Abstract
The interaction between transmembrane helices is of great interest because it directly determines biological activity of a membrane protein. Either destroying or enhancing such interactions can result in many diseases related to dysfunction of different tissues in human body. One much studied form of membrane proteins known as bitopic protein is a dimer containing two membrane-spanning helices associating laterally. Establishing structure-function relationship as well as rational design of new types of drugs targeting membrane proteins requires precise structural information about this class of objects. At present time, to investigate spatial structure and internal dynamics of such transmembrane helical dimers, several strategies were developed based mainly on a combination of NMR spectroscopy, optical spectroscopy, protein engineering and molecular modeling. These approaches were successfully applied to homo- and heterodimeric transmembrane fragments of several bitopic proteins, which play important roles in normal and in pathological conditions of human organism.
Key words: bitopic proteins, transmembrane domain dimer, spatial structure, dynamics, protein-protein interactions, protein-membrane interactions, molecular modeling, NMR
Introduction
Helical membrane proteins are a major class of membrane proteins that are essentially involved in key processes including bioenergetics, signal transduction, ion transmission, catalysis and so on.1 This class of proteins is characterized by the presence of highly hydrophobic stretches of ∼20 amino acids, which span the membrane in an α-helical conformation. Helical membrane proteins can exist as simple structures, with just one or a few TM helices spanning a membrane, as well as large oligomeric complexes with many TM helices. The mechanisms by which helical membrane proteins fold into native structures are beginning to be understood from a confluence of structural and biochemical studies. The fold of a helical membrane protein is largely determined by interactions between membrane-embedded helices. Folding determinants of a membrane protein can be partially understood by dissecting its structure into pairs of interacting helices, which, together with the connecting loops and extramembrane domains, comprise the overall structure. The interaction between TM helices is of a great interest because it directly determines biological activity of a membrane protein, such as ionic channels, G-protein coupled receptors, receptor protein kinases, immune receptors and apoptotic proteins. Either destroying or enhancing such interactions can result in many diseases (developmental, oncogenic, neurodegenerative, immune, cardiovascular and so on) related to dysfunction of different tissues in the human body. Nevertheless, in spite of their significance, only few tens of spatial structures of TM membrane proteins have been obtained so far, while design of new types of drugs targeting membrane proteins requires precise structural information about this class of objects.
Bitopic proteins having only one α-helical TM domain are a class of biologically significant membrane proteins that are the most convenient to study by structural methods. Activity regulation of such proteins is mostly associated with their lateral dimerization in cell membranes. The class includes the majority of receptor kinases and immune receptors, which play a central role in regulating the cell growth, proliferation, differentiation, adhesion, migration and lifespan in the body, thus being directly involved in regulating the development and homeostasis of all tissues in both health and pathology. It is worth mentioning that at the present stage of development of the structural biology methods, obtaining high-resolution structure of full-length receptor kinases and immune receptors is a scientific challenge, and no known recipe exists for solving the problem. X-ray experiences problems with crystallization of membrane proteins and NMR cannot sufficiently deal with high weight and low mobility of large protein-lipid complexes. The crystallographic methods, which recently allowed obtaining high-resolution structure of multi-span transmembrane receptors like G-protein coupled receptors,2 cannot be directly translated to multiple-domains flexible receptors like receptor kinases and immune receptors. Therefore, the structural-dynamic properties of the extracellular, cytoplasmic and membrane parts of such bitopic proteins are still studied separately.
Homo- and heterodimerization of bitopic proteins was earlier thought to involve mostly their extracellular and cytoplasmic domains, but recent studies have been making it increasingly clear that the single-spanning TM domains are also critical for their dimerization and modulation of biological function. Upon bitopic protein activation, ligand-dependent or not, significant intramolecular conformational transitions result in rearrangement of the receptor domains and following receptor dimerization or switching from one dimerization state to another, e.g., ligand-dependent transition from preformed inactive dimeric state into active dimer of ErbB receptor tyrosine kinase.3–6 The so-called “rotation-coupled” and “flexible rotation” activation mechanisms,4–6 which were initially proposed for receptor tyrosine kinases and imply active involvement of TM domains in dimerization and activation of the receptors via proper TM helix-helix packing and rearranging, are possibly widespread among bitopic proteins. However, if biological functions are carried out using only one homo- or heterodimeric state of bitopic protein TM domains, the TM helix-helix interaction can be strong, as in the case of permeabilization of the outer mitochondrial membrane by proapoptotic protein BNip3 in the course of hypoxia-acidosis induced cell death. Furthermore, amino acid polymorphisms and mutations in the TM domain of bitopic proteins have been implicated in numerous human pathological states, including many types of cancers, Alzheimer disease, tissue dysplasias and abnormalities.7,8 It was shown that the mutations affect both the behavior of the isolated transmembrane domains in model lipid bilayers, and the behavior of the full length receptors in the plasma membrane. Most probably, the effects are exerted via yet unknown mutation-induced changes in dimeric structure of the TM domains. Importantly, it was found that isolated TM domains revealed ability not only to homo- and heterodimerize in membrane-like environment, but also to specifically inhibit biological activity of bitopic proteins in cell membrane.7,9,10 So, membrane-spanning segments of bitopic proteins represent a novel class of pharmacologically important targets, whose activity can be modulated by natural or specially designed molecules. Among the most perspective candidates for these purposes are artificial hydrophobic helical peptides, the so-called peptide “interceptors” or “computer helical antimembrane proteins” (CHAMPs),9,11 which are capable of specifically recognizing the target wild-type TM segments of bitopic proteins and interfering with their lateral association in cell membrane. Therefore, understanding of the factors that drive packing of α-helices in membranes has attracted considerable interest of researchers from both scientific and medical communities.
Structural-dynamic information about non-covalently bonded protein oligomers in the membrane is very challenging to obtain. Up to date, there are just a few experimentally solved dimeric structures of the TM domains of bitopic proteins. Several strategies to resolve this problem by various theoretical and physicochemical methods and their combination are currently available, thus providing structural-dynamic information about atomic-scale details of TM helix-helix and helix-membrane interactions. Although the primary objective is structure elucidation, experimental high-resolution structure obtained in a particular membrane mimicking environment usually corresponds to only one of homo- or heterodimeric states of TM domains, which are apparently realized in vivo in the course of bitopic protein activity. ADHESION Molecular modeling, in its turn, allows predicting all possible alternative dimerization interfaces of the bitopic protein TM domains, existence of which in vivo should be verified in experiment. Apparently, thorough understanding of all the aspects of TM helix-helix interaction can only be achieved with multi-disciplinary approach based on a comprehensive set of modeling, biochemical and biophysical tools. This review will discuss the applicable methods, from purely theoretical approaches to direct experimental techniques, which recently allowed describing high-resolution dimeric TM domain structure for several bitopic proteins and understanding some aspects of structure-function relations and their biological activity.
Driving and Stabilizing Forces of TM Helix-Helix Association
Before discussing various approaches to structure investigations, the forces driving and stabilizing TM helix-helix association should be addressed. The folding of an α-helical membrane protein can be conceptualized as a process that occurs in two thermodynamically distinct steps, involving the formation of independently stable TM helices and the subsequent specific TM helix-helix interactions giving rise to higher-order structures,12 in which TM helices are usually more or less tilted with respect to the membrane plane. The mechanism of TM helix association in membrane proteins is clearly different from that of soluble proteins where hydrophobic effect is a dominant driving force for protein folding. The hydrophobic effect vanishes once the helices are inserted in the lipid bilayer and the hydrophobic side chains get exposed to hydrophobic environment. Nevertheless, lipid-protein interactions are most likely indirectly involved in driving the association of TM helices as the entropy term.13,14 While the formation of higher ordered helix oligomers decreases the entropy of the proteins, the entropy of the lipids is greatly increased. After interaction of individual TM helices, a part of “frozen” lipids closely associated with the helices or situated in their immediate neighborhood is released into the membrane lipid pool.15 Therefore, TM helix oligomerization would minimize the protein-lipid interface and thereby increase the entropy of the system, thus contributing to stabilization of membrane proteins. In addition, local lipid composition of the membrane and matching the hydrophobic thickness of lipid bilayer with the hydrophobic length of TM proteins can regulate lipid-protein and protein-protein interactions, e.g., resulting in lipid-mediated protein-protein lateral association into signaling platforms in biomembranes.16–20 Moreover, specific helix-helix interactions require precise mutual orientation of TM helices, imposing certain restrictions on their tilt angle and tilt direction between dimer axis and normal to the membrane, therefore proper hydrophobic matching may influence the specific TM domain association, sorting different active states of dimeric bitopic proteins between lipid phases and microdomains of cell membrane.17–19 Even when helices do not exhibit any tendency for specific association, helix-helix association could still occur as a result of poor packing between the lipids and helices, or from a favorable change in entropy resulting from the release of helix-bound lipids upon helix association.16,17 In these cases, helix association is primarily driven by lipid-protein interactions rather than strongly favorable protein-protein interactions. However, while entropy considerations and hydrophobic matching or mismatching could partly explain the formation of higher ordered TM structures in the membrane, it does not explain the specificity of TM helix interactions.
Other noncovalent forces involved in the formation of TM helix oligomers besides the protein-lipid interactions include van-der-Waals and polar contacts.21,22 Interaction of TM helices often follows a “ridge-into-groove” or a “knob-into-hole” packing.23,24 The ridges or knobs on the surface of one TM helix fit well into grooves or holes on the adjacent helical surface. This geometrical smooth fit allows a very close contact between adjacent TM helices and, as a result, it promotes stabilizing van-der-Waals interactions. In the same time, electrostatic interactions could play a major role in membrane protein folding,25–29 since the strength of such interactions increases with a decreasing dielectric constant of the environment. Electrostatic interactions stabilized folded membrane structures via polar backbone-backbone, backbone-side chain, or side chain-side chain interactions resulting in hydrogen bond formation between adjacent TM helices. Contribution of amino acid residues into interaction energy in the hydrophobic environment is a function of their polarity. Weakly polar amino acids, like glycine, alanine, serine and threonine are characterized by a relatively small electrostatic component of the interaction energy and a complex nature of interaction. In addition to the forming electrostatic interactions, these polar residues with small side chains do also allow two TM helices to come into a close contact and to tightly pack without significant side chain rotamer entropy loss upon dimer formation.30 This does not only facilitate the interhelical hydrogen bonding with participation of polar side chains of serine or threonine, but also allows van-der-Waals interactions between surrounding residues. In addition to polar side chains, the CαH groups of such tightly packed residues are capable of participating in non-canonical hydrogen bonding, e.g., with the opposite carbonyl groups across the helix-helix interface.31 In other words, the marginal polarity of the Cα proton might be sufficient to serve as a hydrogen bond donor in a highly hydrophobic environment. However, although the slightly polar residues could form hydrogen bonds with an adjacent TM helix, they are able to contribute significantly to the specific helix-helix interactions only consisting in an amino acid context, which promotes association of TM helices, for example, by proper packing.28,32–35
Highly polar residues, like histidine, asparagine, aspartic acid, glutamine, glutamic acid, arginine or lysine, can apparently drive noncovalent association of TM helices through strong hydrogen bonding and salt-bridge formation, resulting in very stable helix oligomers. These residues are rarely found in membrane proteins,36 but it has been shown that the presence of a single asparagine, aspartic acid, glutamine or glutamic acid in a TM helix is sufficient to drive stable oligomerization.25,28,29 While highly polar residues can contribute significantly to the stability of the helix-helix interaction, several problems arise when these residues are present in a membrane. Transfer of highly polar residues into a membrane is thermodynamically unfavorable, and only very few of these residues can be tolerated in a single TM helix. Furthermore, in membrane environment, the ionizable side chains of these residues prefer uncharged state and their pKa values can be varied substantially depending on numerous parameters, such as local hydrogen bond network, membrane composition, transmembrane potential, and juxtamembrane environment.37,38 Since highly polar residues could interact with any potential binding partner for hydrogen bonding or salt-bridge formation, which create the danger of non-specific helix-helix association and misfolding,14 the polar substitutions are apparently the most common pathogenic mutations in membrane proteins that cause different human diseases.7,39 On the other hand, for the polar residues located at the level of the lipid headgroups where solubility of charged groups is higher than in the hydrophobic core but the electrostatic shielding is accordingly more effective, the individual interactions are not so formidable and can be modulated by external ligands.40 In addition, arginine and lysine residues are frequently found at the ends of TM helices, where they have a tendency to participate in direct or water-mediated polar-polar interactions with phospholipid headgroups and can modulate the strength of helix-helix dimerization.36,41–43
Other important players participating in TM helix association are π-π and cation-π aromatic interactions, arising either between two aromatic residues or between a basic and an aromatic residue, respectively.44–46 Aromatic rings of tryptophan, phenylalanine, tyrosine and histidine residues and their self-association or interaction with protonated cationic side chains of arginine, lysine and histidine residues have been proposed to consist of van-der-Waals and electrostatic forces contributed by correct packing geometry and interactions with the aromatic ring quadrupole moment. Besides, the indole, phenol and imidazole group of the aromatic residues can participate in hydrogen bonding across TM helix packing interface. Even though weak, CαH-π interactions enhanced in the low dielectric membrane environment can be considered as additional interactions supporting specific TM helix association.45 In addition, aromatic residues have a strong propensity to face phospholipids in the headgroup region and are thought to act as anchors for a membrane protein, influencing the helix tilting and hydrophobic matching in the membrane.42
The TM helix-helix interactions can be roughly grouped on the basis of sequence patterning and interhelical geometry. Since N- and C-termini of α-helical TM domains of bitopic proteins are usually exposed to extracellular and cytoplasmic sides of membrane respectively, such proteins specifically associate into homo- and heterodimers in a parallel manner, in the so-called “head-to-head” orientation. Both right- and left-handed variants of parallel helix-helix dimers with most frequently occurring helix-helix crossing angles near −40° and 20°, respectively, and the distance of 0.7–0.9 nm between helix axes appear to be quite common for TM helix packing in membrane.47 The interfaces of TM helices crossing at negative angles appear to conform to [abcd]n tetrad repeats, in which a and b correspond to interfacial residues.48 Right-handed packing of helix pairs is most often characterized by an i, i + 4 separation of “small” residues, such as glycine, alanine, serine and threonine, along the TM sequence, which alternately termed the small-xxx-small or GG4-like motif firstly exemplified by self-assembling TM domain of glycophorin A.30 In the GG4-like motif, the small residues create a shallow weakly polar groove that complements the surface of an adjacent helix and allows close approach of the helices. The association is stabilized by van-der-Waals contacts resulting from the excellent geometric fit and weak polar interactions, which can contribute to non-canonical hydrogen bonding between CαH and carbonyl groups across helix-helix interface.31 Two GG4-like motifs often follow in tandem, forming the so-called “glycine zipper” motif, which is statistically overrepresented in membrane proteins.49 Positive crossing angles characteristic of left-handed pairs of TM helices result from regular interdigitation of side-chains at a and d positions of an [abcdefg]n heptad repeat motif, whereas e and g positions are located at the periphery of these helix-helix interfaces.23 This heptad pattern was originally identified in water soluble “leucine zipper” interaction domains and gives rise to “knobs-into-holes” packing of side-chains.50 The left-handed TM helix pairings are mostly stabilized along heptad repeats by van-der-Waals contacts of large side chains of valine, leucine and isoleucine residues, while slightly polar interactions of interfacial residues having small side chains, like glycine, alanine and serine, are also important factor for left-handed oligomerization.51–53 In addition, the TM helix-helix dimerization via employment of both tetrad and heptad repeat motifs can be enhanced by π-π, cation-π and CαH-π aromatic interactions across helix packing interface.44,45 Furthermore, interhelical hydrogen bonding with participation of polar residues can work in concert with other helix packing interactions to strongly stabilize both right- and left-handed motifs, which appear to be essential for proper alignment of the polar side chains required for formation of hydrogen bonds.39
In conclusion, TM helix interactions are mostly driven and stabilized by close packing and polar interactions/hydrogen bonding as well as interactions of the helices with the membrane environment. How these forces work together to guarantee specificity and stability of helix-helix interactions is not clear yet and the interplay has to be analyzed in more details in each case. Currently, many unique sequence motifs that are responsible for specific helix-helix association have been identified on the basis of tetrad and heptad repeats, which play primarily a permissive role for close helix-helix interactions (reviewed in refs. 39, 47, 54 and 55). The relative importance of the sequence motifs in stabilizing helix-helix interactions depends on both specific residue content and location of the interactive surfaces relative to the N- and C-termini of α-helical TM segments.56 Besides, the affinity of TM helix association can be modulated by flanking and noninterfacial residues.57 From one to several potential dimerization motifs can be usually identified in each TM region of bitopic proteins which participate in two broad categories of helix-helix interactions.39 In the first, the TM domains form relatively static contacts that are necessary e.g., for the assembly of a functional protein complex or for proper folding and export from endoplasmic reticulum. In the second, the TM domains undergo dynamic conformational changes between alternative dimerization modes important e.g., for signaling process that can involve a change in association state and/or lateral, vertical and rotational motions in the membrane. Most likely, such switchable helix-helix interactions between TM domains do not provide the dominant force regulating protein-protein interactions, but rather fine-tune the system energetics, provide leverage for transmembrane coupling and impose certain restrictions on the allowable conformational transitions undergone by the full length bitopic proteins accomplishing their biological activity.
Molecular Modeling Methods of Predicting Spatial Structure of Dimeric TM Helices
Because of relative simplicity and stability, homo- and heterodimers of TM domains of bitopic proteins represent attractive objects for the development of computational techniques to assess helix-helix interactions in membranes. Methods of molecular modeling provide a reasonably quick and efficient tool for quantitative assessment the mode of helix association in membranes, especially when direct structural methods fail or are prohibitively resource-consuming. In spite of a limited number of experimental spatial structures of TM helical dimers, molecular modeling techniques can already provide quite reasonable atomic-scale models of dimeric structures. In silico approaches can be subdivided into three major categories: molecular docking, Monte Carlo and molecular dynamics simulations. Another important point, which plays a key role in molecular modeling of TM protein-protein interactions, is representation of the membrane. There are three generic ways to take into account the membrane influence upon the membrane protein simulations. In the simplest approach, effect of heterogeneous membrane environment is represented implicitly—in terms of some potential of mean force. Usually, this is done by adding of special terms to the potential energy function of a protein.58 This is a wide group of the so-called implicit or “hydrophobic slab” membrane models.58,59 Though this kind of representation can not provide atomistic details of protein-membrane interactions, it adequately mimics the basic membrane properties, such as membrane transversal hydrophobicity, thickness, curvature and transmembrane voltage. Also, these approaches are quite computationally effective and allow fast sampling of the protein configurational space, thus resulting in reasonable guesses about principal trends of protein behavior in membrane (spatial structure in the membrane-bound state, geometry of binding, etc.,). The second group of modeling techniques employs explicit membrane representation. The simulations are carried out for full-atom hydrated lipid bilayers or detergent micelles with imposed periodic boundary conditions.60 Here, protein-membrane interactions are considered in detail on the atomic level. This class of models is capable of providing the most reliable dimeric structures of TM peptides. Unfortunately, due to large size of the systems (up to 106 particles), such calculations are very time- and resource-consuming. Finally, the third class of membrane models, so-called “coarse-grain” (CG) models, was introduced to avoid excessive computational cost without significantly compromising of atomistic detail and attain the required accuracy without loss of model manageability where it cannot be done with the implicit or explicit representation.61 In CG-models, standard groups of atoms are replaced with “grains,” thus reducing considerably the number of variable degrees of freedom in the protein-membrane systems. Below, we present application of the first two groups of methods to study 3D structure of TM helical dimers.
A group of docking techniques is intended for fast identification of homo- and heterodimeric states of bitopic protein TM domains based on their amino acid sequence.62 Usually, one of the TM monomers is considered as a target, and the other as a ligand, the conformational lability being limited for one or both of the monomers defined with the parameters of the backbone and side chains typical for α-helical TM segments. The membrane is either ignored or modeled implicitly. This method allows quick scanning for spatially complementary surfaces with optimally matched geometrical, hydrophobic/hydrophilic, and electrostatic properties of the interacting TM helices, and thus predicts potential dimerization interfaces and intermonomer hydrogen bonds. However, due to restrictions imposed on the TM helix mobilities and due to the fact that many physical factors of protein-protein and protein-lipid interactions being ignored, docking methods are typically used only for initial characterization of specific helix-helix packing, subsequently investigated with the aid of other methods.
In the group of methods based on Monte Carlo conformational search, both monomers are flexible that permits more careful scanning of the conformational space and thus potentially results in higher resolution of the calculated structures. Moreover, these approaches allow the membrane to be more accurately taken into account, using either implicit or explicit representation. Implicit membrane representation, due to its lower computational cost, results in a more extensive scanning of the conformational space and therefore decreases the probability to miss a realistic helix-helix configuration. On the other hand, explicit membrane models provide more adequate ranking of the predicted dimeric structures. For acceleration of the Monte Carlo conformational search, it was often assumed a priori that the TM helices adopt a proper TM orientation and their backbones were considered “rigid,” and hence, common occurrence of local distortions in TM helices, like kinks and bends, was not taken into account. Under such assumptions, the effects of membrane media on the secondary structure formation and/or stabilization, along with the events accompanying insertion of the peptides, also can not be assessed. However, Monte Carlo protocols without imposing any restraints on the secondary structure and a priori knowledge of the mode of membrane binding for the peptides were recently developed.63,64 Often, Monte Carlo algorithms operate in dihedral angles space, thus reducing dimensionality of the computational task. Usually, Monte Carlo simulations help in delineation of a limited number of low-energy conformational states of TM helical dimers.64 Subsequent analysis of these families of conformers results in very few “native-like” structures, thus facilitating selection of the final models.
Another approach, which is used to predict spatial structures of TM dimers, is molecular dynamics (MD). This is one of the most informative methods, since besides providing the spatial structure it allows estimating the dynamic parameters of interaction, identifying the most important residues, etc. Membrane models of any degree of complexity can be used in MD calculations. One of the key issues in resolving spatial structure with MD methods is selection of the starting state, since this approach has limited capabilities for scanning conformational space—it is computationally resource-intensive and hence the likelihood of arriving at correct structure starting from an essentially wrong one is low. This problem is especially significant in case of calculations in the explicit bilayer. It can be solved by obtaining the starting structure for MD in the full-atomic approximation by other modeling techniques. One of the ways to achieve that is based on generating a set of initial states with different geometries of the dimer packing. Though providing most detailed scanning of the conformational space, this method is often impractical due to unacceptable computational resource requirements, and is essentially limited to implicit membrane calculations. Another way to obtain starting structures for full-atomic MD consists in Monte Carlo conformational search (or docking) in an implicit membrane with subsequent relaxation in the explicit bilayers. An alternative approach consists in preliminary investigation of the dimerization by the CG representation. In this case, the molecules are represented by “grains” (e.g., each of which roughly corresponds to four heavy atoms) that substantially improves the calculation time, so the intervals of up to ∼1 microsecond can be investigated. As was shown, this time scale is sufficient for obtaining a realistic model of the TM dimer, which after MD relaxation in the full atomic representation correlates well with the NMR structure.65
Extensive application of computer modeling methods allowed comprehensive investigation of specific TM dimerization of several bitopic proteins, including the wild type and mutated TM domains of glycophorin A,66 bacteriophage M13 major coat protein,67 proapoptotic protein BNip3,68 erythropoietin receptor,69 amyloid precursor protein APP70 and ErbB receptor tyrosine kinases.3 Most of the methods of molecular modeling of the TM helix specific dimerization have been developed and successfully tested on the TM domain of glycophorin A protein, homodimeric conformation of which was first obtained with high resolution.30 Although a number of successful in silico predictions of TM helix-helix complexes have been reported, the uncertainty of the energy estimate of the final state is still relatively high. Therefore, without employment of additional data it is usually very difficult to choose between several alternative models with close energies, having substantially different geometries. Moreover, if several dimerization modes are actually realized for a protein, computational methods provide little or no information about population and relative stability of the possible modes of helix-helix associates, which can be affected by modeling assumptions in silico as well as by variations of membrane environment and ligand binding in vivo. Partially, such a hypothesis is corroborated by somewhat vague results of mutagenesis studies,66 as well as by NMR71 and MD72,73 data that demonstrate the importance of media effects for stability of helical oligomers and provide examples of their multi-state equilibrium in lipid bilayers and membrane mimics. In real biological membranes, the situation may be more complex—due to nonhomogeneous content of lipid bilayers, their domain structure, variations of physico-chemical characteristics, presence of small molecules (e.g., cholesterol) and so forth.
Combination of Experimental Methods with Molecular Modeling for Obtaining Spatial Structure of TM Domain Dimers
Discrimination between the conformations artificially introduced by computational assumptions and those really occurring in cellular membrane is only possible with the use of additional experimental information, in particular about the TM dimerization interface, see Figure 1. Such an information can be obtained by solid state NMR, site-specific infrared dichroism, mutagenesis in combination with the techniques permitting assessment of dimerization degree (SDS electrophoresis, bioassays in ToxR systems, FRET), Cys scanning (insertion of cysteine residues and analysis of the extent of disulfide bridges formations) and so on (reviewed in refs. 10, 55, 74 and 75). Experimental limitation can be either imposed at the stage of calculations, e.g., in the form of distance restraints for atoms in different monomers, or used for assessing appropriateness of the predicted structures after completion of calculations. Such a combination of experimental and modeling techniques provides important advantages, substantially narrowing the search of dimeric TM structures and simplifying membrane representation and hence significantly accelerating the analysis. Compared to direct structural methods that usually identify only single conformation, this approach gives better credit for a conformational diversity of homo- and heterodimeric TM domain structures, which can occur in vivo during biological activity of bitopic proteins. Effectiveness of such a combination of computational methods with various biophysical and biochemical techniques was proved by its successful applications in a number of studies several of which are presented below.
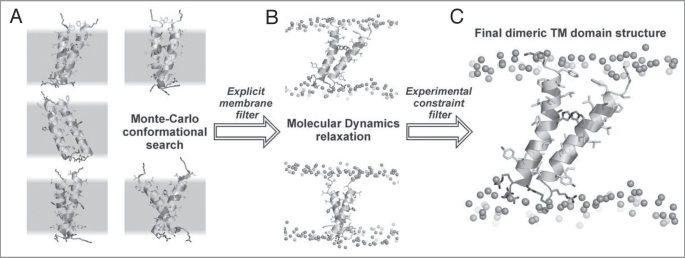
Figure 1.
Spatial structure elucidation of dimeric TM domain of bitopic protein (exemplified by proapoptotic protein BNip3) using computer simulations techniques. (A) Collection of rough models via Monte Carlo conformational search in implicit membranes. Set of possible structure candidates having minimal energy is presented. Peptides are given in ribbon and stick representations. Positions of implicit membrane are selected by gray hatching. (B) Results of MD-relaxation of the MC structures in full-atom hydrated bilayer. Only structures stable in the membrane during MD are selected. Membranes are delineated by position of phosphorus atoms (shown as spheres). Other details were skipped for clarity. (C) The resulting structure selected via comparison with experimental information about dimer interface.
In 1996, Adams et al.76 used global conformational search in vacuum for calculation of the dimeric structure of glycophorin A TM helix. Selection of the proper structure was done based on mutagenesis data. The proposed model of the dimer was in good agreement with the spatial structure obtained by means of NMR spectroscopy in detergent micelles.30 This method was later used for analysis of the glycophorin A TM domain dimerization in lipid bilayers. In this case, the conformational search was done with the distance restraints from solid-state NMR spectroscopy.77,78 Beevers et al.79 suggested a model of the spatial structure of the TM domain of the oncogenic mutant of rat receptor tyrosine kinase Neu by MD calculations in the explicit bilayer with different possible orientations of the monomers. Correctness of the resulting ‘consensus’ structures was assessed based on the information about orientation of the CO groups determined by site-specific infrared dichroism. Vereshaga et al.80 calculated the spatial structure of TM segment dimer of human proapoptotic protein Bnip3. In this case, Monte Carlo conformational search in an implicit membrane with subsequent MD-relaxation of the best models in the full-atom DMPC bilayer was used for identification of the potential structures. Dynamically unstable models were screened out at the stage of MD-relaxation. Correctness of the remaining models was assessed via comparison with the mutagenesis data. As a result, one of the final models consistent with the mutagenesis data was also in good agreement with the NMR-derived structure of dimeric Bnip3 TM domain in lipid bicelles.81 Volynsky et al.82 used modeling methods in combination with ToxR assays to study dimerization of TM segments of ephrin receptor EphA1. A set of spatial structures of the dimer proposed based on Monte Carlo simulations in implicit membrane followed by MD-relaxation in explicit lipid bilayer were employed for rational design of wild-type and mutant genetic constructions for ToxR assays. Such a combined, self-consistent, application of modeling and experimental techniques allowed defining the factors important for dimerization of the TM segment of the EphA1 receptor, providing unambiguous spatial model consistent with the NMR-derived structure of the EphA1 TM dimer in lipid bicelles.38 Moreover, alternative conformations of the dimer were proposed. Metcalf et al.83 reported the models of integrin aIIbβ3 TM heterodimers obtained using a Monte Carlo algorithm that selects conformations by a geometrical filter based on mutagenesis data. The Monte Carlo search for integrin aIIbβ3 TM heterodimers was also carried out with an additional energy term using distance restraints obtained from cysteine-scanning mutagenesis bioassay data.84 In both cases, the proposed heterodimeric models were in good agreement with recently obtained NMR structure of heterodimeric integrin aIIbβ3 TM complex embedded in lipid bicelles.40
Determination of High-Resolution Structureof Dimeric TM Domains of Bitopic Proteins
Despite the fast development of structural biology methods for directly obtaining high-resolution structure of membrane proteins, each new target protein typically poses a new set of challenges. Isolation, purification and handling of membrane proteins in their “native-like” conformations are associated with enormous difficulties and often require expanding the limits of the modern experimental techniques. Besides, tertiary and quaternary structures of membrane proteins are only modestly stabilized and transitions are often observed between conformational substates. Multiple conformations and dynamics considerably complicate characterizing the structure of membrane proteins and their oligomers. For these reasons, despite recent increase in the number of high resolution structures of membrane proteins solved annually, the gap between soluble and membrane protein structures continues to grow. Even among the membrane proteins of known structure, specific oligomeric complexes of small membrane-spanning proteins such as TM domains of bitopic proteins are underrepresented.
Fortunately, oligomeric α-helical TM domains of membrane proteins are amenable to structural-dynamic characterization by heteronuclear NMR spectroscopy. Solution NMR became a major method to determine structures of proteins and protein complexes that are readily soluble in aqueous solution.85 In addition to elucidation of their structures, NMR also offers unique opportunities to probe dynamic processes in such proteins and complexes. Membrane proteins embedded into lipid bilayers cannot be studied by means of solution NMR techniques because their rotations in these environments are slow and highly anisotropic. This leads to unfavorable relaxation and very wide or undectable resonance lines. However, solid-state NMR has been successfully employed to obtain highly resolved spectra of membrane-bound peptides and proteins in such bilayer model systems as liposomes, which can have composition, thickness, surface tension and curvature similar to those of native lipid bilayers and thus adequately mimic cell membranes. Solid-state NMR techniques for membrane protein samples are rapidly evolving, and the structures of several small proteins in lipid bilayers have been already obtained with the aid of these methods.86,87 There are two ways of obtaining high-resolution solid-state NMR spectra, either by performing magic angle spinning (MAS) in order to mimic the rapid tumbling that would naturally occur for a small molecule in solution for averaging the anisotropic interactions in solid-state, or by observing uniformly aligned molecules. Smith and co-workers have used 13C-13C rotational resonance and 13C-15N rotational echo double resonance MAS experiments to measure interhelical distances in the α-helical TM domain dimers of human glycophorin A,77,78 human amyloid precursor protein88 and rat receptor tyrosine kinase Neu (homologue of human ErbB2 receptor) with its constitutively active Val664Glu mutant.89 That allowed developing the structural models for the helix-helix packing in lipid bilayer for these bitopic proteins. The tilt and rotational angles of TM helices can be estimated by analysis of the position, shape and size of the so-called PISA wheels obtained from polarization inversion with spin exchange at the magic angle (PISEMA) experiment acquiring for oriented 15N-labeled membrane proteins.86
An alternative approach to solving high-resolution spatial structures and obtaining dynamic information on membrane proteins is to extract the proteins from their host membranes and disperse them in non-denaturing membrane-mimicking detergent/lipid systems such as micelles, bicelles and nanodiscs, which tumble fast enough to give well-resolved resonance lines when using solution NMR methods. Since, resulting supramolecular membrane protein-detergent/lipid complexes are usually still large on the scale of protein structures that are routinely solved by NMR, the most advanced solution NMR techniques and spectrometers operating at high magnetic fields and equipped with highly sensitive cryoprobes are typically employed to solve high-resolution structure of the membrane proteins. These include labeling the proteins with two or three low-abundant isotopes 2H, 13C and 15N, deuterating of detergents and lipids at least on hydrophobic tails, using transverse relaxation-optimized spectroscopy (TROSY),90 and obtaining structural restraints in addition to those typically obtained from nuclear Overhauser effects (NOE) and chemical shifts, such as restraints obtained from residual dipolar couplings (RDC) and paramagnetic relaxation enhancements (PRE), which can drastically improve both quality and throughput of membrane protein structure determination (reviewed in ref. 91). The accuracy of determining the protein structure is controlled by many factors, including the dynamic properties of the protein itself, as well as the nature and quantity of the experimentally obtained restraints. In case of dimeric TM α-helical proteins, if a well defined structure of monomers is known (particularly side chain conformations and helix bending), just a few restraints can fully determine the structure provided that they define the conformational space in an independent manner. However, since every restraint has an experimental error associated with the precision of measurements and with the accuracy of assignment in case of NOE contacts, having larger number of independently derived consistent restraints greatly increases confidence in the structure of individual TM helices and of the dimer as a whole. In case of underdetermined structures where there are substantial ambiguities in the NMR-derived structural information with only few reliable restraints defining global dimer structure, molecular modeling can allow to resolve the ambiguities in the remaining information in favor of the most physically justifiable model of the dimer. Obviously, this process directly depends on the accuracy of the underlying physical assumptions, i.e., the force fields used in the modeling of the membrane proteins. Given the limited amount of structures obtained in the membrane-mimicking environments, each new experimental structure is of utmost practical and methodological importance.
Micelles formed of soft detergents, short-chain lipids or lysolipids are the smallest among membrane mimicking particles, and are therefore optimal from the standpoint of NMR relaxation, allowing recording spectra with narrow lines and rather good chemical shift dispersion.92,93 A lot of membrane-penetrating peptides, membrane associated peptides and fragments of membrane proteins were studied in micellar solutions by NMR spectroscopy.91,93,94 Most of the structures of helical membrane proteins resolved with NMR spectroscopy were determined in micelles of different types, indicating that there is no universal detergent, applicable for every membrane protein. Therefore, extensive detergent screening is usually made to find a proper environment.93,95,96 Although a majority of membrane proteins maintain native-like structures in micelles and some retain activity, sometimes the detergent providing the best appearance of NMR spectra does not provide proper folding, and the protein dissolved in it remains inactive. Micelles have some disadvantages associated with high curvature of their spherical surfaces. Curvature effects are occasionally observed with small peptides, and the absence of specific phospholipids or mixtures of phospholipids may cause amphiphillic peptides interacting with the membrane surface to have distorted structures in micelles environment.97,98 Integral membrane proteins can also have distorted structure and poor spectrum appearance in micellar solutions, especially when they have structural elements that should lie on the bilayer surface. Both the headgroup region and the hydrocarbon core in a highly curved micelle are packed less orderly and exhibit greater dynamics than in a planar or near-planar lipid bilayer.97,98 The shielding effect of the interfacial headgroup region is less pronounced, and water molecules can penetrate more easily into the micellar core, resulting in a distortion of TM helix structure.99 Importantly, the addition of very modest amounts of phospholipids to micelles can result in dramatic enhancements of NMR spectral quality for some integral membrane proteins.100 This lipid dependence appears to reflect the requirement of some membrane proteins for semi-specific lipid-protein interactions, which cannot be satisfied by detergents only. So, detergent micelles with some amounts of phospholipids offer a valuable compromise for investigating TM peptides in membrane-mimetic systems, combining ease of use and good dissolving properties with an anisotropic environment. Nevertheless, many detergents exert a denaturating effect on membrane proteins and peptides by abrogating helix-helix interactions.101,102 These problems could be overcome by using membrane mimicking particles with elements of flat surface, such as bicelles and nanodiscs.
Nanodiscs were designed on the basis of high-density lipoprotein particles, and consist of fairly large patches of planar lipid bilayers (∼160 lipid molecules) surrounded by the rim formed by apolipoprotein A-I.103–105 The particles have the diameter of about 12 nm and thickness of 4 nm with the overall rotational correlation time of about 80 ns,106 which is rather high for structural NMR studies, but with TROSY90 and CRINEPT107 techniques one can record a readable heteronuclear NMR spectrum and compare with NMR spectra recorded in micelles or bicelles. Nanodiscs have been applied in NMR spectroscopy for a couple of years and only few membrane proteins were studied in this environment so far. However, they proved useful for verifying that other membrane mimicking media provide proper tertiary structure of certain membrane proteins;108 they also have high potential for various bioassay applications.109
Small isotropic bicelles, being a compromise between micelles and nanodiscs, are the most convenient media with excellent bilayer-mimicking properties for NMR structural studies of small membrane proteins and their complexes.91,110 Bicelles are binary mixed micelles, consisting of two types of molecules: long-chain lipids (with long hydrophobic tails) and short-chain lipids or detergents, e.g., dimyristoylphosphatidylcholine (DMPC) mixed with dihexanoylphosphatidylcholine (DHPC) or zwitterionic bile salt derivative CHAPSO.91 A number of bicelle systems have been developed and characterized for their unique liquid-crystal phase behavior. It was shown that under certain conditions bicelles have discoidal shape with a bilayer formed by long-chain lipids and a rim of short-chain lipids.111–115 The shape of the particles is controlled by three parameters: the molar ratio q of long- and short-chain lipid (or detergent) concentrations, total lipid concentration cL, and temperature T; and it can be either disc or perforated bilayer, the dependence being rather complex.111 At q between 0.25 and 0.5, bicelles are tumbling fast, are almost isotropic and can be used for high-resolution structure determination.91,116 The hydrophobic thickness of the aggregates can be controlled by the choice of long-chained lipids, and it was also shown that charged lipids, e.g., with either negative serine or glycerol headgroups, can be incorporated in such particles without loss of stability.117,118 There are a lot of publications on membrane proteins, showing smaller structure-distorting properties of bicellar media.91 Recent determination of the structure of the heterodimeric TM domain of the platelet integrin aIIbβ3 in bicelles provides an elegant example of using this medium to solve an important structural biology problem that proved elusive when conventional micelles were used.40,119 Detergent micelles destabilize the heterodimer to the point where interaction cannot be detected, while the environment provided by bicelles allows at least partial retention of native-like heterodimer avidity. Typical size of supramolecular particles consisting of fast-tumbling bicelles (e.g., DMPC/DHPC bicelle of ∼70 lipid molecules, q of 0.25, cL of 3%, at 40°C) with embedded two bitopic protein TM fragments (∼40 residues including hydrophobic TM segment flanked by polar N- and C-terminal regions) is ∼5 nm corresponding to overall rotational correlation time of ∼18 ns and the effective molecular weight of ∼50 kDa that allows successfully employing the broad capabilities of solution heteronuclear NMR technique for elucidation of protein structural-dynamic properties.38
It is worth mentioning that even if specific interaction of TM helices is weak (e.g., in the case of receptor tyrosine kinase TM domains), low effective ratio of detergent/lipid to protein and restricted mobility within small detergent/lipid particles encourage homo- or heterodimerization. Importantly, typical size of micelles and bicelles allows detecting intermolecular NOE contact network (up to ∼0.6 nm) along TM helix-helix interface that is crucial for obtaining high-resolution structures of homo- and heterodimeric TM domains of bitopic protein. Nevertheless, one of the main problems encountered in structure determination of molecular complexes by NMR spectroscopy is to distinguish between intra- and intermolecular NOE contacts. Concerning self-association of bitopic protein TM domains, if the studied dimers of α-helical TM segments are symmetrical on the NMR time scale, their two monomer chains display similar chemical shifts so that intra- and intermonomeric NOE contacts are indistinguishable in the NMR spectra. Furthermore, small chemical shift dispersion inherent to α-helical structure as well as line broadening owing to large size of the supramolecular system and slow conformational exchange widespread in oligomeric complexes are additional unfavorable factors complicating unambiguous identification of intermonomeric NOE contacts also in the cases of TM heterodimers or asymmetric homodimers.
A computational solution to the symmetry degeneracy problem is the so-called “ambiguous distance restraints” method,120 according to which spatial structure of a symmetrical dimer is calculated in two stages, involving an initial stage the structure refinement of the monomer subunit before proceeding to the dimer. Experimentally identified NOE contacts are interpreted in a conservative manner and only those that are clearly inconsistent with the global fold of the monomer could be assigned as unambiguous intermonomeric NOE contacts. All other NOE contacts are treated as having arisen from either intra- or intermonomer cross-relaxation. In 1997, MacKenzie et al.30 successfully used this strategy in the pioneering work of determining high-resolution structure of homodimeric TM domain of glycophorin A (PDB 1AFO), which was solubilized in DPC micellar media. Glycophorin A, a surface protein marker of human erythrocytes, is widely used as a model protein in developing the experimental and theoretical methods to study the specific dimerization of TM domains of bitopic proteins. In detergent micelles, its membrane-spanning α-helices self-associate in a parallel right-handed manner with crossing angle of −40° via tetrad repeat dimerization pattern L75IxxG79VxxG83VxxT87 including the so-called tandem GG4-like motif (also known as ‘glycine zipper’)49 composed of residues with small side chains allowing close approach of the helices. Along with numerous van-der-Waals interactions, four close polar CαH…O helix-helix contacts, which can be described as non-canonical hydrogen bonds across the dimer interface afforded by GG4-like motif, occur between CαH1 of Gly79 and Gly83 and opposite backbone carbonyls of Ile76 and Val80. The dimer structure also revealed the intramolecular hydrogen bonding of the hydroxyl group of Thr87 with backbone carbonyl group of Gly79. Nevertheless, lately the formation of an intermonomeric hydrogen bond between side chain hydroxyl group of Thr87 and backbone carboxyl group of Val84 was proposed based on several dipolar interaction observed with solid state NMR by Smith et al.77,78 using dry DMPC and POPC lipid bilayers. The work of MacKenzie et al.30 was an important early accomplishment both for technical reasons and because of the insight that the structure provides into membrane protein folding and stability.
For direct search of intermolecular NOE contacts in the dimer interface, the symmetry degeneracy problem can be circumvented by preparing an isotopic “heterodimer,” consisting of 2H, 13C, 15N isotope labeled and natural abundance monomers and by carrying out experiments, which select NOE contacts arising between isotopically bound and nonisotopically bound protons. Besides the case of symmetrical homodimerization, such experiments are useful for directly obtaining interhelical spatial restraints for asymmetric TM dimers (or oligomers) as well for identifying close intermolecular protein-lipid contacts. A simple method to distinguish intermonomer NOE contacts is to produce a 2H/15N-isotopic “heterodimer,” in which one subunit is 15N-labeled and fully deuterated (except NH groups) whereas the other subunit is unlabeled (1H/12C/14N). This method allows directly obtaining interhelical proton-proton restraints from side chain and backbone groups of one subunit to backbone amide groups of the other. Such strategy was successfully used for determination of high-resolution NMR structure of a constitutively disulfide-linked TM domains of the T-cell receptor ξξ-chain homodimer embedded into mixed 5:1 DPC/SDS micelles (PDB 2HAS).121 In detergent micelles, the TM ξξ-chain helices form a left-handed dimer with a crossing angle +23° via extended heptad repeat dimerization pattern C2xxL5D6xxL9xxY12xxxL16T17xxF20xxV23 encompassing almost entire TM segment and making numerous interhelical side chain contacts, several of which are polar. It was shown that the side-chain hydroxyls of Tyr12 and Thr17 form a pair of interhelical hydrogen bonds that create “brackets” defining the lateral edges of the dimer interface. Structural and mutagenesis analysis revealed that two residues Asp6 situated near intersubunit Cys2-Cys2 bridge, which are required for receptor assembly, can form extensive hydrogen-bonding network with several hydrogen-bond donors and acceptors including at least one water molecule, the cysteine carbonyls, the carboxyl side chain and amide groups of aspartic acids themselves. So, the structure of the TM ξξ-chain dimer nicely demonstrated how multiple hydrogen bonding can establish a left-handed TM homodimer.
The strategy of ILV-methyl-selective protonation122 was employed for high-resolution structure determination of the heterodimeric TM domain of intact integrin aIIbβ3 in POPS/POPC/DHPC (q = 0.32) and deuterated DMPC/DHPC (q = 0.30) lipid bicelles (PDB 2K9J).40 The 1H13C3-Ile,Leu,Val;2H/13C/15N-labeled and unlabeled 1:1 mixtures of the aIIb and β3 integrin TM subunits were used for partial side-chain assignments and for identification of intermonomeric proton-proton NOE contacts between methyl groups of one subunits and any groups of the second subunit. Guided by packing interaction with three distinct glycine residues, the integrin TM helices cross at an angle of −25° and connect through tetrad repeat patterns G972xxxG976xxL979L980xxxL984 and V700M701xxI704L705xxG708xxxL712 of aIIb and β3, respectively, forming a TM heterodimer of unique structural complexity. The assembly enables strong electrostatic interactions (as detected by mutagenesis) between side chains of Arg995 and Asp723 of aIIb and β3, respectively, within the relatively low dielectric environment of lipid headgroups. The reported heterodimeric TM structure along with structure-based side-directed mutagenesis of integrin aIIbβ3 provides important insights into the structural basis for integrin signaling in cell membrane, revealing the structural events that underlie the transition from associated to dissociated states upon receptor activation.40
A robust strategy to distinguish intermonomeric NOE contacts in protein dimers was based on producing a 13C/15N-isotopic “heterodimer,” in which one subunit is 13C/15N-labeled and the other subunit is unlabeled. For the direct detection of the intermolecular NOE contacts in such isotopic “heterodimer,” NMR pulse sequences were developed,123,124 employing so-called X-filtering elements to select NOE contacts arising between nonisotopically and isotopically bound protons. Due to fast transverse magnetic relaxation as consequence of relatively big overall correlation time of the studied supramolecular systems, the intermonomeric proton-proton contacts in the 13C/15N-isotopic “heterodimer” are mainly detected from methyl groups (having smallest relaxation rates) to other groups. This approach was successfully applied in our lab for elucidation of structural-dynamic properties of homo- and heterodimeric α-helical TM domains of several biologically different human proteins, including proapoptotic protein BNip3 and representatives of receptor tyrosine kinase ErbB and Eph subfamilies. The high-resolution NMR structures of dimeric TM domains of these bitopic proteins were obtained using DMPC/DHPC (q = 0.25) bicelles consisting of lipids with deuterated hydrophobic tails and lipid/protein molar ratios of ∼35. The resulting NMR structures of the TM domain dimers were subjected to energy relaxation using MD during several ns of MD trajectory in hydrated explicit lipid bilayers with the imposed NMR-derived constraints and then without constraints to study the conformational stability of the dimer in the membrane. The MD-relaxation procedure provided a detailed atomistic picture of the intra- and intermolecular (protein-protein, protein-membrane and protein-water) interactions and allowed estimating the influence of amino acid substitution, including pathogenic TM mutations, on the structural-dynamic properties of bitopic proteins, see Figure 2.
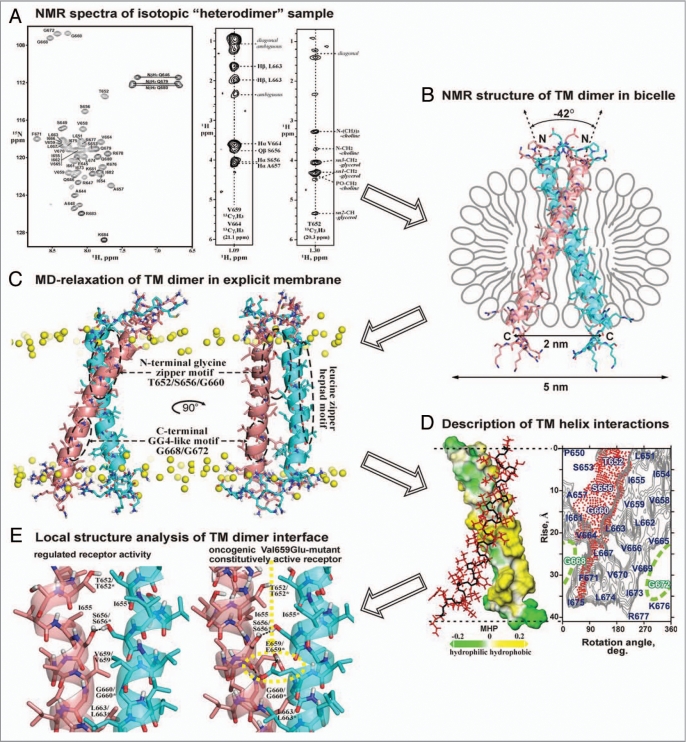
Figures 2.
Spatial structure elucidation of dimeric TM domain of the bitopic protein (exemplified by receptor tyrosine kinase ErbB2)130 using with the aid of heteronuclear solution NMR in lipid bicelles and MD-relaxation in explicit lipid bilayer. (A) Acquisition of NMR spectra of isotopic “heterodimer,” consisting of 13C/15N-isotope labeled and natural abundance ErbB2 TM fragments (residues 641–685) embedded into DMPC/DHPC lipid bicelles. From left to right, 1H-15N HSQC spectrum with amide backbone resonance assignments, two representative 2D strips from the 3D 13C F1-filtered/F3-edited-NOESY spectrum with intermolecular protein-protein and protein-lipid NOE contacts are presented. (B) Determination of high-resolution spatial structure of the right-handed ErbB2 TM homodimer in lipid bicelle using NMR-derived restraints. The obtained N-terminal association mode of the ErbB2 TM dimer via N-terminal dimerization motif corresponds to the receptor active state. (C) MD-relaxation of the ErbB2 TM homodimer in hydrated explicit DMPC lipid bilayer with imposed NMR-derived constraints. Yellow balls show phosphorus atoms of lipid heads. The spatial locations of the three characteristic dimerization motifs of ErbB2tm are marked by dashed ovals. (D) Analysis of interacting surfaces of the ErbB2 TM helices. In left, hydrophobic and hydrophilic (polar) surfaces of one TM helix in the homodimer colored in yellow and green according to the molecular hydrophobicity potential (MHP).132 The second monomer of the dimer is shown with red side chains. In right, hydrophobicity map for ErbB2 TM helix surface with contour isolines encircling hydrophobic regions with high values of MHP is presented with red-point area indicating the helix packing interface via N-terminal glycine zipper motif T652xxxS656xxxG660. The residues composing C-terminal unemployed dimerization GG4-like motif G668xxxG672 are highlighted in green. (E) Local structure analysis of intra and intermolecular interactions in the ErbB2 TM dimer. Comparison of intermonomeric hydrogen bonding (black dotted lines) in the TM helix-helix interface of ErbB2 and its constitutively active Val659Glu-mutant is presented.
BNip3 is a prominent representative of apoptotic Bcl-2 proteins with unique properties initiating an atypical programmed cell death pathway.125 Investigation of spatial structure and internal dynamics of the homodimeric TM domain of human protein BNip3 (PDB 2J5D)81 revealed that in the lipid bicelles the central membrane-spanning α-helices of BNip3 cross at the angle of −45° and form a right-handed parallel symmetric dimer via tetrad repeat pattern S172H173xxA176xxxG180xxxG184. In addition, labile Phe-ring hydrophobic cluster with numerous inter-monomeric stacking interactions between six phenylalanine residues (Phe157/Phe161/Phe165)2 was identified in the interface between short mobile N-terminal helices, flanking the central helices. According to the obtained NMR data supported by MD-relaxation, a hydrophilic motif (Ser172/His173)2 in the centre of BNip3 TM dimerization interface forms a water-accessible His-Ser node of intra- and intermonomeric hydrogen bonds decreasing apparent pKa of the imidazole group below 4. The C-terminal TM part of the BNip3tm dimer is stabilized by van-der-Waals side chain contacts and by weakly hydrophilic backbone contacts of the helices tightly self-associated through a glycine zipper motif, which appears to be essential for proper alignment of the side chains in the His-Ser node required for hydrogen bonding. In the DMPC/DHPC bicelles the His-Ser node undergoes slow conformational exchange with ∼10% occupancy of the minor state probably associated with alternative hydrogen bonding and water permeability. Nevertheless, it was shown that an addition of long chain DPPC lipid to DPC micelles (lipid/detergent ratio of 1:50) with embedded dimeric BNip3 TM domain allows to eliminate the conformational inhomogenity in the dimer interface.100 The revealed structural-dynamic properties of the BNip3 TM domain with a potentially switchable network of hydrogen bonds and water accessibility up to the middle of the membrane appear to enable the protein to form ion-conducting pathway across the membranes. Indeed, the TM domain was shown to induce conductivity of artificial bilayer lipid membrane in a pH-dependent manner.81 These findings and currently available information about phenomenology of programmed cell death allowed us to propose a mechanism of triggering necrosis-like cell death by BNip3 in case of hypoxia-acidosis of human tissues.
Receptor tyrosine kinases conducting biochemical signals across plasma membrane via lateral dimerization play an important role in normal and in pathological conditions of human organism by providing cell signaling, maintaining cellular homeostasis and controlling cell fate.3 Eph receptors are found in a wide variety of cells in developing and mature tissues and represent the largest family of receptor tyrosine kinases regulating cell shape, movement and attachment.126 Because all Eph receptors and their ligand ephrins are cell surface-associated proteins, a direct cell-cell contact is required for receptor activation resulting in cytoskeletal remodeling that underlies cell adhesion, repulsion and motility in both communicating cells. Although the Eph TM segments reveal relatively low amino acid sequence homology, several dimerization motifs, including at least one explicit GG4-like motif, can be identified in each Eph TM region. Structural-dynamic properties of the homodimeric TM domains of the EphA1 and EphA2 receptors were investigated with the aid of solution NMR in lipid bicelles and MD-relaxation in explicit lipid bilayers of different composition. High-resolution spatial structures of homodimeric TM domains of EphA1 (PDB 2K1K and 2K1L) and EphA2 (PDB 2K9Y) embedded into DMPC/DHPC bicelles revealed a right- and left-handed parallel packing of the α-helical TM domains with crossing angle of −45° and +15°, respectively.38,127 The EphA1 TM segment self-associates through the N-terminal glycine zipper motif A550xxxG554xxxG558 whereas the C-terminal GG4-like dimerization motif A560X3G564 is not employed. And vice versa, the EphA2 TM helices interact through the extended heptad repeat motif L535xxxG539xxA542xxxV546xxxL549 assisted by intermolecular stacking interactions of aromatic rings of (FF557)2, whereas the N-terminal glycine zipper motif A536X3G540X3G544 remains vacant. Thus, our studies of the Eph1 and EphA2 receptors demonstrated that the TM domains of different representatives of the same receptor tyrosine kinase family can use alternative dimerization motifs in the same bicellar system, the different motifs possibly being corresponding to active and inactive dimeric state of the receptor. Moreover, in the case of EphA1 TM domain, variations of external pH and lipid composition of the bicelles initiated triggering between the alternative motifs,38 which can be viewed as an argument in favor to the so-called “rotation-coupled” mechanism of the receptor tyrosine kinase activation.4–6 The obtained results indicated also that alternative dimeric conformations of the TM domains can influence the receptor localization in plasma membrane microdomains and signaling platform, such as rafts and caveolae.127
Four human ErbB members of epidermal growth factor receptor family form numerous homo- and heterodimer combinations, recognizing different EGF-related ligands and performing diverse functions in a complex signaling network.128 All the species of the ErbB family are activated by proper ligand-induced dimerization or by reorientation of monomers in preformed receptor dimers after ligand binding that can be widespread among receptor tyrosine kinase family.3,129 So, two possible dimeric conformations of the α-helical ErbB TM segments with interfaces located either at N- or C-terminus were proposed to associate different receptor active states.4–6 According to high-resolution spatial structure of homodimeric ErbB2 TM domain embedded into DMPC/DHPC lipid bicelles (PDB 2JWA),130 the α-helical TM segments of ErbB2 interact with right-handed crossing angle of −42° through the N-terminal glycine zipper motif T652xxxS656xxxG660 (Fig. 2B and C). Polar contact area of this motif is shielded from lipid tails by the side chains of leucine, isoleucine and valine residues, while slightly polar concave surface of the C-terminal GG4-like motif G668xxxG672 is exposed to hydrophobic lipid environment (Fig. 2C and D). In the C-terminal part of the dimeric interface, aromatic rings of the opposite Phe671 residues participate in intermolecular edge-face stacking interaction. Constrained MD-relaxation of the ErbB2tm dimer structure revealed that the (Thr652/Ser656)2 hydrophilic motif in the N-terminal part of the dimerization interface forms a node of switching intra- and intermonomeric hydrogen bonds mediating the ErbB2 TM helix packing (Fig. 2E). Based on the NMR-derived structure it was also shown by molecular modeling that pro-oncogenic Val659Glu mutation (Fig. 2E) leads to overstabilization of the described ErbB2 TM domain conformation which was ascribed to the active state of the tyrosine kinase.130 The assumption that the N-terminal association mode of the ErbB TM dimer corresponds to the receptor active state has been supported by recent structural studies of the juxtamembrane segment and kinase domain dimerization upon kinase activation of the ErbB1 receptor.131 It was shown that folding of the juxtamembrane regions of both monomers in the receptor dimer into an antiparallel helical structure, requiring the spacing between the C-termini of the TM helices to be about 2 nm, is essential for the kinase domain activation.131 The homodimeric ErbB2 TM structure we obtained has exactly the required distance between the C-termini of the TM helices (Fig. 2B), and is thus allowing proper kinase domain activation. Overall these findings enhance understanding of the functional conformational changes of receptor tyrosine kinases during activation of the signaling ligand-receptor complex in cell membranes in normal and pathologic states of human organism.
Concluding Remarks
Many aspects of the specific helix-helix interactions in membranes are yet far from being completely understood and are awaiting detailed investigation, which is only possible through concerted use of various physical-chemical and biological methods supported by molecular modeling. Theoretical and experimental methods to study protein-protein and protein-lipid interactions in membrane are rapidly evolving in a correlated manner. Molecular modeling is used to support interpretation of data about specific TM helix association and vice versa the theoretical modeling parameters are corrected based on the experimentally obtained information. This will likely result, within a few years to come, in detailed description of a large variety of intra- and intermolecular interactions of the TM domains of bitopic proteins and elucidation of the roles of the TM domains in normal and abnormal functioning of the proteins and in their proper localization in cell membranes. The already available information about structural-dynamic properties of the dimeric TM domains of bitopic proteins along with the biophysical and biochemical data provides useful insights into the protein functioning in the human organism at the atomic level. In parallel with TM domain investigations, extensive structural and functional studies of extracellular and cytoplasmic domains of the bitopic proteins are pursued, their results in combination with the information about TM domains already make it possible to produce detailed molecular models of the ligand-receptor complexes in different functional states. Naturally, that does not diminish the current importance and topicality of obtaining the structure of full-length bitopic proteins both separately and in complexes. However, at the present state of development of structural biology this remains quite an ambitious challenge. The most important practical implications of these studies are primarily related to molecular design of pharmaceutical compositions that can affect specific helix-helix association in cell membrane, providing a novel form of therapy of many human diseases related with abnormal activity of the bitopic proteins.
Acknowledgements
Financial support was received from the Russian Foundation for Basic Research, Program MCB RAS and the Russian Funds Investment Group. E.V. Bocharov thanks personally K.A. Beirit for financial support.
Abbreviations
- TM
transmembrane
- MD
molecular dynamics
- NMR
nuclear magnetic resonance
- NOE
nuclear overhauser effect
- DMPC
dimyristoylphosphatidylcholine
- DHPC
dihexanoylphosphatidylcholine
Footnotes
Previously published online: www.landesbioscience.com/journals/celladhesion/article/11930
References
- 1.Ubarretxena-Belandia I, Engelman DM. Helical membrane proteins: diversity of functions in the context of simple architecture. Curr Opin Struct Biol. 2001;11:370–376. doi: 10.1016/s0959-440x(00)00217-7. [DOI] [PubMed] [Google Scholar]
- 2.Cherezov V, Rosenbaum DM, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, et al. High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science. 2007;318:1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2000;281:211–225. doi: 10.1016/s0092-8674(00)00114-8. [DOI] [PubMed] [Google Scholar]
- 4.Moriki T, Maruyama H, Maruyama IN. Activation of preformed EGF receptor dimers by ligand-induced rotation of the transmembrane domain. J Mol Biol. 2001;311:1011–1026. doi: 10.1006/jmbi.2001.4923. [DOI] [PubMed] [Google Scholar]
- 5.Fleishman SJ, Schlessinger J, Ben-Tal N. A putative molecular-activation switch in the transmembrane domain of ErbB2. Proc Natl Acad Sci USA. 2002;99:15937–15940. doi: 10.1073/pnas.252640799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mendrola JM, Berger MB, King MC, Lemmon MA. The single transmembrane domains of ErbB receptors self-associate in cell membranes. J Biol Chem. 2002;277:4704–4712. doi: 10.1074/jbc.M108681200. [DOI] [PubMed] [Google Scholar]
- 7.Li E, Hristova K. Role of receptor tyrosine kinase transmembrane domains in cell signaling and human pathologies. Biochemistry. 2006;45:6241–6251. doi: 10.1021/bi060609y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Selkoe DJ. Alzheimer’s disease: genes, proteins and therapy. Physiol Rev. 2001;81:741–766. doi: 10.1152/physrev.2001.81.2.741. [DOI] [PubMed] [Google Scholar]
- 9.Bennasroune A, Fickova M, Gardin A, Dirrig-Grosch S, Aunis D, Cremel G, et al. Transmembrane peptides as inhibitors of ErbB receptor signaling. Mol Biol Cell. 2004;15:3464–3474. doi: 10.1091/mbc.E03-10-0753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rath A, Johnson RM, Deber CM. Peptides as transmembrane segments: decrypting the determinants for helix-helix interactions in membrane proteins. Biopolymers. 2007;88:217–232. doi: 10.1002/bip.20668. [DOI] [PubMed] [Google Scholar]
- 11.Caputo GA, Litvinov RI, Li W, Bennett JS, Degrado WF, Yin H. Computationally designed peptide inhibitors of protein-protein interactions in membranes. Biochemistry. 2008;47:8600–8606. doi: 10.1021/bi800687h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Popot JL, Engelman DM. Membrane protein folding and oligomerization: the two-stage model. Biochemistry. 1990;29:4031–4037. doi: 10.1021/bi00469a001. [DOI] [PubMed] [Google Scholar]
- 13.Helms V. Attraction within the membrane. Forces behind transmembrane protein folding and supramolecular complex assembly. EMBO Rep. 2002;3:1133–1138. doi: 10.1093/embo-reports/kvf245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schneider D. Rendezvous in a membrane: close packing, hydrogen bonding, and the formation of transmembrane helix oligomers. FEBS Lett. 2004;577:5–8. doi: 10.1016/j.febslet.2004.10.029. [DOI] [PubMed] [Google Scholar]
- 15.Morrow MR, Huschilt JC, Davis JH. Simultaneous modeling of phase and calorimetric behavior in an amphiphilic peptide/phospholipid model membrane. Biochemistry. 1985;24:5396–5406. doi: 10.1021/bi00341a018. [DOI] [PubMed] [Google Scholar]
- 16.Lee AG. How lipids affect the activities of integral membrane proteins. Biochim Biophys Acta. 2004;1666:62–87. doi: 10.1016/j.bbamem.2004.05.012. [DOI] [PubMed] [Google Scholar]
- 17.Nyholm TK, Ozdirekcan S, Killian JA. How protein transmembrane segments sense the lipid environment. Biochemistry. 2007;46:1457–1465. doi: 10.1021/bi061941c. [DOI] [PubMed] [Google Scholar]
- 18.Sparr E, Ash WL, Nazarov PV, Rijkers DT, Hemminga MA, Tieleman DP, Killian JA. Self-association of transmembrane alpha-helices in model membranes: importance of helix orientation and role of hydrophobic mismatch. J Biol Chem. 2005;280:39324–39331. doi: 10.1074/jbc.M502810200. [DOI] [PubMed] [Google Scholar]
- 19.Vidal A, McIntosh TJ. Transbilayer peptide sorting between raft and nonraft bilayers: comparisons of detergent extraction and confocal microscopy. Biophys J. 2005;89:1102–1108. doi: 10.1529/biophysj.105.062380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Meyer FJ, Venturoli M, Smit B. Molecular simulations of lipid-mediated protein-protein interactions. Biophys J. 2008;95:1851–1865. doi: 10.1529/biophysj.107.124164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Senes A, Engel DE, DeGrado WF. Folding of helical membrane proteins: the role of polar, GxxxG-like and proline motifs. Curr Opin Struct Biol. 2004;14:465–479. doi: 10.1016/j.sbi.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 22.Curran AR, Engelman DM. Sequence motifs, polar interactions and conformational changes in helical membrane proteins. Curr Opin Struct Biol. 2003;13:412–417. doi: 10.1016/s0959-440x(03)00102-7. [DOI] [PubMed] [Google Scholar]
- 23.Langosch D, Heringa J. Interaction of transmembrane helices by a knobs-into-holes packing characteristic of soluble coiled coils. Proteins. 1998;31:150–159. doi: 10.1002/(sici)1097-0134(19980501)31:2<150::aid-prot5>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 24.Walther D, Eisenhaber F, Argos P. Principles of helix-helix packing in proteins: the helical lattice superposition model. J Mol Biol. 1996;255:536–553. doi: 10.1006/jmbi.1996.0044. [DOI] [PubMed] [Google Scholar]
- 25.Zhou FX, Cocco MJ, Russ WP, Brunger AT, Engelman DM. Interhelical hydrogen bonding drives strong interactions in membrane proteins. Nat Struct Biol. 2000;7:154–160. doi: 10.1038/72430. [DOI] [PubMed] [Google Scholar]
- 26.Choma C, Gratkowski H, Lear JD, DeGrado WF. Asparagine-mediated self-association of a model transmembrane helix. Nat Struct Biol. 2000;7:161–166. doi: 10.1038/72440. [DOI] [PubMed] [Google Scholar]
- 27.Adamian L, Jackups R, Binkowski TA, Liang J. Higher-order interhelical spatial interactions in membrane proteins. J Mol Biol. 2003;327:251–272. doi: 10.1016/s0022-2836(03)00041-x. [DOI] [PubMed] [Google Scholar]
- 28.Gratkowski H, Lear JD, DeGrado WF. Polar side chains drive the association of model transmembrane peptides. Proc Natl Acad Sci USA. 2001;98:880–885. doi: 10.1073/pnas.98.3.880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou FX, Merianos HJ, Brunger AT, Engelman DM. Polar residues drive association of polyleucine transmembrane helices. Proc Natl Acad Sci USA. 2001;98:2250–2255. doi: 10.1073/pnas.041593698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.MacKenzie KR, Prestegard JH, Engelman DM. A transmembrane helix dimer: structure and implications. Science. 1997;276:131–133. doi: 10.1126/science.276.5309.131. [DOI] [PubMed] [Google Scholar]
- 31.Senes A, Ubarretxena-Belandia I, Engelman DM. The Cα-H…O hydrogen bond: a determinant of stability and specificity in transmembrane helix interactions. Proc Natl Acad Sci USA. 2001;98:9056–9061. doi: 10.1073/pnas.161280798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dawson JP, Weinger JS, Engelman DM. Motifs of serine and threonine can drive association of transmembrane helices. J Mol Biol. 2002;316:799–805. doi: 10.1006/jmbi.2001.5353. [DOI] [PubMed] [Google Scholar]
- 33.Schneider D, Engelman DM. Motifs of two small residues can assist but are not sufficient to mediate transmembrane helix interactions. J Mol Biol. 2004;343:799–804. doi: 10.1016/j.jmb.2004.08.083. [DOI] [PubMed] [Google Scholar]
- 34.Arbely E, Arkin IT. Experimental measurement of the strength of a Calpha-H…O bond in a lipid bilayer. J Am Chem Soc. 2004;126:5362–5363. doi: 10.1021/ja049826h. [DOI] [PubMed] [Google Scholar]
- 35.Mottamal M, Lazaridis T. The contribution of Calpha-H…O hydrogen bonds to membrane protein stability depends on the position of the amide. Biochemistry. 2005;44:1607–1613. doi: 10.1021/bi048065s. [DOI] [PubMed] [Google Scholar]
- 36.Arkin IT, Brunger AT. Statistical analysis of predicted transmembrane alpha-helices. Biochim Biophys Acta. 1998;1429:113–128. doi: 10.1016/s0167-4838(98)00225-8. [DOI] [PubMed] [Google Scholar]
- 37.Smith SO, Smith CS, Bormann BJ. Strong hydrogen bonding interactions involving a buried glutamic acid in the transmembrane sequence of the neu/erbB-2 receptor. Nat Struct Biol. 1996;3:252–258. doi: 10.1038/nsb0396-252. [DOI] [PubMed] [Google Scholar]
- 38.Bocharov EV, Mayzel ML, Volynsky PE, Goncharuk MV, Ermolyuk YS, Schulga AA, et al. Spatial structure and pH-dependent conformational diversity of dimeric transmembrane domain of the receptor tyrosine kinase EphA1. J Biol Chem. 2008;283:29385–29395. doi: 10.1074/jbc.M803089200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moore DT, Berger BW, DeGrado WF. Protein-protein interactions in the membrane: sequence, structural and biological motifs. Structure. 2008;16:991–1001. doi: 10.1016/j.str.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lau TL, Kim C, Ginsberg MH, Ulmer TS. The structure of the integrin aIIbβ3 transmembrane complex explains integrin transmembrane signalling. EMBO J. 2009;28:1351–1361. doi: 10.1038/emboj.2009.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wallin E, Tsukihara T, Yoshikawa S, von Heijne G, Elofsson A. Architecture of helix bundle membrane proteins: an analysis of cytochrome c oxidase from bovine mitochondria. Protein Sci. 1997;6:808–815. doi: 10.1002/pro.5560060407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adamian L, Nanda V, DeGrado WF, Liang J. Empirical lipid propensities of amino acid residues in multispan alpha helical membrane proteins. Proteins. 2005;59:496–509. doi: 10.1002/prot.20456. [DOI] [PubMed] [Google Scholar]
- 43.Peng WC, Lin X, Torres J. The strong dimerization of the transmembrane domain of the fibroblast growth factor receptor (FGFR) is modulated by C-terminal juxtamembrane residues. Protein Sci. 2009;18:450–459. doi: 10.1002/pro.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Johnson RM, Hecht K, Deber CM. Aromatic and cation-pi interactions enhance helix-helix association in a membrane environment. Biochemistry. 2007;46:9208–9214. doi: 10.1021/bi7008773. [DOI] [PubMed] [Google Scholar]
- 45.Unterreitmeier S, Fuchs A, Schäffler T, Heym RG, Frishman D, Langosch D. Phenylalanine promotes interaction of transmembrane domains via GxxxG motifs. J Mol Biol. 2007;374:705–718. doi: 10.1016/j.jmb.2007.09.056. [DOI] [PubMed] [Google Scholar]
- 46.Sal-Man N, Gerber D, Bloch I, Shai Y. Specificity in transmembrane helix-helix interactions mediated by aromatic residues. J Biol Chem. 2007;282:19753–19761. doi: 10.1074/jbc.M610368200. [DOI] [PubMed] [Google Scholar]
- 47.Walters RFS, DeGrado WF. Helix-packing motifs in membrane proteins. Proc Natl Acad Sci USA. 2006;103:13658–13663. doi: 10.1073/pnas.0605878103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Langosch D, Lindner E, Gurezka R. In vitro selection of self-interacting transmembrane segments—membrane proteins approached from a different perspective. IUBMB Life. 2002;54:109–113. doi: 10.1080/15216540214538. [DOI] [PubMed] [Google Scholar]
- 49.Kim S, Jeon TJ, Oberai A, Yang D, Schmidt JJ, Bowie JU. Transmembrane glycine zippers: physiological and pathological roles in membrane proteins. Proc Natl Acad Sci USA. 2005;102:14279–14283. doi: 10.1073/pnas.0501234102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lupas A. Coiled coils: New structures and new functions. Trends Biochem Sci. 1996;21:375–382. [PubMed] [Google Scholar]
- 51.Lear JD, Stouffer AL, Gratkowski H, Nanda V, Degrado WF. Association of a model transmembrane peptide containing gly in a heptad sequence motif. Biophys J. 2004;87:3421–3429. doi: 10.1529/biophysj.103.032839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ruan W, Becker V, Klingmüller U, Langosch D. The interface between self-assembling erythropoietin receptor transmembrane segments corresponds to a membrane-spanning leucine zipper. J Biol Chem. 2004;279:3273–3279. doi: 10.1074/jbc.M309311200. [DOI] [PubMed] [Google Scholar]
- 53.North B, Cristian L, Fu Stowell X, Lear JD, Saven JG, Degrado WF. Characterization of a membrane protein folding motif, the Ser zipper, using designed peptides. J Mol Biol. 2006;359:930–939. doi: 10.1016/j.jmb.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 54.Langosch D, Arkin IT. Interaction and conformational dynamics of membrane-spanning protein helices. Protein Sci. 2009;18:1343–1358. doi: 10.1002/pro.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mackenzie KR. Folding and stability of alpha-helical integral membrane proteins. Chem Rev. 2006;106:1931–1977. doi: 10.1021/cr0404388. [DOI] [PubMed] [Google Scholar]
- 56.Johnson RM, Rath A, Deber CM. The position of the Gly-xxx-Gly motif in transmembrane segments modulates dimer affinity. Biochem Cell Biol. 2006;84:1006–1012. doi: 10.1139/o06-192. [DOI] [PubMed] [Google Scholar]
- 57.Zhang J, Lazaridis T. Transmembrane helix-helix association affinity can be modulated by flanking and noninterfacial residues. Biophys J. 2009;96:4418–4427. doi: 10.1016/j.bpj.2009.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Efremov RG, Nolde DE, Konshina AG, Syrtcev NP, Arseniev AS. Peptides and proteins in membranes: what can we learn via computer simulations? Curr Med Chem. 2004;11:2421–2442. doi: 10.2174/0929867043364496. [DOI] [PubMed] [Google Scholar]
- 59.Feig M, Brooks CL., 3rd Recent advances in the development and application of implicit solvent models in biomolecule simulations. Curr Opin Struct Biol. 2004;14:217–224. doi: 10.1016/j.sbi.2004.03.009. [DOI] [PubMed] [Google Scholar]
- 60.Forrest LR, Sansom MS. Membrane simulations: bigger and better? Curr Opin Struct Biol. 2000;10:174–181. doi: 10.1016/s0959-440x(00)00066-x. [DOI] [PubMed] [Google Scholar]
- 61.Sansom MS, Scott KA, Bond PJ. Coarse-grained simulation: a high-throughput computational approach to membrane proteins. Biochem Soc Trans. 2008;36:27–32. doi: 10.1042/BST0360027. [DOI] [PubMed] [Google Scholar]
- 62.Casciari D, Seeber M, Fanelli F. Quaternary structure predictions of transmembrane proteins starting from the monomer: a docking-based approach. BMC Bioinformatics. 2006;7:340. doi: 10.1186/1471-2105-7-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Efremov RG, Vereshaga YA, Volynsky PE, Nolde DE, Arseniev AS. Association of transmembrane helices: what determines assembling of a dimer? J Comput Aided Mol Des. 2006;20:27–45. doi: 10.1007/s10822-006-9034-6. [DOI] [PubMed] [Google Scholar]
- 64.Vereshaga YA, Volynsky PE, Nolde DE, Arseniev AS, Efremov RG. Helix interactions in membranes: lessons from unrestrained Monte Carlo simulations. J Chem Theory Comput. 2005;1:1252–1264. doi: 10.1021/ct0501250. [DOI] [PubMed] [Google Scholar]
- 65.Psachoulia E, Marshall DP, Sansom MS. Molecular dynamics simulations of the dimerization of transmembrane alpha-helices. Acc Chem Res. 2010;43:388–396. doi: 10.1021/ar900211k. [DOI] [PubMed] [Google Scholar]
- 66.Lemmon MA, Flangan JM, Treutlein HR, Zhang J, Engelman DM. Sequence specificity in the dimerization of transmembrane alpha-helices. Biochemistry. 1992;31:12719–12725. doi: 10.1021/bi00166a002. [DOI] [PubMed] [Google Scholar]
- 67.Melnyk RA, Partridge AW, Deber CM. Transmembrane domain mediated self-assembly of major coat protein subunits from Ff bacteriophage. J Mol Biol. 2002;31:63–72. doi: 10.1006/jmbi.2001.5214. [DOI] [PubMed] [Google Scholar]
- 68.Sulistijo ES, Jaszewski TM, MacKenzie KR. Sequence-specific dimerization of the transmembrane domain of the “BH3-only“ protein BNIP3 in membranes and detergent. J Biol Chem. 2003;278:51950–51956. doi: 10.1074/jbc.M308429200. [DOI] [PubMed] [Google Scholar]
- 69.Constantinescu SN, Keren T, Socolovsky M, Nam H, Henis YI, Lodish HF. Ligand-independent oligomerization of cell-surface erythropoietin receptor is mediated by the transmembrane domain. Proc Natl Acad Sci USA. 2001;98:4379–4384. doi: 10.1073/pnas.081069198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Scheuermann S, Hambsch B, Hesse L, Stumm J, Schmidt C, Beher D, et al. Homodimerization of amyloid precursor protein and its implication in the amyloidogenic pathway of Alzheimer’s disease. J Biol Chem. 2001;276:33923–33929. doi: 10.1074/jbc.M105410200. [DOI] [PubMed] [Google Scholar]
- 71.Gratkowski H, Dai Q, Wand AJ, DeGrado WF, Lear JD. Cooperativity and specificity of association of a designed transmembrane peptide. Biophys J. 2002;83:1613–1619. doi: 10.1016/S0006-3495(02)73930-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Im W, Feig M, Brooks CL., 3rd An implicit membrane generalized born theory for the study of structure, stability and interactions of membrane proteins. Biophys J. 2003;85:2900–2918. doi: 10.1016/S0006-3495(03)74712-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Petrache HI, Grossfield A, MacKenzie KR, Engelman DM, Woolf TB. Modulation of glycophorin A transmembrane helix interactions by lipid bilayers: molecular dynamics calculations. J Mol Biol. 2000;302:727–746. doi: 10.1006/jmbi.2000.4072. [DOI] [PubMed] [Google Scholar]
- 74.Li E, Merzlyakov M, Lin J, Searson P, Hristova K. Utility of surface-supported bilayers in studies of transmembrane helix dimerization. J Struct Biol. 2009;168:53–60. doi: 10.1016/j.jsb.2009.03.005. [DOI] [PubMed] [Google Scholar]
- 75.Schneider D, Finger C, Prodöhl A, Volkmer T. From interactions of single transmembrane helices to folding of alpha-helical membrane proteins: analyzing transmembrane helix-helix interactions in bacteria. Curr Protein Pept Sci. 2007;8:45–61. doi: 10.2174/138920307779941578. [DOI] [PubMed] [Google Scholar]
- 76.Adams PD, Engelman DM, Brünger AT. Improved prediction for the structure of the dimeric transmembrane domain of glycophorin A obtained through global searching. Proteins. 1996;26:257–261. doi: 10.1002/(SICI)1097-0134(199611)26:3<257::AID-PROT2>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 77.Smith SO, Eilers M, Song D, Crocker E, Ying W, Groesbeek M, et al. Implications of threonine hydrogen bonding in the glycophorin A transmembrane helix dimer. Biophys J. 2002;82:2476–2486. doi: 10.1016/S0006-3495(02)75590-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Smith SO, Song D, Shekar S, Groesbeek M, Ziliox M, Aimoto S. Structure of the transmembrane dimer interface of glycophorin A in membrane bilayers. Biochemistry. 2001;40:6553–6558. doi: 10.1021/bi010357v. [DOI] [PubMed] [Google Scholar]
- 79.Beevers AJ, Kukol A. The transmembrane domain of the oncogenic mutant ErbB-2 receptor: a structure obtained from site-specific infrared dichroism and molecular dynamics. J Mol Biol. 2006;361:945–953. doi: 10.1016/j.jmb.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 80.Vereshaga YA, Volynsky PE, Pustovalova JE, Nolde DE, Arseniev AS, Efremov RG. Specificity of helix packing in transmembrane dimer of the cell death factor BNIP3: a molecular modeling study. Proteins. 2007;69:309–325. doi: 10.1002/prot.21555. [DOI] [PubMed] [Google Scholar]
- 81.Bocharov EV, Pustovalova YE, Pavlov KV, Volynsky PE, Goncharuk MV, Ermolyuk YS, et al. Unique dimeric structure of BNip3 transmembrane domain suggests membrane permeabilization as a cell death trigger. J Biol Chem. 2007;282:16256–16266. doi: 10.1074/jbc.M701745200. [DOI] [PubMed] [Google Scholar]
- 82.Volynsky PE, Mineeva EA, Goncharuk MV, Ermolyuk YS, Arseniev AS, Efremov RG. Computer simulations and modeling-assisted ToxR screening in deciphering 3D structures of transmembrane α-helical dimers: ephrin receptor A1. Phys Biol. 2010;7:16014. doi: 10.1088/1478-3975/7/1/016014. [DOI] [PubMed] [Google Scholar]
- 83.Metcalf DG, Kulp DW, Bennett JS, DeGrado WF. Multiple approaches converge on the structure of the integrin aIIbβ3 transmembrane heterodimer. J Mol Biol. 2009;392:1087–1101. doi: 10.1016/j.jmb.2009.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhu J, Luo BH, Barth P, Schonbrun J, Baker D, Springer TA. The structure of a receptor with two associating transmembrane domains on the cell surface: integrin aIIbβ3. Mol Cell. 2009;34:234–249. doi: 10.1016/j.molcel.2009.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wüthrich K. NMR of Proteins and Nucleic Acids. New York: Wiley,; 1986. [Google Scholar]
- 86.Opella SJ, Marassi FM. Structure determination of membrane proteins by NMR spectroscopy. Chem Rev. 2004;104:3587–3606. doi: 10.1021/cr0304121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Andronesi OC, Becker S, Seidel K, Heise H, Young HS, Baldus M. Determination of membrane protein structure and dynamics by magic-angle-spinning solid-state NMR spectroscopy. J Am Chem Soc. 2005;127:12965–12974. doi: 10.1021/ja0530164. [DOI] [PubMed] [Google Scholar]
- 88.Sato T, Tang T, Reubins G, Fei JZ, Taiki F, Kienlen-Campar P, et al. A helix-to-coil transition at the ɛ-cut site in the transmembrane dimer of the amyloid precursor protein is required for proteolysis. Proc Natl Acad Sci USA. 2008;106:1421–1426. doi: 10.1073/pnas.0812261106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Smith SO, Smith C, Shekar S, Peersen O, Ziliox M, Aimoto S. Transmembrane interactions in the activation of the Neu receptor tyrosine kinase. Biochemistry. 2002;41:9321–9332. doi: 10.1021/bi012117l. [DOI] [PubMed] [Google Scholar]
- 90.Pervushin K, Riek R, Wider G, Wüthrich K. Attenuated T2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc Natl Acad Sci USA. 1997;94:12366–12371. doi: 10.1073/pnas.94.23.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kim HJ, Howell SC, Van Horn WD, Jeon YH, Sanders CR. Recent advances in the application of solution NMR spectroscopy to multi-span integral membrane proteins. Prog Nucl Magn Reson Spectrosc. 2009;55:335–360. doi: 10.1016/j.pnmrs.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gautier A, Kirkpatrick JP, Nietlispach D. Solution-state NMR spectroscopy of a seven-helix transmembrane protein receptor: backbone assignment, secondary structure and dynamics. Angew Chem Int Ed Engl. 2008;47:7297–7300. doi: 10.1002/anie.200802783. [DOI] [PubMed] [Google Scholar]
- 93.Krueger-Koplin RD, Sorgen PL, Krueger-Koplin ST, Rivera-Torres IO, Cahill SM, Hicks DB, et al. An evaluation of detergents for NMR structural studies of membrane proteins. J Biomol NMR. 2004;28:43–57. doi: 10.1023/B:JNMR.0000012875.80898.8f. [DOI] [PubMed] [Google Scholar]
- 94.Sanders CR, Sönnichsen F. Solution NMR of membrane proteins: practice and challenges. Magn Reson Chem. 2006;44:24–40. doi: 10.1002/mrc.1816. [DOI] [PubMed] [Google Scholar]
- 95.Page RC, Moore JD, Nguyen HB, Sharma M, Chase R, Gao FP, et al. Comprehensive evaluation of solution nuclear magnetic resonance spectroscopy sample preparation for helical integral membrane proteins. J Struct Funct Genomics. 2006;7:51–64. doi: 10.1007/s10969-006-9009-9. [DOI] [PubMed] [Google Scholar]
- 96.Maslennikov I, Kefala G, Johnson C, Riek R, Choe S, Kwiatkowski W. NMR spectroscopic and analytical ultracentrifuge analysis of membrane protein detergent complexes. BMC Struct Biol. 2007;7:74. doi: 10.1186/1472-6807-7-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lindberg M, Biverståhl H, Gräslund A, Mäler L. Structure and positioning comparison of two variants of penetratin in two different membrane mimicking systems by NMR. Eur J Biochem. 2003;270:3055–3063. doi: 10.1046/j.1432-1033.2003.03685.x. [DOI] [PubMed] [Google Scholar]
- 98.Chou JJ, Kaufman JD, Stahl SJ, Wingfield PT, Bax A. Micelle-induced curvature in a water-insoluble HIV-1 Env peptide revealed by NMR dipolar coupling measurement in stretched polyacrylamide gel. J Am Chem Soc. 2002;124:2450–2451. doi: 10.1021/ja017875d. [DOI] [PubMed] [Google Scholar]
- 99.Bordag N, Keller S. Alpha-helical transmembrane peptides: a “divide and conquer” approach to membrane proteins. Chem Phys Lipids. 2010;163:1–26. doi: 10.1016/j.chemphyslip.2009.07.009. [DOI] [PubMed] [Google Scholar]
- 100.Sulistijo ES, Mackenzie KR. Structural basis for dimerization of the BNIP3 transmembrane domain. Biochemistry. 2009;48:5106–5120. doi: 10.1021/bi802245u. [DOI] [PubMed] [Google Scholar]
- 101.Melnyk RA, Partridge AW, Deber CM. Retention of native-like oligomerization states in transmembrane segment peptides: application to the Escherichia coli aspartate receptor. Biochemistry. 2001;40:11106–11113. doi: 10.1021/bi010642e. [DOI] [PubMed] [Google Scholar]
- 102.Therien AG, Deber CM. Oligomerization of a peptide derived from the transmembrane region of the sodium pump gamma subunit: effect of the pathological mutation G41R. J Mol Biol. 2002;322:550–583. doi: 10.1016/s0022-2836(02)00781-7. [DOI] [PubMed] [Google Scholar]
- 103.Borch J, Hamann T. The nanodisc: a novel tool for membrane protein studies. Biol Chem. 2009;390:805–814. doi: 10.1515/BC.2009.091. [DOI] [PubMed] [Google Scholar]
- 104.Nath A, Atkins WM, Sligar SG. Applications of phospholipid bilayer nanodiscs in the study of membranes and membrane proteins. Biochemistry. 2007;46:2059–2069. doi: 10.1021/bi602371n. [DOI] [PubMed] [Google Scholar]
- 105.Ritchie TK, Grinkova YV, Bayburt TH, Denisov IG, Zolnerciks JK, Atkins WM, et al. Reconstitution of membrane proteins in phospholipid bilayer nanodiscs. Methods Enzymol. 2009;464:211–231. doi: 10.1016/S0076-6879(09)64011-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lyukmanova EN, Shenkarev ZO, Paramonov AS, Sobol AG, Ovchinnikova TV, Chupin VV, et al. Lipid-protein nanoscale bilayers: a versatile medium for NMR investigations of membrane proteins and membrane-active peptides. J Am Chem Soc. 2008;130:2140–2141. doi: 10.1021/ja0777988. [DOI] [PubMed] [Google Scholar]
- 107.Riek R, Wider G, Pervushin K, Wüthrich K. Polarization transfer by cross-correlated relaxation in solution NMR with very large molecules. Proc Natl Acad Sci USA. 1999;96:4918–4923. doi: 10.1073/pnas.96.9.4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Shenkarev ZO, Lyukmanova EN, Paramonov AS, Shingarova LN, Chupin VV, Kirpichnikov MP, et al. Lipid-Protein nanodiscs as reference medium in detergent screening for high-resolution NMR studies of integral membrane proteins. J Am Chem Soc. 2010;132:5628–5629. doi: 10.1021/ja9097498. [DOI] [PubMed] [Google Scholar]
- 109.Borch J, Hamann T. The nanodisc: a novel tool for membrane protein studies. Biol Chem. 390:805–814. doi: 10.1515/BC.2009.091. 200. [DOI] [PubMed] [Google Scholar]
- 110.Poget SF, Girvin ME. Solution NMR of membrane proteins in bilayer mimics: small is beautiful, but sometimes bigger is better. Biochim Biophys Acta. 2007;1768:3098–3106. doi: 10.1016/j.bbamem.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Vold RR, Prosser RS, Deese AJ. Isotropic Solutions of phospholipid micelles: A new membrane mimetic for high-resolution NMR studies of polypeptides. J Biomol NMR. 1997;9:329–335. doi: 10.1023/a:1018643312309. [DOI] [PubMed] [Google Scholar]
- 112.Lee D, Walter KF, Brückner AK, Hilty C, Becker S, Griesinger C. Bilayer in small bicelles revealed by lipid-protein interactions using NMR spectroscopy. J Am Chem Soc. 2008;130:13822–13823. doi: 10.1021/ja803686p. [DOI] [PubMed] [Google Scholar]
- 113.Van Dam L, Karlsson G, Edwards K. Direct observation and characterization of DMPC/DHPC aggregates under conditions relevant for biological solution NMR. Biochim Biophys Acta. 2004;1664:241–256. doi: 10.1016/j.bbamem.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 114.Glover KJ, Whiles JA, Wu G, Yu N, Deems R, Struppe JO, et al. Structural evaluation of phospholipid bicelles for solution-state studies of membrane-associated biomolecules. Biophys J. 2001;81:2163–2171. doi: 10.1016/s0006-3495(01)75864-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Luchette PA, Vetman TN, Prosser RS, Hancock RE, Nieh MP, Glinka CJ, et al. Morphology of fast-tumbling bicelles: a small angle neutron scattering and NMR study. Biochim Biophys Acta. 2001;1513:83–94. doi: 10.1016/s0005-2736(01)00358-3. [DOI] [PubMed] [Google Scholar]
- 116.Prosser RS, Evanics F, Kitevski JL, Al-Abdul-Wahid MS. Current applications of bicelles in NMR studies of membrane-associated amphiphiles and proteins. Biochemistry. 2006;45:8453–8465. doi: 10.1021/bi060615u. [DOI] [PubMed] [Google Scholar]
- 117.Lind J, Nordin J, Mäler L. Lipid dynamics in fast-tumbling bicelles with varying bilayer thickness: effect of model transmembrane peptides. Biochim Biophys Acta. 2008;1778:2526–2534. doi: 10.1016/j.bbamem.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 118.Struppe J, Whiles JA, Vold RR. Acidic phospholipid bicelles: a versatile model membrane system. Biophys J. 2000;78:281–289. doi: 10.1016/S0006-3495(00)76591-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lau TL, Partridge AW, Ginsberg MH, Ulmer TS. Structure of the integrin beta3 transmembrane segment in phospholipid bicelles and detergent micelles. Biochemistry. 2008;47:4008–4016. doi: 10.1021/bi800107a. [DOI] [PubMed] [Google Scholar]
- 120.Nilges M, O’Donoghue SI. Ambiguous NOEs and automated NOE assignment. Prog Nucl Magn Reson Spectrosc. 1998;32:107–139. [Google Scholar]
- 121.Call ME, Schnell JR, Xu C, Lutz RA, Chou JJ, Wucherpfennig KW. The structure of the zetazeta transmembrane dimer reveals features essential for its assembly with the T cell receptor. Cell. 2006;127:355–368. doi: 10.1016/j.cell.2006.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Tugarinov V, Kay LE. Methyl groups as probes of structure and dynamics in NMR studies of high-molecular-weight proteins. Chembiochem. 2005;6:1567–1577. doi: 10.1002/cbic.200500110. [DOI] [PubMed] [Google Scholar]
- 123.Zwahlen C, Legault P, Vincent SJF, Greenblatt J, Konrat R, Kay LE. Methods for Measurement of Intermolecular NOEs by Multinuclear NMR Spectroscopy: Application to a Bacteriophage λ N-Peptide/boxB RNA Complex. J Am Chem Soc 1. 1997;119:6711–6721. [Google Scholar]
- 124.Stuart AC, Borzilleri KA, Withka JM, Palmer AG., 3rd Compensating for variations in 1H-13C scalar coupling constants in isotope-filtered NMR experiments. J Am Chem Soc. 1999;121:5346–5347. [Google Scholar]
- 125.Chen G, Cizeau J, Vande Velde C, Park JH, Bozek G, Bolton J, et al. Nix and Nip3 form a subfamily of pro-apoptotic mitochondrial proteins. J Biol Chem. 1999;274:7–10. doi: 10.1074/jbc.274.1.7. [DOI] [PubMed] [Google Scholar]
- 126.Pasquale EB. Eph receptor signalling casts a wide net on cell behaviour. Nat Rev Mol Cell Biol. 2005;6:462–475. doi: 10.1038/nrm1662. [DOI] [PubMed] [Google Scholar]
- 127.Bocharov EV, Mayzel ML, Mineev KS, Tkach EN, Ermolyuk YS, Schulga AA, et al. Left-handed dimer of EphA2 transmembrane domain: helix packing diversity among receptor tyrosine kinases. Biophysical J. 2010;98:881–889. doi: 10.1016/j.bpj.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Warren CM, Landgraf R. Signaling through ERBB receptors: multiple layers of diversity and control. Cell Signal. 2006;18:923–933. doi: 10.1016/j.cellsig.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 129.Tao RH, Maruyama IN. All EGF (ErbB) receptors have preformed homo- and heterodimeric structures in living cells. J Cell Sci. 2008;121:3207–3217. doi: 10.1242/jcs.033399. [DOI] [PubMed] [Google Scholar]
- 130.Bocharov EV, Mineev KS, Volynsky PE, Ermolyuk YS, Tkach EN, Sobol AG, et al. Spatial structure of the dimeric transmembrane domain of the growth factor receptor ErbB2 presumably corresponding to the receptor active state. J Biol Chem. 2008;283:6950–6956. doi: 10.1074/jbc.M709202200. [DOI] [PubMed] [Google Scholar]
- 131.Jura N, Endres NF, Engel K, Deindl S, Das R, Lamers MH, et al. Mechanism for activation of the EGF receptor catalytic domain by the juxtamembrane segment. Cell. 2009;137:1293–1307. doi: 10.1016/j.cell.2009.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Efremov RG, Vergoten G. Hydrophobic nature of membrane-spanning α-helical peptides as revealed by Monte Carlo simulations and molecular hydrophobicity potential analysis. J Phys Chem. 1995;99:10658–10666. [Google Scholar]