Abstract
The transmembrane (TM) domains of receptor tyrosine kinases (RTKs) play an active role in signaling. They contribute to the stability of full-length receptor dimers and to maintaining a signaling-competent dimeric receptor conformation. In an exciting new development, two structures of RTK TM domains have been solved, a break-through achievement in the field. Here we review these structures, and we discuss recent studies of RTK TM domain dimerization energetics, possible synergies between domains, and the effects of pathogenic RTK TM mutations on structure and dimerization.
Key words: transmembrane domain, dimerization thermodynamics, receptor tyrosine kinases, pathogenic mutations, dimer structure
Introduction
RTKs are single-pass transmembrane (TM) proteins that play a critical role in cell growth, differentiation and motility. Their N-terminal extracellular domains, containing characteristic arrays of structural motifs, are involved in ligand (growth factor) binding. The single TM domain is followed by a juxtamembrane region and a catalytic domain, related to that of soluble tyrosine kinases.1–3
RTKs mediate signal transduction across the plasma membrane via lateral dimerization in the membrane plane. While monomers are inactive, dimeric RTKs have catalytic activity and cross-phosphorylate each other within the dimer. Phosphorylation of the catalytic domains in turn triggers signaling cascades.4–6 Thus, the dimerization of RTKs serves as a regulator of their activity. The ligand affects the monomer-dimer equilibrium, by stabilizing the dimeric state via a conformational change in the extracellular domain.7
The RTK family is subdivided into several sub-families, including the epidermal growth factor receptors (ErbBs), the fibroblast growth factor receptors (FGFRs), the insulin and the insulin-like growth factor receptors (IR and IGFR), the platelet-derived growth factor receptors (PDGFRs), the vascular endothelial growth factor receptors (VEGFRs), the hepatocyte growth factor receptors (HGFRs), the nerve growth factor receptors (NGFRs), and the erythropoietin-producing (Eph) receptors.8 While the Eph sub-family is the largest RTK family, the ErbB sub-family is the best characterized one. Indeed, many of the general principles of RTK signaling have emerged from studies of ErbBs.3
In recent years, a consensus has been emerging that RTK TM domains play an active role in signaling, with ample evidence coming from biophysical and cellular studies. In 2006, we reviewed research which demonstrates that RTK TM domains contribute to (1) stability of full-length dimers and to (2) maintaining a signaling-competent structure.9 These general concepts for the role of RTK TM domains in signaling have not changed, and new experimental evidence has emerged that further supports them. In this review, we discuss experimental results published since 2006.
Despite many studies of RTKs and their TM domains, the exact mechanism of signal transduction from the extracellular to the intracellular domains is unknown. One of the major experimental challenges in these studies is the expression of full-length RTKs in sufficient quantities for biophysical and structural characterization. While there have been some important advances in RTK overexpression, such as preparation of large quantities of pure EGFR,10 and long-term Neu/ErbB2 expression in engineered cells expressing anti-apoptotic proteins,11 biophysical and structural characterization of interactions is carried out predominantly with the isolated domains, either extracellular, catalytic or TM. Thus, knowledge about the synergy between these RTK domains in signaling is lacking, and it is not yet clear how exactly ligand binding and structural changes in the extracellular domains are coupled to phosphorylation in the catalytic domains. While the TM domains are expected to be critical in this process, most probably this issue will not be completely resolved until high resolution structures of full-length RTKs are available.
While solving structures of full-length RTK dimers remains a formidable challenge, high resolution structures of RTK TM domain dimers are now available. Since 2006, Bocharov and colleagues have determined two such structures, a break-through achievement in the field.
Structure of RTK TM Dimers
The first solved structure was the structure of the human ErbB2 TM domain dimer.12 The structure was solved in bicelles composed of dimyristoylphosphatidylcholine (DMPC) and dihexanoylphosphatidylcholine (DHPC). To address the effect of the bicelle environment, the authors performed circular dichroism measurements in the bicelles and in lipid vesicles and demonstrated that the secondary structure is the same. The peptide to lipid ratio in the bicelles was high, 1–35. At such high peptide concentrations, all proteins were in the dimeric state, making structure determination possible.
The ErbB2 TM dimer structure that Borcharov and his colleagues solved is believed to correspond to the active state of the dimer. The ErbB2 TM domain is believed to have two dimerization motifs, one close the N-terminus and one close to the C terminus, and is thus capable of forming active and inactive dimers and switching between the two conformations. The solved ErbB2 TM dimer ultilizes the N terminal dimerization motif, while the C terminus does not participate in the interface.
The ErbB2 TM domains are highly helical, and the helices pack into a symmetrical parallel dimer. The two helices cross at −42° angle, forming a right-handed dimer. The amino acids participating in the dimer interface are Thr652, Ser656 and Gly660. Molecular dynamics simulations suggest that transient hydrogen bonds may form between the side chains of Ser656 and Thr652. The authors emphasize that these are transient hydrogen bonds, such that the TM domain dimer can undergo a structural transition into a second, presumably inactive state, which uses the C-terminal dimerization motif.
There are two known pathogenic mutations in ErbB2 TM domain, and the solved ErbB2 dimer structure is consistent with current knowledge about these mutations. One of them, Ile665Val, is associated with an increased risk of cancer. The ErbB2 dimer structure suggests that this mutation can induce tighter dimer packing and overstabilize the mutant dimer. The second one is the oncogenic Val659Glu mutation, and the structure is consistent with the formation of stabilizing Glu-mediated hydrogen bonds that do not distort the active TM dimer structure.
ErbB2 TM domain and its pathogenic mutants have been researched intensively,13–17 and it is remarkable how well the structure explains the biochemical data and confirms the structural predictions. For instance, a prediction that a GxxxG-like motif would be a part of the dimer interface has been confirmed. One surprise is that the solved structure corresponds to the active dimeric state, suggesting that the active state is the more stable one. Previously, the active state was believed to be the less stable one, based on general arguments about signaling and on results from computational studies.18 It thus appears that computational methods are not always reliable for correctly predicting dimer stabilities. However, it is not yet clear if the active dimer structure is the stable one within the context of the full-length ErbB2 dimer. It is very likely that the catalytic, and especially the extracellular domains, also impact dimer stability.
The second structure solved by Bocharov and colleagues is the TM domain dimer of the erythropoietin-producing A1 2 (EphA1) receptor.19 The large Eph subfamily of RTKs is unique, with the active Eph dimer mediating communications between two cells and bridging two cellular membranes. Upon cell-cell contact, the extracellular domains of two Eph receptors bind two ligands that are tethered to the opposing membrane, to form the signaling complex. Eph signaling is critical during development and patterning. In adults, Eph signaling maintains its importance, playing a role in cell motility, wound healing and tumor metastasis.
The EphA1 TM dimer structure was solved in lipid bicelles, just as the ErbB2 TM dimer structure. Unlike ErbB2, however, different EphA1 TM dimer conformations were observed to coexist in the bicelles. The major conformation is a right-handed dimer with crossing angle of −44°. The crossing is close to the N-terminus, and contacts are mediated by Ala550, Gly554 and Gly558. While the high resolution structure of the second conformation could not be solved, the measured chemical shifts suggest that the dimer interface is likely composed of Leu557, Ala560, Gly564 and Val567. The minor conformation is thus expected to be a left-handed dimer with a crossing angle of about 30°. Bocharov and colleagues suggest a pivotal point close to residues Gly558, Ala559 and Ala560, around which a transition between the two conformations may occur. The transition between the two configurations may be facilitated by changes in the protonation state of Glu547, which is positioned close to the N-terminus. Indeed, the minor conformation is observed only at high pH when Glu547 is deprotonated. The pKa of Glu547 is shifted from the typical Glu value in solution, and is around five. The protonation leads to considerable local rearrangements, affecting the structure around residue 550 (helix melting). These structural changes are accompanied by increased water penetration inside the dimer interface in this region, and by slight bending of the helices in the region of the major dimerization motif. These structural changes are enough to destabilize the major TM dimer structure such that the minor one becomes observable too. Over-all, the EphA1 TM dimer structure supports the idea that RTK TM domains play an active dynamic role in signal transduction.
Bocharov and colleagues speculate that the major EphA1 TM dimer conformation is the active structure, because this structure is characterized by rigid TM helices that can effectively connect the intracellular and extracellular domains during signal transduction. EphA1 is expressed in epithelial tissues, where the pH is known to be as low as 4, and thus the authors further speculate that EphA1 is uniquely suited to transduce signals in this acid environment (the other Eph receptors do not have ionizable membrane embedded residues).
For years, interactions between RTK TM domains were believed to be very weak, a belief that was partially based on dimerization measurements in detergents.20,21 The weak interactions had raised concerns that structure determination for RTK TM dimers would be impossible. Now the structures of ErbB2 and EphA1 TM domain dimers refute these concerns, and we can look forward to more high resolution structures of RTK TM domains in the near future.
Thermodynamics of RTK TM Domain Dimerization
Along with structural studies, characterization of dimerization thermodynamics is crucial for understanding the role of RTK TM domains in signaling. The interactions between RTK TM domains are often studies in the inner E. coli membrane using genetic two-hybrid methods.16,17,22–24 These assays (ToxR, TOXCAT, GALLEX) measure the interaction of membrane spanning helices linking a periplasmic maltose binding protein (MBP) with a cytosolic DNA-binding domain that is activated upon dimerization. These assays can report on both homodimer and heterodimer stabilities. A method to measure thermodynamics of homo and heterodimerization of TM helices in lipid bilayers is FRET.25,26 Experimental details associated with such measurements were recently reviewed.27 The advantage of this method over the bacterial assays is that it can directly measure dimer fractions and yield dimerization free energies. So far, three different TM domains from three different RTK subfamilies have been characterized in terms of their homodimerization energetics in lipid bilayers, the TM domains of ErbB1, FGFR3 and EphA1.28–30 The strengths of interactions for these three RTK TM domains are very similar, about −3 kcal/mole.
A question of interest is whether the interactions between RTK TM domains are similar, or whether their strength varies within subfamilies. Varying interaction strengths may be a means to achieve specificity of response to particular ligands, as mediated by particular receptors. So far a hierarchy of dimerization strengths has been measured for the ErbB family of receptors in detergents.15 Detergent systems, however, are not optimal for characterization of RTK TM domain interactions,21,31,32 and thus we are looking forward to detailed characterization of homodimer and heterodimer RTK TM domain stabilities in bilayers and in E. coli membranes in the near future.
Pathogenic Mutations in RTK TM Domains
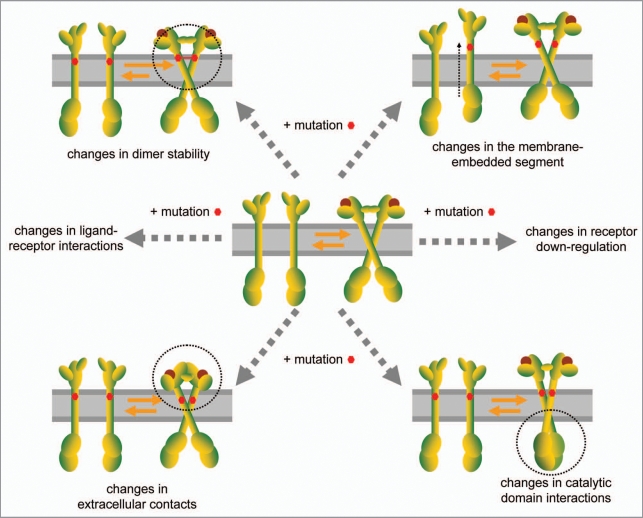
A list of known pathogenic mutations is given in reference 9, and no new mutations were identified during the past 3 years (to the best of our knowledge). Pathogenic TM domain mutations can affect RTK function in different ways (see Fig. 1). If the TM domain mutations induce structural changes that propagate to the other RTK domains, they can affect ligand binding, extracellular contacts, or the structure or orientation of the kinase domains. Furthermore, pathogenic mutations have been shown to interfere with the downregulation and internalization of the activated dimeric receptors. Examples, such as the effect of the achondroplasia Gly380Arg mutation on FGFR3 downregulation, were reviewed in reference 9. A more recent study addressed FGFR3 TM mutations to cysteine residues, Tyr373Cys, Ser371Cys and Gly370Cys, all linked to thanatophoric dysplasia I (TD I).33 These residues are most likely in the bilayer headgroup region of the membrane, and are sometimes assigned as part of FGFR3 extracellular domain. The mutations cause a downregulation defect, which is currently believed to be the major determinant of the pathology.33 In addition, mutations to Cys, such as the TD mutations described above and the rare Gly375Cys FGFR3 mutation identified in some achondroplasia cases, may promote the formation of disulfide bonds, which over-stabilize the FGFR3 dimer and cause unregulated signaling.34,35
Figure 1.
Pathogenic single amino acid mutations in RTK TM domains may affect signaling through different mechanisms.
Some RTK TM domain mutations impact the energetics of dimerization, stabilizing the active dimeric state of the receptors without introducing structural changes and affecting the soluble domains. Often these mutations substitute aliphatic amino acids that participate in the dimer interface with Glu or Asp, which have hydrogen bonding capabilities. One example is the Ala391Glu mutation in FGFR3, which has been identified as a somatic mutation in bladder cancer36 and as a germ-line mutation in Crouzon syndrome with acanthosis nigricans,37 an autosomal dominant disorder characterized by the following three phenotypic features: (1) mild disturbances of the growth plate of the long bones, (2) premature ossification of the skull (craniosynostosis) and (3) skin hyperpigmentation and hyperkeratosis. A second example is the Val664Glu mutation in rat Neu which has been shown to be oncogenic.38,39 The Val664Glu mutation in rat Neu corresponds to the Val659Glu mutation in human ErbB2, discussed above. Recently, the effects of the Ala391Glu and the Val664Glu mutations on receptor dimerization in mammalian cells were investigated using western blotting.40 In this study, the activation of Neu and a Neu chimera consisting of the extracellular and intracellular domains of Neu and FGFR3 TM domain was measured, and the effect of the mutations on dimerization was determined. The authors developed a quantitative physical-chemical description of receptor activation in terms of dimerization free energies, and generated mathematical predictions of active fractions as a function of receptor expression. The mathematical predictions were tested by comparing them to western blot measurements of active fractions of Neu and chimeric Neu_FGFR3 receptors in CHO cells. The predictions described the experimental data, yielding a quantitative measure of receptor over-activation due to the two studied mutations. In CHO cells, the Val664Glu mutation increased the Neu activation propensity by about −1.1 kcal/mole, while the increase due to the Ala391Glu mutation is about −0.7 kcal/mole. The measured effect of the order of −1 kcal/mole is similar to results obtained for a model system comprising the isolated FGFR3 TM domains in bilayers of different composition, demonstrating that the Ala391Glu mutation stabilizes the FGFR3 TM domain dimer by −1.3 ± 0.2 kcal/mole.41 In addition, computational and experimental studies of the structure of the isolated Neu/ErbB2 and FGFR3 TM domain dimers demonstrate dimer stabilization via Glu-mediated hydrogen bonding.9,12,14,41–43 Taken together, these results strongly suggest that hydrogen bonds stabilize the mutant Neu/Val664Glu and Neu_FGFR3/Ala391Glu receptors in mammalian cells, a mechanism that was originally proposed 18 years ago,42 but has been debated ever since. Furthermore, the results suggest that an increase of the order of −1 kcal/mole may be sufficient to transform normal RTK signaling processes into pathogenic processes.
The quantitative measurements of Neu and Neu_FGFR3 activation discussed above have provided new insights into the pathology mechanism due to the two investigated mutations. In the study, it was possible to directly link changes in dimerization to changes in activation, because no ligand is required for Neu activation. Neu/ErbB2 is the only RTK that does not have a ligand, and thus future physical-chemical models of RTK activation should account for ligand binding.
RTK TM domain mutations can also affect the positioning of RTK TM domains within the membrane, by inducing a shift in the membrane-embedded segment, as shown in reference 44, see Figure 1. In this paper the authors demonstrated a shift in the membrane-embedded segment of FGFR3 due to the Gly380Arg mutation, linked to achondroplasia, using a neutron diffraction technique that yields structural information in fluid lipid bilayers. The authors selectively deuterated amino acids in both wild-type and mutant FGFR3 TM domains and determined their depths of penetration in 1-palmytoyl-2-oleoylphospatidylcholine (POPC) bilayers. While Gly380 in wild-type is positioned at ∼6 Å from the bilayer center, the mutant Arg380 is at about 11 Å from the bilayer center. The observed shift in the membrane-embedded segment of FGFR3 may be the underlying cause for the compromised downregulation of FGFR3 in achondroplasia.
Juxtamembrane Domains
During the past three years, researchers have also investigated the role of RTK juxtamembrane (JM) domains in signaling. The JM domain connects the TM domain with the catalytic domain, and likely works synergistically with the TM domain in signal transduction.
The juxtamembrane domain is usually of 40–80 residues long, and contains several basic residues (Lys and Arg) located close to the membrane surface. Amino acids in this region have been shown to serve as binding and phosphorylation sites for signaling molecules.45–50 The importance of the JM domain in signaling is evident from several mutations, deletions and insertions in this domain that can lead to cancer.51–53 In addition, recent studies have provided evidence for the active role of the JM domains in regulating RTK activity.
While the JM domain in the insulin receptor is known as a binding site for regulatory proteins, the JM domains in most RTKs appear to also regulate kinase activity by functioning as an autoinhibitory segment.53 Biochemical studies of Eph, PDGF β, FLT3 and c-Kit receptors have shown that phosphorylation of Tyr residues in the JM domain is essential for ligand-dependent kinase activity, probably by releasing the inhibitory control exerted by the JM domain through contacts with the kinase domain.54–57
In ErbB1, however, the role of the JM domain is not autoinhibitory. On the contrary, the JM domain has been shown to be essential in the activation of the kinase.50 Recent studies have suggested that the role of the JM domain is to stabilize the activated asymmetric kinase dimer by forming an antiparallel dimer with the JM segment of the other receptor.58,59 Furthermore, quantitative studies of ligand binding as a function of receptor expression have shown that the binding affinity of the ligand to the extracellular domain is affected by the JM domain. Based on these observations, an “inside-out” signaling mechanism for ErbB1 was proposed.60 It can be expected that the TM domains play an active role in this process of “inside-out” signaling. Thus, it is preferable that future investigations of the role of RTK domains are not restricted to isolated RTK TM domains and utilize longer RTK constructs.
Conclusion and Outlook
As emphasized in our previous review,9 the TM domains play two crucial roles in RTK signaling, a thermodynamic and a structural role. In their thermodynamic role, RTK TM domains have a propensity to form sequence-specific dimers and thus they contribute to the over-all stability of the full-length RTK dimers. In their structural role, the TM dimers control the orientation of the catalytic domains and establish the signaling-competent conformation of the full-length dimeric receptors. Since 2006, these concepts about the role of the TM domains in signaling have not changed, and new data published since then have provided further support for this view. In the future, emphasis should be placed on investigations of different RTK families, and on identifying similarities and differences within families and between families. These studies should provide insights into the specificity of the response mediated by different RTKs.
One of the most exciting advancements during the past three years was the determination of two RTK TM domain dimer structures.12,19 We anticipate more such homodimer structures and we hope to see the first heterodimer structure in the near future. The role of heterodimerization within RTK subfamilies is now well established, and yet biophysical studies of heterodimers in membranes are rare.17 Quantitative measurements of TM heterodimerization strengths for different RTK subfamilies are on the wish list.
Studies during the past three years have suggested that there may be an intricate connection between the different RTK domains during signaling.60 Thus, in the future the role of RTK TM domains should be investigated within the context of full-length RTKs. So far, differences in TM domain strengths of homodimerization have been assessed for the full-length Neu receptors in mammalian cells,40 but the approach is yet to be applied to ligand-binding receptors.
While no new TM domain mutations have been reported during the past three years (to the best of our knowledge), the molecular mechanism of pathology due to some of the known mutations has been elucidated. Specifically, mutations in RTK TM domains have been shown to (1) stabilize full-length RTK dimers in the plasma membrane,40 (2) change the membrane-embedded segment,44 and (3) affect downregulation,33,61,62 see Figure 1. It can be expected that TM domain mutations affect signaling in multiple ways. For instance, structural changes occurring due to mutations in the TM domains may propagate to the extracellular and catalytic domains (Fig. 1). Such effects will be observable only for full-length receptors, and will likely be controversial until high-resolution structures of full-length wild-type and mutant receptor dimers are eventually solved.
Footnotes
Previously published online: www.landesbioscience.com/journals/celladhesion/article/10725
References
- 1.Fantl WJ, Johnson DE, Williams LT. Signaling by receptor tyrosine kinases. Annu Rev Biochem. 1993;62:453–481. doi: 10.1146/annurev.bi.62.070193.002321. [DOI] [PubMed] [Google Scholar]
- 2.L’Horte CGM, Knowles MA. Cell responses to FGFR3 signaling: growth, differentiation and apoptosis. Experim Cell Res. 2005;304:417–431. doi: 10.1016/j.yexcr.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 3.Linggi B, Carpenter G. ErbB receptors: new insights on mechanisms and biology. Trends Cell Biol. 2006;16:649–656. doi: 10.1016/j.tcb.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 4.Eswarakumar VP, Lax I, Schlessinger J. Cellular signaling by fibroblast growth factor receptors. Cytokine Growth Factor Rev. 2005;16:139–149. doi: 10.1016/j.cytogfr.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 5.Schlessinger J. Common and distinct elements in cellular signaling via EGF and FGF receptors. Science. 2004;306:1506–1507. doi: 10.1126/science.1105396. [DOI] [PubMed] [Google Scholar]
- 6.Zhang XW, Gureasko J, Shen K, Cole PA, Kuriyan J. An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell. 2006;125:1137–1149. doi: 10.1016/j.cell.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 7.Schlessinger J. Ligand-induced, receptor-mediated dimerization and activation of EGF receptor. Cell. 2002;110:669–672. doi: 10.1016/s0092-8674(02)00966-2. [DOI] [PubMed] [Google Scholar]
- 8.van der Geer P, Hunter T, Lindberg RA. Receptor protein-tyrosine kinases and their signal transduction pathways. Annu Rev Cell Biol. 1994;10:251–337. doi: 10.1146/annurev.cb.10.110194.001343. [DOI] [PubMed] [Google Scholar]
- 9.Li E, Hristova K. Role of receptor tyrosine kinase transmembrane domains in cell signaling and human pathologies. Biochemistry. 2006;45:6241–6251. doi: 10.1021/bi060609y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qiu C, Tarrant MK, Boronina T, Longo PA, Kavran JM, Cole RN, et al. In vitro enzymatic characterization of near full length EGFR in activated and inhibited states. Biochemistry. 2009;48:6624–6632. doi: 10.1021/bi900755n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O’Connor S, Li E, Majors BS, He L, Placone J, Baycin D, et al. Increased expression of the integral membrane protein ErbB2 in Chinese hamster ovary cells expressing the anti-apoptotic gene Bcl-xL. Protein Expr Purif. 2009;67:41–47. doi: 10.1016/j.pep.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bocharov EV, Mineev KS, Volynsky PE, Ermolyuk YS, Tkach EN, Sobol AG, et al. Spatial structure of the dimeric transmembrane domain of the growth factor receptor ErbB2 presumably corresponding to the receptor active state. J Biol Chem. 2008;283:6950–6956. doi: 10.1074/jbc.M709202200. [DOI] [PubMed] [Google Scholar]
- 13.Weiner DB, Liu J, Cohen JA, Williams WV, Greene MI. A point mutation in the Neu oncogene mimics ligand induction of receptor aggregation. Nature. 1989;339:230–231. doi: 10.1038/339230a0. [DOI] [PubMed] [Google Scholar]
- 14.Smith SO, Smith C, Shekar S, Peersen O, Ziliox M, Aimoto S. Transmembrane interactions in the activation of the Neu receptor tyrosine kinase. Biochemistry. 2002;41:9321–9332. doi: 10.1021/bi012117l. [DOI] [PubMed] [Google Scholar]
- 15.Duneau JP, Vegh AP, Sturgis JN. A dimerization hierarchy in the transmembrane domains of the HER receptor family. Biochemistry. 2007;46:2010–2019. doi: 10.1021/bi061436f. [DOI] [PubMed] [Google Scholar]
- 16.Mendrola JM, Berger MB, King MC, Lemmon MA. The single transmembrane domains of ErbB receptors self-associate in cell membranes. J Biol Chem. 2002;277:4704–4712. doi: 10.1074/jbc.M108681200. [DOI] [PubMed] [Google Scholar]
- 17.Escher C, Cymer F, Schneider D. Two GxxxG-like motifs facilitate promiscuous interactions of the human ErbB transmembrane domains. J Mol Biol. 2009;389:10–16. doi: 10.1016/j.jmb.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 18.Fleishman SJ, Schlessinger J, Ben-Tal N. A putative molecular-activation switch in the transmembrane domain of erbB2. Proc Nat Acad Sci USA. 2002;99:15937–15940. doi: 10.1073/pnas.252640799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bocharov EV, Mayzel ML, Volynsky PE, Goncharuk MV, Ermolyuk YS, Schulga AA, et al. Spatial structure and pH-dependent conformational diversity of dimeric transmembrane domain of the receptor tyrosine kinase EphA1. J Biol Chem. 2008;283:29385–29395. doi: 10.1074/jbc.M803089200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stanley AM, Fleming KG. The transmembrane domains of ErbB receptors do not dimerize strongly in micelles. J Mol Biol. 2005;347:759–772. doi: 10.1016/j.jmb.2005.01.059. [DOI] [PubMed] [Google Scholar]
- 21.Matthews EE, Zoonens M, Engelman DM. Dynamic helix interactions in transmembrane signaling. Cell. 2006;127:447–450. doi: 10.1016/j.cell.2006.10.016. [DOI] [PubMed] [Google Scholar]
- 22.Langosch D, Brosig B, Kolmar H, Fritz HJ. Dimerisation of the glycophorin a transmembrane segment in membranes probed with the ToxR transcription activator. J Mol Biol. 1996;263:525–530. doi: 10.1006/jmbi.1996.0595. [DOI] [PubMed] [Google Scholar]
- 23.Ruan WM, Becker V, Klingmuller U, Langosch D. The interface between self-assembling erythropoietin receptor transmembrane segments corresponds to a membrane-spanning leucine zipper. J Biol Chem. 2004;279:3273–3279. doi: 10.1074/jbc.M309311200. [DOI] [PubMed] [Google Scholar]
- 24.Gerber D, Sal-Man N, Shai Y. Two motifs within a transmembrane domain, one for homodimerization and the other for heterodimerization. J Biol Chem. 2004;279:21177–21182. doi: 10.1074/jbc.M400847200. [DOI] [PubMed] [Google Scholar]
- 25.You M, Li E, Wimley WC, Hristova K. FRET in liposomes: measurements of TM helix dimerization in the native bilayer environment. Anal Biochem. 2005;340:154–164. doi: 10.1016/j.ab.2005.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Merzlyakov M, You M, Li E, Hristova K. Transmembrane helix heterodimerization in lipids bilayers: probing the energetics behind autosomal dominant growth disorders. J Mol Biol. 2006;358:1–7. doi: 10.1016/j.jmb.2006.01.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Merzlyakov M, Hristova K. Forster resonance energy transfer measurements of transmembrane helix dimerization energetics. Methods Enzymol. 2008;450:107–127. doi: 10.1016/S0076-6879(08)03406-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li E, You M, Hristova K. SDS-PAGE and FRET suggest weak interactions between FGFR3 TM domains in the absence of extracellular domains and ligands. Biochemistry. 2005;44:352–360. doi: 10.1021/bi048480k. [DOI] [PubMed] [Google Scholar]
- 29.Chen L, Merzlyakov M, Cohen T, Shai Y, Hristova K. Energetics of ErbB1 transmembrane domain dimerization in lipid bilayers. Biophys J. 2009;96:4622–4630. doi: 10.1016/j.bpj.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Artemenko EO, Egorova NS, Arseniev AS, Feofanov AV. Transmembrane domain of EphA1 receptor forms dimers in membrane-like environment. Biochim Biophys Acta. 2008;1778:2361–2367. doi: 10.1016/j.bbamem.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 31.Soong R, Merzlyakov M, Hristova K. Hill coefficient analysis of transmembrane helix dimerization. J Membr Biol. 2009;230:49–55. doi: 10.1007/s00232-009-9185-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walkenhorst WF, Merzlyakov M, Hristova K, Wimley WC. Polar residues in transmembrane helices can decrease electrophoretic mobility in polyacrylamide gels without causing helix dimerization. Biochim Biophys Acta. 2009;1788:1321–1331. doi: 10.1016/j.bbamem.2009.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bonaventure J, Gibbs L, Horne WC, Baron R. The localization of FGFR3 mutations causing thanatophoric dysplasia type I differentially affects phosphorylation, processing and ubiquitylation of the receptor. FEBS J. 2007;274:3078–3093. doi: 10.1111/j.1742-4658.2007.05835.x. [DOI] [PubMed] [Google Scholar]
- 34.Adar R, Monsonego-Ornan E, David P, Yayon A. Differential activation of cysteine-substitution mutants of fibroblast growth factor receptor 3 is determined by cysteine localization. J Bone Miner Res. 2002;17:860–868. doi: 10.1359/jbmr.2002.17.5.860. [DOI] [PubMed] [Google Scholar]
- 35.You M, Spangler J, Li E, Han X, Ghosh P, Hristova K. Effect of pathogenic cysteine mutations on FGFR3 transmembrane domain dimerization in detergents and lipid bilayers. Biochemistry. 2007;46:11039–11046. doi: 10.1021/bi700986n. [DOI] [PubMed] [Google Scholar]
- 36.van Rhijin B, van Tilborg A, Lurkin I, Bonaventure J, de Vries A, Thiery JP, et al. Novel fibroblast growth factor receptor 3 (FGFR3) mutations in bladder cancer previously identified in non-lethal skeletal disorders. Eur J Hum Genet. 2002;10:819–824. doi: 10.1038/sj.ejhg.5200883. [DOI] [PubMed] [Google Scholar]
- 37.Meyers GA, Orlow SJ, Munro IR, Przylepa KA, Jabs EW. Fibroblast-growth-factor-receptor-3 (Fgfr3) transmembrane mutation in Crouzon-syndrome with acanthosis nigricans. Nat Genet. 1995;11:462–464. doi: 10.1038/ng1295-462. [DOI] [PubMed] [Google Scholar]
- 38.Schechter AL, Stern DF, Vaidyanathan L, Decker SJ, Drebin JA, Greene MI, et al. The Neu oncogene—an Erb-B-related gene encoding a 185,000-Mr tumor-antigen. Nature. 1984;312:513–516. doi: 10.1038/312513a0. [DOI] [PubMed] [Google Scholar]
- 39.Bargmann CI, Hung MC, Weinberg RA. Multiple independent activations of the Neu Ooncogene by a point mutation altering the transmembrane domain of P185. Cell. 1986;45:649–657. doi: 10.1016/0092-8674(86)90779-8. [DOI] [PubMed] [Google Scholar]
- 40.He L, Hristova K. Pathogenic activation of receptor tyrosine kinases in mammalian membranes. J Mol Biol. 2008;384:1130–1142. doi: 10.1016/j.jmb.2008.10.036. [DOI] [PubMed] [Google Scholar]
- 41.Li E, You M, Hristova K. FGFR3 dimer stabilization due to a single amino acid pathogenic mutation. J Mol Biol. 2006;356:600–612. doi: 10.1016/j.jmb.2005.11.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sternberg MJE, Gullick WJ. A sequence motif in the transmembrane region of growth factor receptors with tyrosine kinase activity mediates dimerization. Protein Eng. 1990;3:245–248. doi: 10.1093/protein/3.4.245. [DOI] [PubMed] [Google Scholar]
- 43.Smith SO, Smith CS, Bormann BJ. Strong hydrogen bonding interactions involving a buried glutamic acid in the transmembrane sequence of the neu/erbB-2 receptor. Nature Struct Biol. 1996;3:252–258. doi: 10.1038/nsb0396-252. [DOI] [PubMed] [Google Scholar]
- 44.Han X, Mihailescu M, Hristova K. Neutron diffraction studies of fluid bilayers with transmembrane proteins: Structural consequences of the achondroplasia mutation. Biophys J. 2006;91:3736–3747. doi: 10.1529/biophysj.106.092247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hunter T, Ling N, Cooper JA. Protein kinase-C phosphorylation of the Egf receptor at a threonine residue close to the cytoplasmic face of the plasma-membrane. Nature. 1984;311:480–483. doi: 10.1038/311480a0. [DOI] [PubMed] [Google Scholar]
- 46.Mori S, Ronnstrand L, Yokote K, Engstrom A, Courtneidge SA, Claessonwelsh L, et al. Identification of 2 Juxtamembrane Autophosphorylation sites in the Pdgf beta-receptor—involvement in the interaction with Src family tyrosine kinases. EMBO J. 1993;12:2257–2264. doi: 10.1002/j.1460-2075.1993.tb05879.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aifa S, Johansen K, Nilsson UK, Liedberg B, Lundstrom I, Svensson SPS. Interactions between the juxtamembrane domain of the EGFR and calmodulin measured by surface plasmon resonance. Cell Signal. 2002;14:1005–1013. doi: 10.1016/s0898-6568(02)00034-7. [DOI] [PubMed] [Google Scholar]
- 48.Meyer AN, Gastwirt RF, Schlaepfer DD, Donoghue DJ. The cytoplasmic tyrosine kinase Pyk2 as a novel effector of fibroblast growth factor receptor 3 activation. J Biol Chem. 2004;279:28450–28457. doi: 10.1074/jbc.M403335200. [DOI] [PubMed] [Google Scholar]
- 49.Sato T, Pallavi P, Golebiewska U, McLaughlin S, Smith SO. Structure of the membrane reconstituted transmembrane-juxtamembrane peptide EGFR622–660 and its interaction with Ca2+/calmodulin. Biochemistry. 2006;45:12704–12714. doi: 10.1021/bi061264m. [DOI] [PubMed] [Google Scholar]
- 50.Thiel KW, Carpenter G. Epidermal growth factor receptor juxtamembrane region regulates allosteric tyrosine kinase activation. Proc Nat Acad Sci USA. 2007;104:19238–19243. doi: 10.1073/pnas.0703854104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Casteran N, De Sepulveda P, Beslu N, Aoubala M, Letard S, Lecocq E, et al. Signal transduction by several KIT juxtamembrane domain mutations. Oncogene. 2003;22:4710–4722. doi: 10.1038/sj.onc.1206587. [DOI] [PubMed] [Google Scholar]
- 52.Lennartsson J, Jelacic T, Linnekin D, Shivakrupa R. Normal and oncogenic forms of the receptor tyrosine kinase kit. Stem Cells. 2005;23:16–43. doi: 10.1634/stemcells.2004-0117. [DOI] [PubMed] [Google Scholar]
- 53.Hubbard SR. Juxtamembrane autoinhibition in receptor tyrosine kinases. Nat Rev Mol Cell Biol. 2004;5:464–470. doi: 10.1038/nrm1399. [DOI] [PubMed] [Google Scholar]
- 54.Griffith J, Black J, Faerman C, Swenson L, Wynn M, Lu F, et al. The structural basis for autoinhibition of FLT3 by the juxtamembrane domain. Mol Cell. 2004;13:169–178. doi: 10.1016/s1097-2765(03)00505-7. [DOI] [PubMed] [Google Scholar]
- 55.Binns KL, Taylor PP, Sicheri F, Pawson T, Holland SJ. Phosphorylation of tyrosine residues in the kinase domain and juxtamembrane region regulates the biological and catalytic activities of eph receptors. Mol Cell Biol. 2000;20:4791–4805. doi: 10.1128/mcb.20.13.4791-4805.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ma YS, Cunningham ME, Wang XM, Ghosh I, Regan L, Longley BJ. Inhibition of spontaneous receptor phosphorylation by residues in a putative alpha-helix in the KIT intracellular juxtamembrane region. J Biol Chem. 1999;274:13399–13402. doi: 10.1074/jbc.274.19.13399. [DOI] [PubMed] [Google Scholar]
- 57.Baxter RM, Secrist JP, Vaillancourt RR, Kazlauskas A. Full activation of the platelet-derived growth factor beta-receptor kinase involves multiple events. J Biol Chem. 1998;273:17050–17055. doi: 10.1074/jbc.273.27.17050. [DOI] [PubMed] [Google Scholar]
- 58.Brewer MR, Choi SH, Alvarado D, Moravcevic K, Pozzi A, Lemmon MA, et al. The juxtamembrane region of the EGF receptor functions as an activation domain. Mol Cell. 2009;34:641–651. doi: 10.1016/j.molcel.2009.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jura N, Endres NF, Engel K, Deindl S, Das R, Lamers MH, et al. Mechanism for activation of the EGF receptor catalytic domain by the juxtamembrane segment. Cell. 2009;137:1293–1307. doi: 10.1016/j.cell.2009.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Macdonald-Obermann JL, Pike LJ. The intracellular juxtamembrane domain of the epidermal growth factor (EGF) receptor is responsible for the allosteric regulation of EGF binding. J Biol Chem. 2009;284:13570–13576. doi: 10.1074/jbc.M109.001487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Monsonego-Ornan E, Adar R, Rom E, Yayon A. FGF receptors ubiquitylation: dependence on tyrosine kinase activity and role in downregulation. FEBS Lett. 2002;528:83–89. doi: 10.1016/s0014-5793(02)03255-6. [DOI] [PubMed] [Google Scholar]
- 62.Cho JY, Guo CS, Torello M, Lunstrum GP, Iwata T, Deng CX, et al. Defective lysosomal targeting of activated fibroblast growth factor receptor 3 in achondroplasia. Proc Nat Acad Sci USA. 2004;101:609–614. doi: 10.1073/pnas.2237184100. [DOI] [PMC free article] [PubMed] [Google Scholar]