Fig. 6.
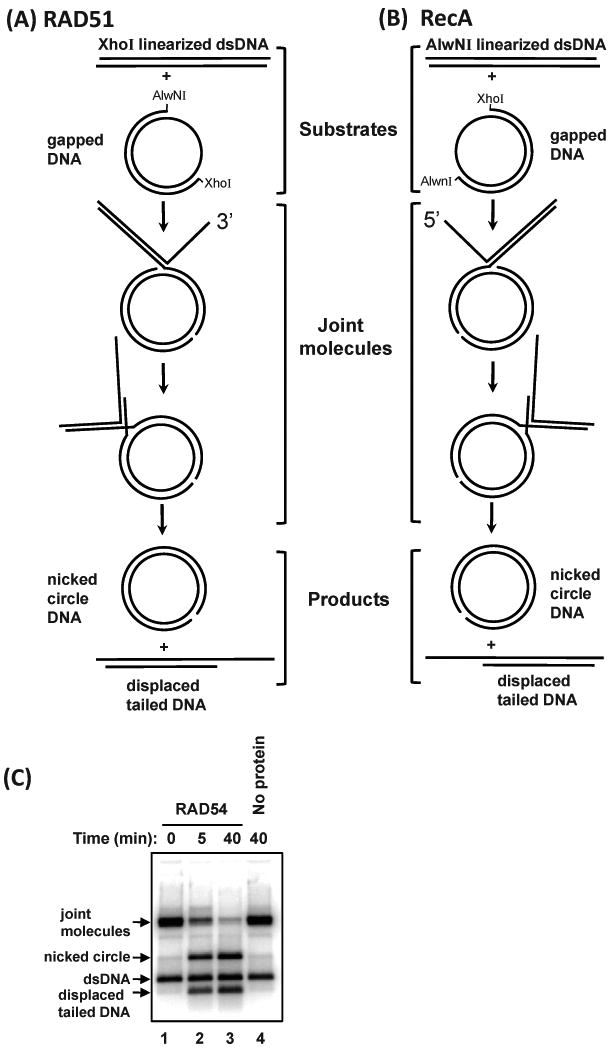
Production of joint molecules used to test the branch migration activity of proteins. (A) Joint molecules with a 3′ displaced ssDNA tail are produced by RAD51 using gapped DNA and pBS II K (+) dsDNA linearized with XhoI. (B) Joint molecules with a 5′ displaced ssDNA tail are produced by RecA using gapped DNA and pBS II K (+) dsDNA linearized with AlwNI. Joint molecules are deproteinized and used as substrates to test the branch migration activity of proteins of interest. (C) The branch migration of joint molecules with a 3′ displaced ssDNA tail (0.5 nM, molecules) by RAD54 (200 nM) at 30 °C. In the control reaction (Lane 4), RAD54 was replaced with storage buffer and carried out on for 40 min. Over the time course of the experiment, the amount of joint molecules decreases, while nicked circle and displaced tailed DNA increase.
