Abstract
Cell surface receptors of the integrin family are pivotal to cell adhesion and migration. The activation state of heterodimeric αβ integrins is correlated to the association state of the single-pass α and β transmembrane domains. The association of integrin αIIbβ3 transmembrane domains, resulting in an inactive receptor, is characterized by the asymmetric arrangement of a straight (αIIb) and tilted (β3) helix relative to the membrane in congruence to the dissociated structures. This allows for a continuous association interface centered on helix-helix glycine-packing and an unusual αIIb(GFF) structural motif that packs the conserved Phe-Phe residues against the β3 transmembrane helix, enabling αIIb(D723)β3(R995) electrostatic interactions. The transmembrane complex is further stabilized by the inactive ectodomain, thereby coupling its association state to the ectodomain conformation. In combination with recently determined structures of an inactive integrin ectodomain and an activating talin/β complex that overlap with the αβ transmembrane complex, a comprehensive picture of integrin bi-directional transmembrane signaling has emerged.
Key words: cell adhesion, membrane protein, integrin, platelet, transmembrane complex, transmembrane signaling
The communication of biological signals across the plasma membrane is fundamental to cellular function. The ubiquitous family of integrin adhesion receptors exhibits the unusual ability to convey signals bi-directionally (outside-in and inside-out signaling), thereby controlling cell adhesion, migration and differentiation.1–5 Integrins are Type I heterodimeric receptors that consist of large extracellular domains (>700 residues), single-pass transmembrane (TM) domains, and mostly short cytosolic tails (<70 residues). The activation state of heterodimeric integrins is correlated to the association state of the TM domains of their α and β subunits.6–10 TM dissociation initiated from the outside results in the transmittal of a signal into the cell, whereas dissociation originating on the inside results in activation of the integrin to bind ligands such as extracellular matrix proteins. The elucidation of the role of the TM domains in integrin-mediated adhesion and signaling has been the subject of extensive research efforts, perhaps commencing with the demonstration that the highly conserved GFFKR sequence motif of α subunits (Fig. 1), which closely follows the first charged residue on the intracellular face, αIIb(K989), constrains the receptor to a default low affinity state.11 Despite these efforts, an understanding of this sequence motif had not been reached until such time as the structure of the αIIb TM segment was determined.12 In combination with the structure of the β3 TM segment13 and available mutagenesis data,6,9,10,14,15 this has allowed the first correct prediction of the overall association of an integrin αβ TM complex.12 The predicted association was subsequently confirmed by the αIIbβ3 complex structure determined in phospholipid bicelles,16 as well as by the report of a similar structure based on molecular modeling using disulfide-based structural constraints.17 In addition to the structures of the dissociated and associated αβ TM domains, their membrane embedding was defined12,13,16,18,19 and it was experimentally recognized that, in the context of the native receptor, the TM complex is stabilized by the inactive, resting ectodomain.16 These advances in integrin membrane structural biology are complemented by the recent structures of a resting integrin ectodomain and an activating talin/β cytosolic tail complex that overlap with the αβ TM complex,20,21 allowing detailed insight into integrin bi-directional TM signaling.
Figure 1.

Amino acid sequence of integrin αIIb and β3 transmembrane segments and flanking regions. Membrane-embedded residues12,13,16,18,19 are enclosed by a gray box. Residues 991–995 constitute the highly conserved GFFKR sequence motif of integrin α subunits.
Structure and Membrane Embedding of Integrin α and β Transmembrane Segments
In order to understand the structure, association and membrane embedding of the αIIb and β3 TM segments, it is useful to first discuss their amino acid sequences, whose principal features are well conserved among the 18 α and 8 β human subunits.16 A stretch of 23 hydrophobic residues, commencing at αIIb(I966)/β3(I693) on the extracellular side, is terminated prior to αIIb(K989)/β3(K716) on the intracellular face, and is succeeded by four and five mostly hydrophobic residues, respectively (Fig. 1). Assuming α-helical conformation for the 23-residue hydrophobic segments preceding αIIb(K989)/β3(K716), the conformation and membrane embedding of these four and five residue segments will govern the TM helix tilt and, thus, the default orientation at which αIIb and β3 will face each other.
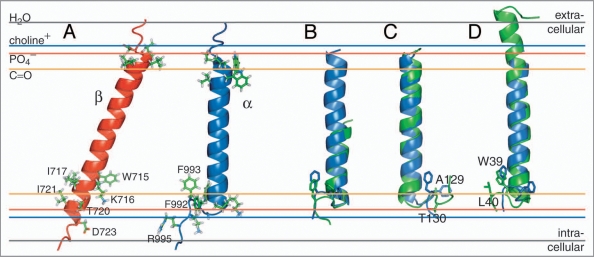
For β3, a 29-residue linear membrane-embedded helix, encompassing I693-I721, was revealed in phospholipid bicelle model membranes by NMR spectroscopy.13 This conformation embeds β3(K716) and indicates a pronounced TM helix tilt that minimizes the distance of K716’s positive side chain ɛ-NH3+ moiety to a negatively charged species such as a lipid’s PO4− group; i.e., β3(K716) points towards the cytosol (Fig. 2A). With 3.6 residues per helix turn, this points the most polar residue of the five-residue stretch, T720, towards the cytosol, whereas the remaining hydrophobic residues can orient their side chains mostly towards the membrane. An equivalent arrangement appears difficult for αIIb, which carries Phe residues at positions +3 and +4 from αIIb(K989) (Fig. 1), as do all 18 human α subunits.12 With this realization, the unusual structure of the αIIb TM segment (Fig. 2A), determined in phospholipid bicelles by NMR,12 is readily rationalized. A non-helical Gly-Phe-Phe conformation, with glycine efficiently abrogating helical tendencies and phenylalanine immersion in the membrane, is energetically preferable over a helical conformation wherein one phenylalanine invariably would have to face the cytosol. As a result, the αIIb TM segment can maintain a straight helix orientation in the membrane.12 Thus, the dissociated αIIb/β3 TM segments face each other in a straight/tilted helix-helix arrangement (Fig. 2A). Despite the existence of relevant mutagenesis data6 and the plethora of proposed integrin TM signaling models, this arrangement had been an overlooked possibility prior to the structural αIIb/β3 TM studies.12,13 Furthermore, the combination of the αIIb/β3 TM structures with TM mutagenesis data6,9,10,14,15 resulted in the first correct prediction of the structure of the associated αIIbβ3 TM state.12
Figure 2.
NMR structures of the individual integrin αIIb and β3 transmembrane segments. (A) Structures of the monomeric αIIb and β3 TM segments (PDB entries 2k1a and 2rmz, respectively) and their estimated membrane embedding.12,13,18,19 Selected side chains are shown in ball-and-stick representation. (B) An αIIb-homologous GFF structural motif is found in glycogen phosphorylase b (GPb; PDB entry 1a8i;22 residues 748–750). The αIIb TM segment and pertinent GPb helix are shown in blue and green, respectively. (C) For αIIb (blue), structural homology was found for a TM helix of cytochrome c oxidase (PDB entry 1xme, chain A), shown in green. The side chains of αIIb(F992–F993) and 1xme-A (A129-T130) are shown in ball-and-stick representation. F992 and A129 are structurally homologous. (D) The β peptide from the light-harvesting protein B-800/850 (homologous PDB entries 2fkw, chain S; 1nkz, chain F; and 1kzu, chain B also exhibits homology to αIIb. Shown is the structural alignment of integrin αIIb(P965–R995), in blue, with chain F of 1nkz, in green. The side chains of αIIb(F992–F993) and 1nkz-F(W39–L40) are shown in ball-and-stick representation. F993 and W39 are structurally homologous. The experimentally determined average positions of the lipid carbonyl, phosphate and choline groups (16, 20 and 22 Å from the center of the membrane) are depicted for 1,2-Dioleoyl-sn-Glycero-3-Phosphocholine.54 The bulk water phase is assumed to be fully established at 28 Å.55
The structure of the αIIb TM segment is noteworthy for its rarity. There are only a few examples of a GFF turn motif in relation to an α-helix in water soluble proteins. A fine example is found within the structure of glycogen phosphorylase b (GPb) of Oryctolagus cuniculus22 (Fig. 2B). Reminiscent of a membrane-water interface, the GFF motif of GPb is localized at the protein surface-water interface (c.f. PDB entry 1a8i; residues 748–750). In membrane proteins, no direct homologue has thus far been reported to my knowledge; however, some similarities to a TM helix of cytochrome c oxidase from Thermus thermophilus,23 and the β peptide (chain F) of the light-harvesting complex II from Rps. acidophila,24 are discernible (Fig. 2C and D). Like αIIb, the helix of cytochrome c oxidase is followed by a left-handed turn initiated by N127 that localizes A129 at a similar position to αIIb(F992) (Fig. 2C). The free energy gain of an Ala compared to a Phe membrane insertion is obviously limited. For the β peptide of the light-harvesting complex II, a right-handed turn is induced by P38 that localizes W39 at a similar position to αIIb(F993). W39 and L40 are thus able to re-immerse into the membrane (Fig. 2D). Finally, it is noted that the VFFA sequence near the N-terminus of the C-terminal TM helix of the amyloid precursor protein (APP) also appears to be membrane immersed, albeit in helical conformation, probably residing along the membrane surface.25
Structural Basis of Integrin αIIbβ3 Transmembrane Association
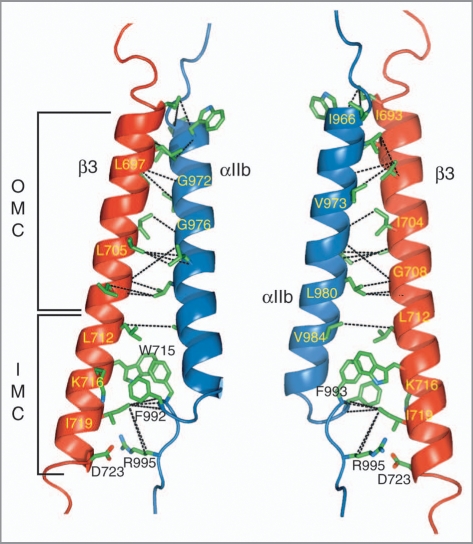
The NMR structure determination of the αIIbβ3 TM complex was carried out in bilayer-forming phospholipid bicelles26 employing non-covalently associated TM segments to achieve physiological conditions and to avoid any bias of the association interface, respectively. The use of sophisticated isotope-labeling patterns and deuterated lipids allowed the observation of close side chain distances between αIIb and β3, while avoiding the suppression of contaminating intramolecular protein and lipid signals at high lipid-to-protein ratios.16 Intersubunit distances were thus obtained along the entire dimerization interface (Fig. 3), which is characterized by protein backbone conformations that closely followed the dissociated αIIb and β3 states.16 The association within the extracellular membrane leaflet is most conspicuously defined by the packing of aliphatic side chains against three glycine residues, αIIb(G972), αIIb(G976) and β3(G708), as is common for TM helix associations, defining an outer membrane clasp (OMC; Fig. 3). From the TM sequences alone (Fig. 1), such an arrangement would be difficult to deduce. However, remarkably accurate predictions of the OMC had been achieved previously by molecular modeling studies.27–29 Within the intracellular membrane leaflet, αIIbβ3 association defines a novel packing motif, referred to as the integrin inner membrane clasp (IMC; Fig. 3).16 Residues αIIb(F992) and αIIb(F993) pack into a groove formed by β3(L712), β3(W715), β3(K716) and β3(I719). In addition to hydrophobic interactions, electrostatic contributions arising from β3(W715/ɛ-NH)-αIIb(F993/CO) and β3(K716/ɛ-NH3+)-αIIb(F992/CO) interactions would be compatible with the obtained structural ensemble and permissive side chain rotamers. However, competing interactions with lipid molecules may lead K716 to again prefer to fully neutralize its positive charge by engaging a lipid molecule. The NMR intersubunit distance restraints juxtapose positively charged αIIb(R995) on average to negatively charged β3(D723), enabling the formation of a mutually beneficial electrostatic interaction that completes the IMC (Fig. 3). Based on the strongly dissociating effects of altering either residue to alanine in the context of phospholipid bicelles and CHO membranes,16 coupled with the ability of R↔D charge reversal variants to maintain an inactive receptor,6 it is appropriate to favor ensemble structures that exhibit 245 a close αIIb(R995)-β3(D723) side chain distance16 to represent the fully associated αIIbβ3 TM complex. The lipid embedding of the αIIbβ3 TM complex was found essentially unchanged to its constituent monomeric peptides (Fig. 2A),16 showing the absence of an intrinsic tendency to alter its membrane topology.
Figure 3.
NMR structure of the integrin αIIbβ3 transmembrane complex. The locations of the outer and inner membrane clasps, OMC and IMC, respectively, are indicated. The structure illustrates the intersubunit distance (NOE) restraints used to calculate the αIIbβ3 TM complex.16 For clarity, interproton NOE connectivities are denoted by dotted black lines between the carbon atoms that are covalently bonded to the hydrogen nuclei, giving rise to the NOEs. The structure shown is the average structure of an ensemble of 20 structures calculated without an αIIb(R995)-β3(D723) structural constraint. It exhibits a negligible backbone r.m.s.d. of 0.32 Å to the average structure calculated with such a restraint (PDB entry 2k9j). The depicted orientations are related by a rotation of ∼180° about the y axis.
For the NMR structure determinations of the associated and dissociated integrin αIIbβ3 TM states,12,13,16 care was taken to ensure a physiological lipid environment. It was shown that an asymmetric lipid bilayer distribution is not essential to a study of the β3 TM segment;13 the aggregation states of the bicelle-embedded αIIb and β3 peptides were assessed to be predominantly monomeric;12,13 the invariance of the structures to changes in lipid composition was confirmed;12,13,16 and the comparison of membrane substitutes for β3 revealed the superiority of phospholipid bicelles over dodecylphosphocholine (DPC) micelles.13 This is also an extension of careful earlier work which revealed that, when fused to a coiled-coil construct as TM helix substitute, the GFFKR motif of αIIb failed to specifically interact with β3,30 providing an early indication that GFFKR does not have a tendency to adopt helical conformation, as had been reported in some later studies.31–33 In particular, the recent report of an αIIbβ3 TM structure, performed in 50%/50% CD3CN/H2O solution,33 lacked basic physiological conditions (e.g., a lipid-water interface), and invoked severe distortions in helical geometry of both αIIb and β3 to juxtapose αIIb(R995)-β3(D723). Since platelet membranes contain a lipid composition34 that would be difficult to completely reproduce in vitro, extensive mutagenesis data in mammalian cells were also useful in further confirming the asymmetry of the αIIbβ3 association encoded by the IMC.6,16,17,35
Subsequent to the publication of the αIIb and β3 TM structures,12,13 modeling predictions for the IMC improved conspicuously.17,36 A particularly exhaustive study was based on αIIb-β3 disulfide cross-linking efficiencies fed into an extension of the Rosetta sparse-data sampling approach.17 In this approach, the overall IMC arrangement was predicted with relative consistency, but with quite a considerable degree of heterogeneity, perhaps owing to the absence of any experimental restraints on its central αIIb(GFF) motif. Notably, out of 5,000 Rosetta solutions, only 52 structures were ultimately selected;17 for membrane protein modeling approaches to become independent tools of structure determination, a more robust convergence of structures appears to be desirable.
Mechanism of Bi-Directional Integrin αIIbβ3 Transmembrane Signaling
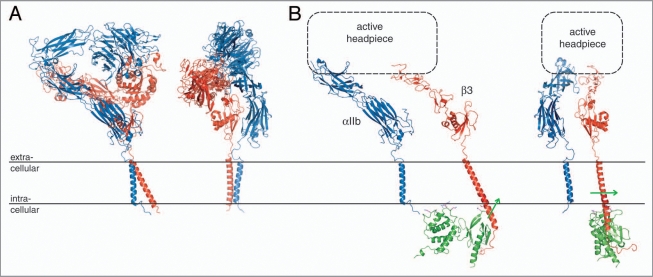
Just two short years ago, integrin transmembrane signaling had been a little understood, perhaps even misunderstood, area of integrin biology. In addition to the novel information regarding the TM segments discussed above, the recently determined structures of a resting integrin ectodomain, and of an activating talin/β cytosolic tail complex that overlap with the TM structures,20,21 have profoundly changed this scenario. The C-terminal extension of the original, monumental αVβ3 ectodomain crystal structure37 has provided an excellent fusion partner for an αVβ3 TM model based on the αIIbβ3 TM complex structure,16,20 thus yielding a model of a complete, resting integrin (Fig. 4A). Another recently determined ectodomain structure of integrin αIIbβ3,38 which succeeded the original αVβ3 structure by seven years, incorporates a non-physiological disulfide bond at the membrane interface. Based upon the ability of the ectodomain to stabilize the association of a TM complex that carries destabilizing mutations, a mutual thermodynamic coupling of the resting ectodomain conformation and TM complex has been demonstrated.16 The structures of the resting αVβ3 and αIIbβ3 ectodomains reveal a defined, well-packed membrane-proximal αβ stalk interface (Fig. 4A), which is further demonstrated by the findings of a thorough mutagenesis study39 that can serve to verify such a stabilization. Thus, outside-in signaling appears to be based on ectodomain rearrangements that disrupt the αβ stalk interface, which, in turn, destabilizes the TM complex.16 An ensuing dissociation of the TM complex and accompanying separation of the cytoplasmic tails may indeed occur.7,8 It is also noted that Mn2+-mediated activation does not require separation of the αβ TM and cytoplasmic segments.7,15
Figure 4.
Integrin bi-directional signaling. (A) Model of a resting integrin receptor. The αVβ3 ectodomain crystal structure was fused to an αVβ3 TM model based on the αIIbβ3 TM complex NMR structure.16,20 The α and β subunits are shown in blue and red, respectively. (B) Model of an active integrin conformation. The α(Calf-1/2) and β(TD/IE2-4) domains of the integrin ectodomain37 are shown dissociated with a schematic active headpiece. The dissociated β TM segment was overlaid with the activating talin2/β1D crystal structure (PDB entry 3g9w). Talin2 is shown in green and the direction of its effected β TM segment reorientation21 is indicated by green arrows. Two orientations, which are related by a rotation of ∼90° about the y axis, are shown for both models.
In contrast to the invariably symmetric arrangement of homodimeric TM complexes,40–43 the asymmetric nature of heterodimeric integrins allows for two distinct association motifs (OMC and IMC), perhaps reflective of their ability to signal outside-in as well as inside-out. Moreover, the extension of the IMC into the lipid headgroup region allows for direct structural interferences by ligands that are capable of integrin activation. Binding of the F3 domain from the talin N-terminal head domain to integrin β tails is sufficient for integrin activation44 (i.e., dissociation of the TM complex); however, additional talin domains contribute to activation.45 Detailed insight into talin-mediated activation has emerged from a recent talin2/β1D cytosolic tail complex structure and its biochemical characterization.21 In addition to direct competition for the conserved αIIb(R995)-β3(D723) salt bridge,6,21,46,47 a membrane orientation patch (MOP) on the F2 domain has been identified that can effect a reorientation of the β TM helix tilt as a consequence of talin interaction with the intracellular membrane surface (Fig. 4B). The altered helix tilt creates a mismatch between the orientation of αIIb and β3 found within their complex,21 thereby favoring the dissociated state. In turn, the dissociation of the TM complex destabilizes the membrane-proximal αβ stalk interface, thereby initiating a sequence of structural rearrangements that results in an active, adhesive receptor. There are two principal models for this rearrangement. The switchblade model suggests an extension of the extracellular domains,48 whereas, in the deadbolt model, the ectodomain retains a compact structure.49 Electron microscopy provides a range of compact and extended ectodomain structures,38 but techniques such as fluorescent lifetime imaging microscopy (FLIM) and cryoelectron tomography suggest the absence of ectodomain extension as a direct consequence of integrin activation.20,50
Outlook
Having established the structural basis of αIIbβ3 integrin TM signaling, a deeper understanding of integrin TM subunit associations in other integrin receptors is felt to be desirable. Promising work in this area has focused on integrin αLβ2,51,52 and an initial mutagenesis study16 suggests varying affinities between different subunit combinations. From a therapeutic standpoint, the design of peptides, or the discovery of small molecules that can compete for native αβ TM interactions and thereby modulate the receptor’s activation state,53 are of further interest.
Acknowledgements
I thank Amin Arnaout for providing the coordinates of the αVβ3-cTM model and Nick Anthis for providing the coordinates of the talin2/β1D complex. I am indebted to Michael Stallcup for his helpful comments and to Diana Gegala for critically reading the manuscript. T.S.U. gratefully acknowledges support from the American Heart Association, the National Institutes of Health (HL089726), and The John Douglas French Alzheimer’s Foundation.
Footnotes
Previously published online: www.landesbioscience.com/journals/celladhesion/article/10592
References
- 1.Hynes RO. Integrins: Bidirectional, allosteric signaling machines. Cell. 2002;110:673–687. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- 2.Askari JA, Buckley PA, Mould AP, Humphries MJ. Linking integrin conformation to function. J Cell Sci. 2009;122:165–170. doi: 10.1242/jcs.018556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harburger DS, Calderwood DA. Integrin signalling at a glance. J Cell Sci. 2009;122:159–163. doi: 10.1242/jcs.018093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ginsberg MH, Partridge A, Shattil SJ. Integrin regulation. Curr Opin Cell Biol. 2005;17:509–516. doi: 10.1016/j.ceb.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 5.Bennett JS, Berger BW, Billings PC. The structure and function of platelet integrins. J Thromb Haemost. 2009;7:200–205. doi: 10.1111/j.1538-7836.2009.03378.x. [DOI] [PubMed] [Google Scholar]
- 6.Hughes PE, DiazGonzalez F, Leong L, Wu CY, McDonald JA, Shattil SJ, et al. Breaking the integrin hinge-A defined structural constraint regulates integrin signaling. J Biol Chem. 1996;271:6571–6574. doi: 10.1074/jbc.271.12.6571. [DOI] [PubMed] [Google Scholar]
- 7.Kim M, Carman CV, Springer TA. Bidirectional transmembrane signaling by cytoplasmic domain separation in integrins. Science. 2003;301:1720–1725. doi: 10.1126/science.1084174. [DOI] [PubMed] [Google Scholar]
- 8.Zhu JQ, Carman CV, Kim M, Shimaoka M, Springer TA, Luo BH. Requirement of alpha and beta subunit transmembrane helix separation for integrin outside-in signaling. Blood. 2007;110:2475–2483. doi: 10.1182/blood-2007-03-080077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li W, Metcalf DG, Gorelik R, Li RH, Mitra N, Nanda V, et al. A push-pull mechanism for regulating integrin function. Proc Natl Acad Sci USA. 2005;102:1424–1429. doi: 10.1073/pnas.0409334102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Partridge AW, Liu SC, Kim S, Bowie JU, Ginsberg MH. Transmembrane domain helix packing stabilizes integrin aIIbbeta3 in the low affinity state. J Biol Chem. 2005;280:7294–7300. doi: 10.1074/jbc.M412701200. [DOI] [PubMed] [Google Scholar]
- 11.Otoole TE, Katagiri Y, Faull RJ, Peter K, Tamura R, Quaranta V, et al. Integrin cytoplasmic domains mediate inside-out signal-transduction. J Cell Biol. 1994;124:1047–1059. doi: 10.1083/jcb.124.6.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lau T-L, Dua V, Ulmer TS. Structure of the integrin alphaIIb transmembrane segment. J Biol Chem. 2008;283:16162–16168. doi: 10.1074/jbc.M801748200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lau T-L, Partridge AP, Ginsberg MH, Ulmer TS. Structure of the Integrin beta3 Transmembrane Segment in Phospholipid Bicelles and Detergent Micelles. Biochemistry. 2008;47:4008–4016. doi: 10.1021/bi800107a. [DOI] [PubMed] [Google Scholar]
- 14.Luo BH, Carman CV, Takagi J, Springer TA. Disrupting integrin transmembrane domain heterodimerization increases ligand binding affinity, not valency or clustering. Proc Natl Acad Sci USA. 2005;102:3679–3684. doi: 10.1073/pnas.0409440102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luo BH, Springer TA, Takagi J. A specific interface between integrin transmembrane helices and affinity for ligand. PLoS Biol. 2004;2:776–786. doi: 10.1371/journal.pbio.0020153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lau T-L, Kim C, Ginsberg MH, Ulmer TS. The structure of the integrin alphaIIbbeta3 transmembrane complex explains integrin transmembrane signalling. EMBO J. 2009;28:1351–1361. doi: 10.1038/emboj.2009.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu J, Luo BH, Barth P, Schonbrun J, Baker D, Springer TA. The structure of a receptor with two associating transmembrane domains on the cell surface: Integrin alpha(IIb)beta(3) Mol Cell. 2009;34:234–249. doi: 10.1016/j.molcel.2009.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stefansson A, Armulik A, Nilsson IM, von Heijne G, Johansson S. Determination of N- and C-terminal borders of the transmembrane domain of integrin subunits. J Biol Chem. 2004;279:21200–21205. doi: 10.1074/jbc.M400771200. [DOI] [PubMed] [Google Scholar]
- 19.Armulik A, Nilsson I, von Heijne G, Johansson S. Determination of the border between the transmembrane and cytoplasmic domains of human integrin subunits. J Biol Chem. 1999;274:37030–37034. doi: 10.1074/jbc.274.52.37030. [DOI] [PubMed] [Google Scholar]
- 20.Xiong JP, Mahalingham B, Alonso JL, Borrelli LA, Rui XL, Anand S, et al. Crystal structure of the complete integrin alphaVbeta3 ectodomain plus an alpha/beta. transmembrane fragment. J Cell Biol. 2009;186:589–600. doi: 10.1083/jcb.200905085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anthis NJ, Wegener KL, Ye F, Kim C, Goult BT, Lowe ED, et al. The structure of an integrin/talin complex reveals the basis of inside-out signal transduction. EMBO J. 2009;28:3623–3632. doi: 10.1038/emboj.2009.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gregoriou M, Noble MEM, Watson KA, Garman EF, Krulle TM, De la Fuente C, et al. The structure of a glycogen phosphorylase glucopyranose spirohydantoin complex at 1.8 angstrom resolution and 100 K: The role of the water structure and its contribution to binding. Protein Sci. 1998;7:915–927. doi: 10.1002/pro.5560070409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hunsicker-Wang LM, Pacoma RL, Chen Y, Fee JA, Stout CD. A novel cryoprotection scheme for enhancing the diffraction of crystals of recombinant cytochrome ba(3) oxidase from Thermus thermophilus. Acta Crystallogr Sect D-Biol Crystallogr. 2005;61:340–343. doi: 10.1107/S0907444904033906. [DOI] [PubMed] [Google Scholar]
- 24.Papiz MZ, Prince SM, Howard T, Cogdell RJ, Isaacs NW. The structure and thermal motion of the B800–850 LH2 complex from Rps. acidophila at 2.0 (A)over-circle resolution and 100 K: New structural features and functionally relevant motions. J Mol Biol. 2003;326:1523–1538. doi: 10.1016/s0022-2836(03)00024-x. [DOI] [PubMed] [Google Scholar]
- 25.Beel AJ, Mobley CK, Kim HJ, Tian F, Hadziselimovic A, Jap B, et al. Structural studies of the transmembrane C-terminal domain of the amyloid precursor protein (APP): Does APP function as a cholesterol sensor? Biochemistry. 2008;47:9428–9446. doi: 10.1021/bi800993c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee D, Walter KFA, Bruckner A-K, Hilty C, Becker S, Griesinger C. Bilayer in small bicelles revealed by lipid-protein interactions using NMR spectroscopy. J Am Chem Soc. 2008;130:13822–13823. doi: 10.1021/ja803686p. [DOI] [PubMed] [Google Scholar]
- 27.Hoefling M, Kessler H, Gottschalk KE. The Transmembrane Structure of integrin alphaIIbbeta3: Significance for signal transduction. Angew Chem-Int Edit. 2009;48:6590–6593. doi: 10.1002/anie.200902016. [DOI] [PubMed] [Google Scholar]
- 28.Gottschalk KE, Adams PD, Brunger AT, Kessler H. Transmembrane signal transduction of the alpha(IIb)beta(3) integrin. Protein Sci. 2002;11:1800–1812. doi: 10.1110/ps.4120102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin X, Tan SM, Law SKA, Torres J. Unambiguous prediction of human integrin transmembrane heterodimer interactions using only homologous sequences. Proteins. 2006;65:274–279. doi: 10.1002/prot.21072. [DOI] [PubMed] [Google Scholar]
- 30.Ulmer TS, Yaspan B, Ginsberg MH, Campbell ID. NMR analysis of structure and dynamics of the cytosolic tails of integrin alphaIIbbeta3 in aqueous solution. Biochemistry. 2001;40:7498–7508. doi: 10.1021/bi010338l. [DOI] [PubMed] [Google Scholar]
- 31.Vinogradova O, Velyvis A, Velyviene A, Hu B, Haas TA, Plow EF, et al. A structural mechanism of integrin alpha(IIb)beta(3) “inside-out” activation as regulated by its cytoplasmic face. Cell. 2002;110:587–597. doi: 10.1016/s0092-8674(02)00906-6. [DOI] [PubMed] [Google Scholar]
- 32.Weljie AM, Hwang PM, Vogel HJ. Solution structures of the cytoplasmic tail complex from platelet integrin alphaIIb- and beta3-subunits. Proc Natl Acad Sci USA. 2002;99:5878–5883. doi: 10.1073/pnas.092515799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang J, Ma YQ, Page RC, Misra S, Plow EF, Qin J. Structure of an integrin alphaIIbbeta3 transmembrane-cytoplasmic heterocomplex provides insight into integrin activation. Proc Natl Acad Sci USA. 2009;106:17729–17734. doi: 10.1073/pnas.0909589106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.García-Guerra R, García-Domínguez JA, González-Rodríguez J. A new look at the lipid composition of the plasma membrane of human blood platelets relative to the GPIIb/IIIa (integrin alphaIIbbeta3) content. Platelets. 1996;7:195–205. doi: 10.3109/09537109609023579. [DOI] [PubMed] [Google Scholar]
- 35.Kim C, Lau T-L, Ulmer TS, Ginsberg MH. Interactions of platelet integrin alphaIIb and beta3 transmembrane domains in mammalian cell membranes and their role in integrin activation. Blood. 2009;113:4747–5473. doi: 10.1182/blood-2008-10-186551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Metcalf DG, Kulp DW, Bennett JS, Degrado WF. Multiple approaches converge on the structure of the integrin alphaIIb/beta3 transmembrane heterodimer. J Mol Biol. 2009;392:1087–1101. doi: 10.1016/j.jmb.2009.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xiong JP, Stehle T, Diefenbach B, Zhang RG, Dunker R, Scott DL, et al. Crystal structure of the extracellular segment of integrin alphaVbeta3. Science. 2001;294:339–345. doi: 10.1126/science.1064535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu J, Luo BH, Xiao T, Zhang C, Nishida N, Springer TA. Structure of a complete integrin ectodomain in a physiologic resting state and activation and deactivation by applied forces. Mol Cell. 2008;32:849–861. doi: 10.1016/j.molcel.2008.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kamata T, Handa M, Sato Y, Ikeda Y, Aiso S. Membrane-proximal alpha/beta stalk interactions differentially regulate integrin activation. J Biol Chem. 2005;280:24775–24783. doi: 10.1074/jbc.M409548200. [DOI] [PubMed] [Google Scholar]
- 40.MacKenzie KR, Prestegard JH, Engelman DM. A transmembrane helix dimer: Structure and implications. Science. 1997;276:131–133. doi: 10.1126/science.276.5309.131. [DOI] [PubMed] [Google Scholar]
- 41.Call ME, Schnell JR, Xu CQ, Lutz RA, Chou JJ, Wucherpfennig KW. The structure of the zeta zeta transmembrane dimer reveals features essential for its assembly with the T cell receptor. Cell. 2006;127:355–368. doi: 10.1016/j.cell.2006.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bocharov EV, Mayzel ML, Volynsky PE, Goncharuk MV, Ermolyuk YS, Schulga AA, et al. Spatial Structure and pH-dependent Conformational Diversity of Dimeric Transmembrane Domain of the Receptor Tyrosine Kinase EphA1. J Biol Chem. 2008;283:29385–29395. doi: 10.1074/jbc.M803089200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bocharov EV, Pustovalova YE, Pavlov KV, Volynsky PE, Goncharuk MV, Ermolyuk YS, et al. Unique dimeric structure of BNip3 transmembrane domain suggests membrane permeabilization as a cell death trigger. J Biol Chem. 2007;282:16256–16266. doi: 10.1074/jbc.M701745200. [DOI] [PubMed] [Google Scholar]
- 44.Calderwood DA, Yan B, de Pereda JM, Alvarez BGYF, Liddington RC, et al. The Phosphotyrosine Binding-like Domain of Talin Activates Integrins. J Biol Chem. 2002;277:21749. doi: 10.1074/jbc.M111996200. [DOI] [PubMed] [Google Scholar]
- 45.Bouaouina M, Lad Y, Calderwood DA. The N-terminal domains of talin cooperate with the phosphotyrosine binding-like domain to activate beta1 and beta3 integrins. J Biol Chem. 2008;283:6118–6125. doi: 10.1074/jbc.M709527200. [DOI] [PubMed] [Google Scholar]
- 46.Imai Y, Park EJ, Peer D, Peixoto A, Cheng GY, von Andrian UH, et al. Genetic perturbation of the putative cytoplasmic membrane-proximal salt bridge aberrantly activates alpha(4) integrins. Blood. 2008;112:5007–15. doi: 10.1182/blood-2008-03-144543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ghevaert C, Salsmann A, Watkins NA, Schaffner-Reckinger E, Rankin A, Garner SF, et al. A nonsynonymous SNP in the ITGB3 gene disrupts the conserved membrane-proximal cytoplasmic salt bridge in the alpha(IIb)beta(3) integrin and cosegregates dominantly with abnormal proplatelet formation and macrothrombocytopenia. Blood. 2008;111:3407–14. doi: 10.1182/blood-2007-09-112615. [DOI] [PubMed] [Google Scholar]
- 48.Takagi J, Petre BM, Walz T, Springer TA. Global conformational rearrangements in integrin extracellular domains in outside-in and inside-out signaling. Cell. 2002;110:599–611. doi: 10.1016/s0092-8674(02)00935-2. [DOI] [PubMed] [Google Scholar]
- 49.Xiong JP, Stehle T, Goodman SL, Arnaout MA. New insights into the structural basis of integrin activation. Blood. 2003;102:1155–9. doi: 10.1182/blood-2003-01-0334. [DOI] [PubMed] [Google Scholar]
- 50.Ye F, Liu J, Winkler H, Taylor KA. Integrin alpha(IIb)beta(3) in a membrane environment remains the same height after Mn2+ activation when observed by cryoelectron tomography. J Mol Biol. 2008;378:976–986. doi: 10.1016/j.jmb.2008.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vararattanavech A, Lin X, Torres J, Tan SM. Disruption of the integrin alphaLbeta2 transmembrane domain interface by beta2 Thr-686 mutation activates alphaLbeta2 and promotes micro-clustering of the alphaL subunits. J Biol Chem. 2009;284:3239–3249. doi: 10.1074/jbc.M802782200. [DOI] [PubMed] [Google Scholar]
- 52.Vararattanavech A, Tang ML, Li HY, Wong CH, Law SKA, Torres J, et al. Permissive transmembrane helix heterodimerization is required for the expression of a functional integrin. Biochem J. 2008;410:495–502. doi: 10.1042/BJ20071218. [DOI] [PubMed] [Google Scholar]
- 53.Caputo GA, Litvinov RI, Li W, Bennett JS, DeGrado WF, Yin H. Computationally designed peptide inhibitors of protein-protein interactions in membranes. Biochemistry. 2008;47:8600–8606. doi: 10.1021/bi800687h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wiener MC, White SH. Structure Of A Fluid Dioleoylphosphatidylcholine Bilayer Determined By Joint Refinement Of X-Ray And Neutron-Diffraction Data.3. Complete Structure. Biophys J. 1992;61:434–447. doi: 10.1016/S0006-3495(92)81849-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhao W, Rog T, Gurtovenko AA, Vattulainen I, Karttunen M. Atomic-scale structure and electrostatics of anionic palmitoyloleoylphosphatidylglycerol lipid bilayers with Na+ counterions. Biophys J. 2007;92:1114–1124. doi: 10.1529/biophysj.106.086272. [DOI] [PMC free article] [PubMed] [Google Scholar]