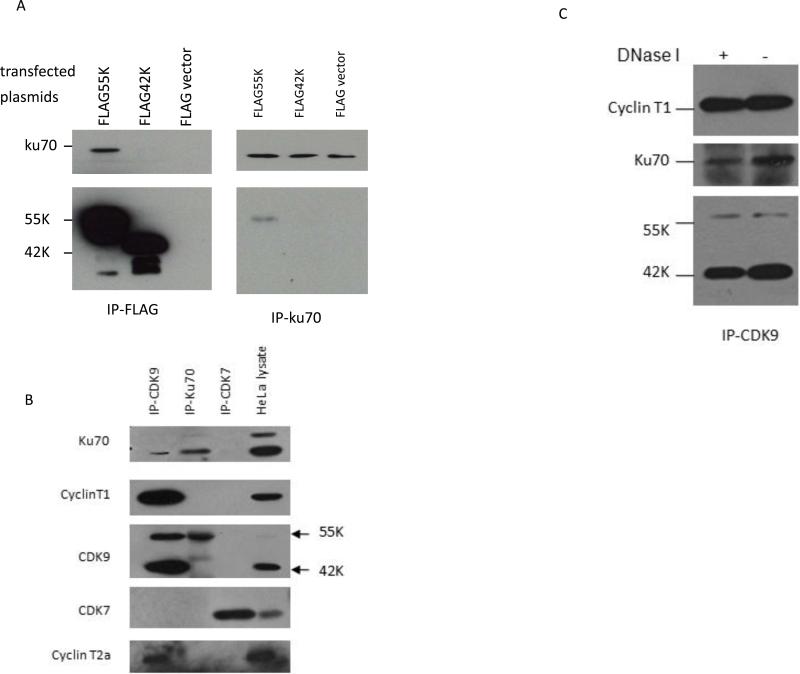
To verify that Ku70 specifically associates with the 55K CDK9 protein, we transfected the Flag-tagged 55K and 42K plasmid vectors into HeLa cells and performed immunoprecipitations with the Flag antibody and a Ku70 antiserum (Fig. 3A). In Flag immunoprecipitations, Ku70 was co-immunoprecipitated with the Flag-55K protein but not the Flag-42 protein or the parental Flag vector. In Ku70 immunoprecipitations, the Flag-55K protein but not the Flag-42K protein was co-immunoprecipitated with Ku70 (Fig. 3A). We also used antiserum against CDK9, Ku70, and CDK7 to assess the specificity of interaction between the endogenous CDK9 and Ku70 proteins (Fig. 3B). The antiserum against CDK9 was able to co-immunoprecipitate Ku70 as well as the Cyclin T1 and Cyclin T2a regulatory subunits of CDK9. Because of its lower expression level relative to the more abundant 42K CDK9 protein, the 55K protein is barely visible in the HeLa cell lysate lane in the exposure of the immunoblot shown in Figure 3B. The antiserum against Ku70 was able to co-immunoprecipitate the 55K but not 42K CDK9 proteins. The CDK7 antiserum did not co-immunoprecipitate Ku70 or the Cyclin T1 or T2a proteins as expected. It is notable that the Ku70 antiserum did not co-immunoprecipitate either Cyclin T1 or Cyclin T2a with the 55K CDK9 protein. We attempted to determine if the Ku70 antiserum could immunoprecipitate the Cyclin K regulatory subunit of CDK9, but we were unable to obtain unequivocal data due to the low expression level of Cyclin K and background in immunoblots. We also treated a HeLa cell extract with DNase I to determine if the association between the 55K protein and Ku70 was dependent upon DNA bridges (Fig. 3C). Control experiments were performed with plasmid DNA to verify that the DNase I was highly active under these reaction conditions. The DNase I treatment did not affect the ability of Ku70 to co-immunoprecipitate with CDK9, indicating that the 55K-Ku70 association does not involve a DNA bridge. In summary, the data shown in Figure 3 demonstrate that the 55K CDK9 protein is specifically associated with Ku70 in HeLa cells.
Figure 3. Ku70 specifically associates with the 55K CDK9 protein.
A. Plasmid expression vectors for Flag-tagged 55K and 42K CDK9 proteins and parental Flag vector were transfected into HeLa cells and immunoprecipitations were performed with the Flag antibody (left panel) or a Ku70 antiserum (right panel). Proteins present in immunoprecipitations were examined by immunoblotting. B. Cell extracts were prepared from HeLa cells and immunoprecipitations were performed with antisera against CDK9, Ku70, and CDK7 as indicated. The presence of the indicated proteins in immunoprecipitations was examined in immunoblots; HeLa cell lysate is shown on right lane. C. A HeLa cell extract was treated with and without DNase as indicated, and immunoprecipitations were performed with a CDK9 antiserum. The indicated proteins in the immunoprecipitates were examined in an immunoblot.
shRNA depletion of 55K CDK9 protein induces H2AX phosphorylation
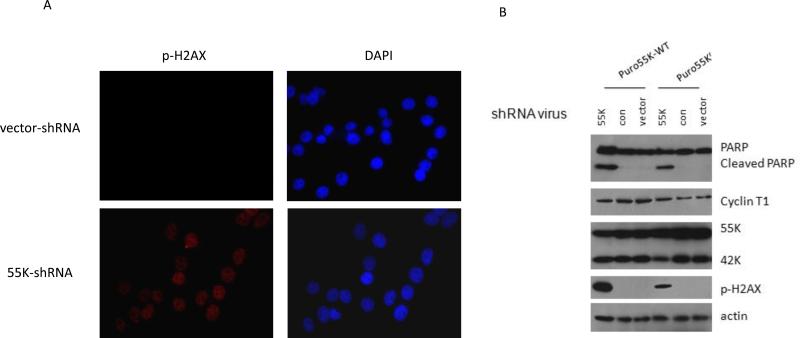
The association of Ku70 with the 55K CDK9 proteins suggests that the 55K protein may be involved in repair of DNA double-strand breaks (DSBs). We therefore examined if the shRNA depletion of the 55K protein induced DSBs. HeLa cells were infected with the 55K-shRNA or parental shRNA vector. To monitor DSBs, we used an indirect immunofluorescent assay to examine phosphorylation of histone H2AX (γ-H2Ax) at two days post-infection with these vectors, as phosphorylation of four residues at the carboxyl terminus of histone H2AX is a sensitive marker for DSBs (17). Cells depleted for the 55K protein demonstrated an elevated level of γ-H2AX relative to cells transduced with the vector (Fig. 4A). We obtained similar results as those shown in Figure 4A with a flow cytometry assay to measure γ-H2AX in 55K-depleted cells (data not shown). These results suggest that DSBs induced by depletion of the 55K CDK9 protein may trigger apoptosis as observed in Figure 1C.
Figure 4. Depletion of 55K CDK9 protein induces DSBs.
A. HeLa cells were infected with the 55K-shRNA or Vector-shRNA lentiviruses and at two days post-transduction were processed for indirect immunofluorescence to detect γ-H2AX to monitor DSBs and stained with DAPI to visualize DNA. B. Pools of puromycin-resistant HeLa cells over-expressing the wild type 55K CDK9 (puro55K-WT) or the shRNA-resistant CDK9 protein (Puro55Kr) were infected with the 55K-shRNA, Con-shRNA, or parental lentiviral vector At day four post-infection cell extracts were prepared and the indicated proteins evaluated in immunoblots.
To determine whether depletion of the 55K protein and not an off-target effect of the 55K shRNA was the cause of DSBs, we generated pools of cells that over-express either the wild type 55K protein or a variant resistant to the shRNA termed 55Kr (see Fig. 1A). These cell pools were infected with the 55K-shRNA, Con-shRNA, or Vector-shRNA lentiviruses and cell extracts were prepared at four days post-infection. We examined cleavage of Poly(ADP-ribose) Polymerase (PARP) as a marker of apoptosis in this experiment. The pool of 55K-WT cells displayed a high level of PARP cleavage in the infection with the 55K-shRNA vector, and this was significantly lower in cells that expressed the 55Kr protein (Fig. 4B). Similarly, the pool of 55K-WT cells infected with the 55K-shRNA vector displayed a high level of DSBs as indicated by γ-H2AX and this was significantly lower in the pool of cells that expressed the 55Kr protein. Additionally, in other experiments that used flow cytometry assays to measure sub-2N DNA content, we observed that a cell pool which over-expressed the 55Kr protein demonstrated lower levels of apoptosis than a cell pool that over-expressed the wild type 55K protein. Taken together, our data suggest that depletion of the 55K protein, rather than an off-target effect by the shRNA vector, induces DSBs and apoptosis.
Discussion
We found in this study that the 55K CDK9 protein is essential for cell viability in HeLa cells, and it specifically associates with Ku70 and may play a role in DNA repair. This CDK9 function appears to be distinct from the role of CDK9 as the catalytic subunit of P-TEFb, a general RNAP II elongation factor required for the expression of most, if not all, protein coding genes. Using co-immunoprecipitations with a Ku70 antiserum, we were unable to identify the cyclin partner(s) of 55K CDK9 proteins that are associated with Ku70 (Fig. 3B). The cyclin partner does not appear to be Cyclin T1 or T2 and it is therefore possible that it is Cyclin K. However, we were unable to detect Cyclin K in Ku70 immunoprecipitates due to a high background in immunoblots with two commercially available antisera against Cyclin K. Additional work will be required to resolve this issue, although it is possible that the 55K protein does not contain a cyclin partner when associated with Ku70.
We used DNA microarrays to carry out a transcriptional profile analysis of HeLa cells depleted of the 55K protein (see Supplemental data). When RNA from the 55K depleted cells was compared to the RNA from cells infected with the Con-shRNA vector, 1,712 transcripts were down-regulated ≥ 2-fold, while 910 transcripts were up-regulated ≥ 2-fold. It is not clear whether these changes in gene expression are the result of direct effects on RNAP II transcriptional elongation following depletion of the 55K protein from P-TEFb complexes, rather than indirect effects due to induction of DNA damage and apoptosis. We performed a Gene Ontology (GO) analysis of transcripts that were repressed ≥ 2-fold by the shRNA depletion (Supplemental Figure 1). Genes significantly over-represented by the 55K depletion include those involved in the response to DNA damage, DNA repair, and programmed cell death.
Ku70 is found in a heterodimer complex with Ku80 and was originally identified as an autoantigen of patients suffering from polymyositis-scleroderma overlap syndrome (18). The Ku70/80 heterodimer has multiple cellular functions, including DNA replication, telomere maintenance, regulation of transcription, and inhibition of apoptosis [reviewed in (19)]. The role of Ku70/80 in complexes with XRCC4/ligase IV and DNA-PKcs has been extensively studied as they are involved in the nonhomologous end-joining (NHEJ) of double-strand DNA breaks. Ku70/80 are regulatory subunits of DNA-PKcs and function to stabilize and bring together DNA ends for ligation [reviewed in (16)]. Because depletion of the 55K CDK9 protein induces DSBs, it is possible that the 55K protein plays a role in NHEJ and the DSB repair pathway.
Finally, CDK9 has been actively studied in HIV research because P-TEFb mediates the transcriptional activation function of the viral Tat protein (20;21). Tat associates with P-TEFb through a direct protein-protein interaction with the Cyclin T1 regulatory subunit. The Tat/P-TEFb complex binds to the TAR RNA element that forms in nascent viral transcripts, whereupon CDK9 phosphorylates a number of substrates in the RNAP II complex to activate transcriptional elongation. A defining step in the HIV life cycle is integration of the viral cDNA into a cellular chromosome, a process that involves cleavage of cellular DNA and ligation to viral cDNA. Single-strand gaps are generated during proviral integration and these gaps are repaired by cellular enzymes. Additionally, unintegrated viral cDNA is self-ligated to generate two types of circular DNA, termed 1 or 2-LTR circles and formation of these circular DNAs requires cellular enzymes. It has been proposed that DSB repair enzymes are involved in post-integration repair of viral DNA and the circularization of viral LTR circles (22;23). It is therefore possible that the 55K/Ku70 complex may play a role in the repair of proviral integration or circularization of viral LTR circles, adding another role for CDK9 in the HIV life cycle.
Supplementary Material
Acknowledgements
This work was funded by grants from the National Institutes of Health (AI035381 to A.P.R and CA100420 to L.A.D.). We thank Jun Qin and Sun Yun Jung for mass spectrometry and Alison Bertuch for helpful discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature Cited
- 1.Zhou Q, Yik JH. Microbiol Mol Biol Rev. 2006;70:646–659. doi: 10.1128/MMBR.00011-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peterlin BM, Price DH. Mol Cell. 2006;23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 3.Michels AA, Bensaude O. Biotechnol J. 2008;3:1022–1032. doi: 10.1002/biot.200800104. [DOI] [PubMed] [Google Scholar]
- 4.Yang Z, Yik JH, Chen R, He N, Jang MK, Ozato K, Zhou Q. Mol Cell. 2005;19:535–545. doi: 10.1016/j.molcel.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 5.Jang MK, Mochizuki K, Zhou M, Jeong HS, Brady JN, Ozato K. Mol Cell. 2005;19:523–534. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 6.Shore SM, Byers SA, Maury W, Price DH. Gene. 2003;307:175–182. doi: 10.1016/s0378-1119(03)00466-9. 175-82. [DOI] [PubMed] [Google Scholar]
- 7.Liu H, Herrmann CH. J Cell Physiol. 2005;203:251–260. doi: 10.1002/jcp.20224. [DOI] [PubMed] [Google Scholar]
- 8.Sung TL, Rice AP. Retrovirology. 2006;3:66. doi: 10.1186/1742-4690-3-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shore SM, Byers SA, Dent P, Price DH. Gene. 2005;350:51–58. doi: 10.1016/j.gene.2005.01.015. [DOI] [PubMed] [Google Scholar]
- 10.Andersen JS, Lam YW, Leung AK, Ong SE, Lyon CE, Lamond AI, Mann M. Nature. 2005;433:77–83. doi: 10.1038/nature03207. [DOI] [PubMed] [Google Scholar]
- 11.Yu W, Wang Y, Shaw CA, Qin XF, Rice AP. Retrovirology. 2006;3:32. doi: 10.1186/1742-4690-3-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qin XF, An DS, Chen IS, Baltimore D. Proc Natl Acad Sci U S A. 2003;100:183–188. doi: 10.1073/pnas.232688199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herrmann CH, Carroll RG, Wei P, Jones KA, Rice AP. J Virol. 1998;72:9881–9888. doi: 10.1128/jvi.72.12.9881-9888.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frese KK, Latorre IJ, Chung SH, Caruana G, Bernstein A, Jones SN, Donehower LA, Justice MJ, Garner CC, Javier RT. EMBO J. 2006;25:1406–1417. doi: 10.1038/sj.emboj.7601030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu W, Ramakrishnan R, Wang Y, Chiang K, Sung TL, Rice AP. PLoS ONE. 2008;3:e3146. doi: 10.1371/journal.pone.0003146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morio T, Kim H. Int J Biochem Cell Biol. 2008;40:598–603. doi: 10.1016/j.biocel.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 17.Bonner WM, Redon CE, Dickey JS, Nakamura AJ, Sedelnikova OA, Solier S, Pommier Y. Nat Rev Cancer. 2008;8:957–967. doi: 10.1038/nrc2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mimori T, Akizuki M, Yamagata H, Inada S, Yoshida S, Homma M. J Clin Invest. 1981;68:611–620. doi: 10.1172/JCI110295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Riha K, Heacock ML, Shippen DE. Annu Rev Genet. 2006;40:237–277. doi: 10.1146/annurev.genet.39.110304.095755. [DOI] [PubMed] [Google Scholar]
- 20.Barboric M, Peterlin BM. PLoS Biol. 2005;3:e76. doi: 10.1371/journal.pbio.0030076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rice AP, Herrmann CH. Curr HIV Res. 2003;1:395–404. doi: 10.2174/1570162033485159. [DOI] [PubMed] [Google Scholar]
- 22.Smith JA, Daniel R. ACS Chem Biol. 2006;1:217–226. doi: 10.1021/cb600131q. [DOI] [PubMed] [Google Scholar]
- 23.Skalka AM, Katz RA. Cell Death Differ. 2005;12(Suppl-8) doi: 10.1038/sj.cdd.4401573. [DOI] [PubMed] [Google Scholar]
- 24.Liu H, Rice AP. Gene. 2000;252:51–59. doi: 10.1016/s0378-1119(00)00215-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.