Abstract
Cellular DNA undergoes constant assault from a wide range of genotoxic stress. In order to maintain genome integrity, cells develop a repertoire of sophisticated systems to detect DNA damage and mediate cellular responses to DNA damage. Defects in the DNA damage response have been implicated in a variety of disorders including aging and cancer. Tumor suppressor p53 is a key intermediate in DNA damage response by inducing cell cycle arrest to allow repair or promoting apoptosis to eliminate irreparably damaged cells. A recent study described a novel layer of p53-mediated cellular response to DNA damage, i.e., modulation of cell adhesion and motility. JMY, a p53 co-factor, was demonstrated to be a multifunctional protein that coordinates cell adhesion and motility with nuclear p53 response. These results suggest that abnormal JMY activity and/or localization could contribute to tumor invasion and reveal JMY as a potential therapeutic target.
Key words: p53, JMY, DNA damage, actin, cell motility, cell adhesion
The preservation of genome integrity is crucial for cell survival and function, which is an intimidating task due to constant assault on DNA by a variety of genotoxic stress. While exogenous genotoxic agents such as ultraviolet light, ionizing radiation, oxidative stress, and chemical mutagens make major contributions to DNA lesions, endogenous DNA damage is constantly derived from cellular metabolism or routine errors in DNA replication. In order to combat these threats that challenge genome stability, cells have evolved a repertoire of sophisticated systems to detect DNA damage and mediate cellular responses to DNA damage. Defects in the DNA damage response have been implicated in a variety of disorders including aging and cancer.1
It is well established that DNA damage triggers two major cellular responses in eukaryotic cells. The first is cell cycle arrest that would block cell cycle progression to allow time for DNA damage repair before the damage could be passed on to daughter cells. The second is the induction of apoptosis as a means of eliminating irreparably damaged cells when the level of damage is particularly severe. Tumor suppressor p53 is a key intermediate in cellular response to genotoxic stress by actively engaging in these two branches of DNA damage response. In addition, recent data demonstrate that if DNA damage is severe, especially if it is chronic, cells have another choice by entering a protracted cell cycle arrest that is termed cellular senescence, in which damaged cells remain alive but are unable to proliferate.2
Are there any other cellular processes to be discovered that DNA damage could impinge on? The answer is yes. Most recently, Rodier et al. identified inflammation as a new response to persistent DNA damage. Persistent but not transient, DNA damage responses initiate increased secretion of inflammatory cytokines such as interleukin-6. This response is associated with cellular senescence, but also occurs in damaged cycling cells that are near, or have bypassed, senescence.3 This study revealed a surprising novel consequence of DNA damage response wherein damaged cells communicate their compromised state to the surrounding tissue and impact cell microenvironment.3
Now Coutts et al. gave us another surprise by demonstrating a novel layer of p53-mediated cellular response to DNA damage, i.e., modulation of cell adhesion and motility. Furthermore, they established that this fresh aspect of DNA damage response is critically dependent on the coordination of a multifunctional protein JMY (junction-mediating and regularity protein).4
Originally identified as a new p300-interacting protein by yeast two-hybrid approach, JMY cooperates with p300 to regulate p53-dependent transcription and plays important role in DNA damage response.5 In cells depleted of JMY, p53 activity is compromised. Moreover, a variety of damage agents such as ultraviolet light, etoposide and actinomycin D lead to the accumulation of JMY, indicating that JMY is a DNA-damage responsive protein.6 Intriguingly JMY contains a potential Arp2/3-activating sequence,7 indicating the possible involvement of JMY in cell motility. Detailed analysis revealed that JMY combines two separate actin nucleation promoting activities by both activating Arp2/3 and assembling filaments directly using a Arp2/3-independent, Spire-like mechanism.7 Furthermore, in motile cells JMY co-localizes with actin filaments at the leading edge and both overexpression and knockdown experiments demonstrated that JMY promotes cell migration in wound-healing assay.7 Thus JMY plays a novel role in controlling cell motility besides nuclear function in p53 response.
Based on their previous findings partially summarized above, in a paper recently published,4 Coutts et al. went further to demonstrate that JMY also impacts cell adhesion and coordinates cell adhesion and motility with nuclear p53 response in response to DNA damage. Intrigued by the observed abnormal cell morphology upon JMY depletion, the authors wondered whether alterations in cell adhesion are responsible for the phenotype. Therefore, they examined E-cadherin and N-cadherin protein levels and found their upregulation upon JMY depletion by siRNA in cells. Furthermore, simultaneous depletion of both JMY and E-cadherin or N-cadherin rescued the decrease of cell motility upon JMY depletion alone, confirming that JMY regulates cell motility via a novel mechanism that modulates cadherin stability, although the details of this regulation remain unexplored by the authors.
Next the authors investigated how JMY activity is regulated upon DNA damage by UV irradiation. They found that upon DNA damage endogenous as well as ectopic JMY undergoes nuclear accumulation. This raises the interesting possibility that nuclear translocalization of JMY upon DNA damage may abolish the ability of JMY to modulate cell adhesion and motility in the cytoplasm. Using scratch wound assay as readout of cell motility, the authors observed that UV damage eliminated the augment of cell motility by ectopic JMY expression. Moreover, loss of JMY by siRNA-mediated depletion had much less impact on cell motility in UV damaged cells than in untreated cells. In other words, nuclear translocation of JMY induced by DNA damage partially phenocopys the loss of JMY in terms of cell motility. To substantiate their findings that DNA damage reduces cell motility by modulating JMY localization, the authors elegantly made use of a JMY tagged with nuclear localization signal (NLS), JMY-NLS, and found that cells expressing JMY-NLS displayed reduced motility compared to cells expressing wild-type JMY. Importantly, upon DNA damage JMY-NLS-expressing cells and wild-type JMY-expressing cells displayed similar defects in cell motility, providing strong support that DNA damage induces nuclear translocation of JMY to disrupt its ability to promote cell motility.4
Given the previous established role of JMY as nuclear p53 transcriptional co-activator, the authors next examined the impact of JMY nuclear localization on p53 transcriptional activity. They found that in JMY-NLS-expressing cells p53 transcriptional activity was increased further compared to wild-type JMY-expressing cells. This result suggested that nuclear JMY augments p53 activity during DNA damage response.4
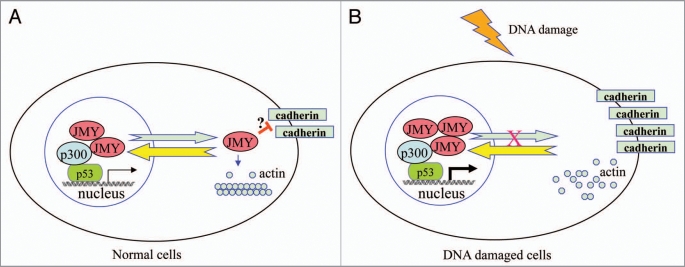
As one of the most important tumor suppressors, p53 has been extensively implicated in preventing tumor initiation by controlling cell cycle progression, inducing apoptosis and maintaining genome stability. Nevertheless, a flurry of recent evidence supports the emerging role of p53 in restraining cell migration and invasion in tumor progression.8 Very recently, two reports provided mechanistic insight into how p53 controls cancer cell invasion by inducing the MDM2-mediated degradation of Slug9 or regulating the expression of caldesmon, an actin-binding protein that inhibits podosome/invadopodium formation.10 In both cases, the transcriptional activity of p53 is crucially involved.9,10 In contrast, the latest findings presented by Coutts et al.4 unravel a completely different mode of p53 action in controlling cell adhesion and migration that may be independent of p53 transcriptional activity. In this scenario, upon DNA damage p53 may simply and quickly sequester JMY in the nucleus and inhibit its export to the cytoplasm where JMY could modulate cell adhesion and motility (Fig. 1).
Figure 1.
Proposed model for the function of JMY in normal cells and DNA damaged cells. (A) In normal cells, JMY undergoes constant shutting between the cytoplasm and the nucleus. While JMY localizes predominantly in the nucleus ready for p53 response, a fraction of JMY gets exported to the cytoplasm, where it would promote actin filament formation and disrupt the stability of cadherin via unclear mechanisms (marked with ?). As a result cells maintain a certain extent of motility. (B) Upon DNA damage, JMY is recruited by p53 to participate in p53 response in the nucleus and its export to the cytoplasm gets blocked. Consequently, cell adhesion is enhanced resulting from stabilized cadherin while actin filament formation is inhibited, contributing to the loss of cell motility.
In conclusion, Coutts et al. provide the first evidence for a potential novel mechanism by which p53 and p53 co-factor JYM modulate cell motility in response to DNA damage. Controlling the cellular localization of JMY seems to be the key for the coordination of cell adhesion and motility with nuclear p53 response by this multifunctional protein. This suggests that abnormal JMY cellular localization could contribute to tumor invasion and indicates JMY as a therapeutic target. Indeed, the jmy gene has been localized to the long arm of chromosome 5 in band 5q 13.2, where diverse chromosomal aberrations have been identified in a range of malignancies, and investigations are under way to determine the direct involvement of the jmy gene in these chromosomal aberrations.5
As often happens with scientific endeavors, more questions are now raised than answered. Given the surprising finding that JMY regulates cell adhesion, one obvious question is the mechanistic details underlying the modulation of cadherin stability by JMY. Although the transcriptional role of JMY could not be excluded, it is possible that cytoplasmic JMY interacts with Rho GTPase family to inhibit the endocytic recycling of cadherin, thus leading to reduced level of cadherin on the plasma membrane. For instance, it was demonstrated recently that abnormal integrin recycling accounts for mutant p53 induced cell invasion.11 In cells subjected to oncogenic stress, p53 is known to be activated. In this case, does JMY also get sequestered in the nucleus? How would these cells behave with respect to cell adhesion and motility? How about JMY expression and subcellular localization in invasive cells as well as tumor samples? What signals and/or events are responsible for the shutting of JMY between the cytoplasm and the nucleus and how nuclear accumulation of JMY induced by DNA damage is achieved? Undoubtedly, future investigations addressing these questions will further our understanding of various facets of JMY personality on different stages such as the cytoplasm and the nucleus.
Footnotes
Previously published online: www.landesbioscience.com/journals/celladhesion/article/11368
References
- 1.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.d’Adda di Fagagna F. Living on a break: cellular senescence as a DNA-damage response. Nat Rev Cancer. 2008;8:512–522. doi: 10.1038/nrc2440. [DOI] [PubMed] [Google Scholar]
- 3.Rodier F, Coppé JP, Patil CK, Hoeijmakers WA, Muñoz DP, Raza SR, et al. Persistent DNA damage signalling triggers senescence-associated inflammatory cytokine secretion. Nat Cell Biol. 2009;11:973–979. doi: 10.1038/ncb1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coutts AS, Weston L, La Thangue NB. A transcription co-factor integrates cell adhesion and motility with the p53 response. Proc Natl Acad Sci USA. 2009;106:19872–19877. doi: 10.1073/pnas.0906785106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shikama N, Lee CW, France S, Delavaine L, Lyon J, Krstic-Demonacos M, La Thangue NB. A novel cofactor for p300 that regulates the p53 response. Mol Cell. 1999;4:365–376. doi: 10.1016/s1097-2765(00)80338-x. [DOI] [PubMed] [Google Scholar]
- 6.Coutts AS, Boulahbel H, Graham A, La Thangue NB. Mdm2 targets the p53 transcription cofactor JMY for degradation. EMBO Rep. 2007;8:84–90. doi: 10.1038/sj.embor.7400855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zuchero JB, Coutts AS, Quinlan ME, Thangue NB, Mullins RD. p53-cofactor JMY is a multifunctional actin nucleation factor. Nat Cell Biol. 2009;11:451–459. doi: 10.1038/ncb1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roger L, Gadea G, Roux P. Control of cell migration: a tumour suppressor function for p53? Biol Cell. 2006;98:141–152. doi: 10.1042/BC20050058. [DOI] [PubMed] [Google Scholar]
- 9.Wang SP, Wang WL, Chang YL, Wu CT, Chao YC, Kao SH, et al. p53 controls cancer cell invasion by inducing the MDM2-mediated degradation of Slug. Nat Cell Biol. 2009;11:694–704. doi: 10.1038/ncb1875. [DOI] [PubMed] [Google Scholar]
- 10.Mukhopadhyay UK, Eves R, Jia L, Mooney P, Mak AS. p53 suppresses Src-induced podosome and rosette formation and cellular invasiveness through the upregulation of caldesmon. Mol Cell Biol. 2009;29:3088–3098. doi: 10.1128/MCB.01816-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Muller PAJ, Caswell PT, Doyle B, Iwanicki MP, Tan EH, et al. Mutant p53 drives invasion by promoting integrin recycling. Cell. 2009;139:1327–1341. doi: 10.1016/j.cell.2009.11.026. [DOI] [PubMed] [Google Scholar]