Abstract
Among the many transmembrane receptor classes, the receptor tyrosine kinases represent an important superfamily, involved in many cellular processes like embryogenesis, development and cell division. Deregulation and dysfunctions of these receptors can lead to various forms of cancer and other diseases. Mostly, only fragmented knowledge exists about functioning of the entire receptors, and many studies have been performed on isolated receptor domains. In this review we focus on the function of the ErbB family of receptor tyrosine kinases with a special emphasis on the role of the transmembrane domain and on the mechanisms underlying regulated and deregulated signaling. Many general aspects of ErbB receptor structure and function have been analyzed and described. All human ErbBs appear to form homo- and heterodimers within cellular membranes and the single transmembrane domain of the receptors is involved in dimerization. Additionally, only defined structures of the transmembrane helix dimer allows signaling of ErbB receptors.
Key words: ErbB, EGFR, receptor, receptor-tyrosine kinase, transmembrane proteins, signaling, helix-helix interaction
How Do Cells “Get the Message”?
From simple prokaryotes to complex eukaryotes, signal transduction in between cells is vital to organize populations of individual cells up to extremely complex multicellular organisms. Cellular signaling not only allows cells to communicate with each other but also allows cells to sense their inanimate environment and to interact and communicate with it. Signals arriving at a certain cell can be highly diverse and can range from single photons or mere physical force to ions and complex signaling molecules that have been produced by other cells. The extreme diversity of signals arising from or reaching a cell has led to a similar complexity on the part of the entities that recognize them—the receptors. Although individual signals can be rather diverse, all signals typically have to cross a cellular membrane, which separates the extracellular milieu from the more ordered intracellular environment. Some of the incoming signals are able to cross a membrane on their own, such as photons or hydrophobic signaling molecules. However, since a biological membrane does not allow the passage of charged or large hydrophilic molecules per se, cells have evolved a diverse repertoire of membrane integral receptors, which either selectively enable a signal to cross a membrane or convert the information of an extracellular signal into a different (intracellular) form that is then subsequently processed by a cell.
Since many diseases are directly related to a dysfunction of such membrane integral receptors and the resulting successive transduction of misinformation,1 the principles guiding receptor function have received much attention in the past decades. An intimate knowledge about the structure and function of transmembrane (TM) receptors may offer the opportunity to manipulate disturbed cellular signaling processes by the development of new and advanced pharmaceuticals.
This review focuses on the question how eukaryotic cell surface receptors transduce incoming signals to the intracellular space via their transmembrane domains (TMDs) with a special emphasis on the role of the TMDs in signaling by members of the tyrosine kinase receptor superfamily, the epidermal growth factor receptors (EGFR), also known as HER or ErbB receptors.
Principles of Transmembrane Signaling
Membrane proteins make up to 30% of all genes in pro- and eukaryotes2 and about 50% of all currently available drugs target membrane integral proteins, such as cell surface receptors.3
Enzyme-linked receptors are α-helical membrane proteins that often consist of an extracellular ligand binding domain, a TMD and an intracellular domain, which mediates downstream signal transduction within a cell. Based on the enzymatic activity linked to the receptor, six types of enzyme-linked TM receptors are primarily distinguished:4 (1) Receptor guanylyl cyclases; (2) receptor serine/threonine kinases; (3) histidine kinase associated receptors; (4) receptor-like tyrosine phosphatases; (5) tyrosine kinase associated receptors and (6) receptor tyrosine kinases (RTKs).
Although the functional implications are largely elusive, some of these receptors are suggested to form homo- and/or heterooligomers in the presence or the absence of a ligand, and it has already been shown in some cases that individual TMDs can contribute to oligomerization (as further outlined below). Ligand binding to a receptor is assumed to result in a structural re-arrangement of a signaling incompetent inactive to an active state. A structural rearrangement in a signal receiver domain is transmitted across the membrane by the TMD resulting in receptor activation and downstream signaling events. But how are the signals transmitted across a membrane by a single TM segment?
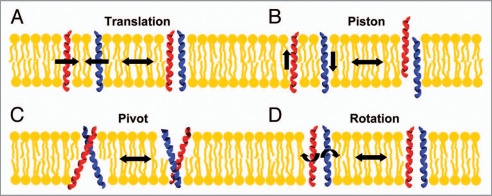
The exact motions of individual TM helices occurring during signal transduction processes are still only rudimentarily understood but TM signal transduction must involve changes in the topography of individual TMDs with respect to the membrane and to each other. Four kinds of potential movements of individual TM helices were recently described,5,6 which could be involved in TM signaling without counteracting restrictions defined by the lipid bilayer (see Fig. 1). (1) Individual helices can move in the membrane plane and form transient interactions (translational motion) and such interactions could involve defined interaction motifs (see below). (2) Individual TM helices can move perpendicular to the membrane (piston motion). (3) Transverse helix movements along an axis parallel to the membrane (pivot motion) result in tilting of individual helices or in changes in the tilt angle. (4) Rotation of the helices along an axis perpendicular to the membrane results in a reorientation of intra- and extracellular domains, as do all movements described above. Structural reorientations of the soluble domains are key for signal transduction across a membrane.
Figure 1.
Principles of TM signaling. (A) Two TM helices can form transient interactions in a membrane (translational motion), which results in interactions of receptor intra- and extracellular domains. (B) Movements of interacting TM helices perpendicular to the membrane plane (piston motion) result in structural changes in the contacts between intra- and extracellular domains. (C) A change in the crossing angle of two TM helices (pivot motion) also leads to a reorientation of water soluble receptor domains. (D) Rotations around the helix axis of two interacting TM helices lead to a rotation of the outer-membrane regions and positions the soluble domains differently. All these motions of TM helices would enable the transfer of a signal from one to the other surface of a membrane.
Structural changes in signal receiver domains, which are induced by ligand binding, subsequently activate signaling cascades at the other side of the membrane, which can be highly diverse. Often, the simply stunning complexity of the triggered signaling network and its spatio-temporal organization in the cell can only be processed and illustrated with computer built models.7 Nevertheless, due to various technical difficulties associated with analyses of the structure and function of membrane proteins,8 even the principles governing signal transduction via simple single-span TM proteins are only poorly understood today. Because of the many technical problems associated with the analyses of TM receptors (and TM proteins in general), often only isolated receptor domains are characterized rather than the full length receptor. However, research on isolated receptor parts only leads to a fragmented knowledge about the receptor function, and it typically remains an open question if the isolated domains would behave the same in the full length receptor context in vivo.
In the following paragraphs we will briefly discuss advances in understanding the function of different families of TM receptors with a single TM α-helix. Information about the functioning of other single-span TM receptors, which have been characterized to some extent in the past, are very helpful to understand and evaluate the principles governing human ErbB receptor function. Remarkably, the principles underlying signal transduction via human ErbB receptors (and human RTKs in general) appear to be similar to the principles guiding signal transduction via other single-span TM receptors, such as cadherins or the Epo receptor.
Examples of Transmembrane Signaling: Single-Spanning Transmembrane Receptors
In the few cases where detailed structural information is available about the TMDs of single-spanning TM receptors, these structures are devoid of other domains, and the in vivo mechanisms can only be deduced by a complementary approach where different studies with individual domains are compared and combined. Typically, it is essentially not understood how individual members of receptor families with a single TM helix transmit signals across a membrane via the TMD, but the following examples highlight two representatives where the mechanism of signal transduction is understood to some extent.
The role of the TMD in functioning of human cadherins, where an intracellular ligand binding event translates to cell-cell adhesion, is partly understood. Classical or type 1 cadherins represent single spanning TM glycoproteins that mediate cell-cell contacts in a calcium dependent manner.9 Cadherin induced tissue formation depends on the clustering of cells in a regulated and defined way. Proteins of the classical cadherin subfamily participate in adherent junctions and desmosomes, and they all possess conserved extracellular cadherin domains. Expression of individual cadherins is tissue specific and cadherins are involved in adhering cells to form and maintain a tissue during and after embryogenesis. Furthermore, cadherins appear to also be involved in certain forms of cancer (reviewed in ref. 10). Cadherins usually consist of an extracellular calcium binding domain, a single TM α-helix and an unstructured intracellular domain. The extracellular domain is unable to facilitate cell-cell adhesion unless calcium is present, and the intracellular domain is interacting with members of the catenin family of proteins, which facilitate interactions with the cytoskeleton.11 If the intracellular part is activated by interaction with catenins and the cytoskeleton,12 a signal is transferred to the extracellular domain resulting in lateral dimerization of the cadherins (translational movement, Fig. 1) and in subsequent cell-cell adhesion, which involves formation of antiparallel tetramers composed of two dimers from adjacent cells.13 Interestingly, removal of the intracellular domains results in lateral dimer formation of cadherins and in subsequent cellular adhesion.14 Therefore, the intracellular domains appear to negatively regulate the adhesion event and prevent the rest of the protein from dimer formation. How this negative regulation is achieved and which receptor part of the remaining protein drives dimerization is, however, unknown. The extracellular cadherin domain alone has an intrinsic tendency to form dimers in solution.15 However, the single TM α-helix of cadherins is able to form oligomers in vivo16 and is essential for dimerization of truncated cadherin variants.17,18 Thus, interactions of the single TM α-helix are critical for cadherin signaling and the intracellular cadherin domain prevents individual cadherins from dimer formation, promoted by defined TM helix-helix interactions.
The human erythropoietin receptor (EpoR) is a representative of the family of human type one cytokine receptors. The EpoR is activated by erythropoietin (Epo) and is involved in survival of red blood cells progenitors.19 Structurally, the receptor consists of an extracellular Epo-binding domain, a single TM helix and an intracellular domain,20 which is associated with a Janus kinase that mediates downstream signal transduction via the Jak-STAT pathway.21 Although only the structure of the extracellular EpoR domain with a bound Epo-ligand has been solved,22 the potential involvement of the TMD in signal transduction is also described. The EpoR exists as a preformed inactive dimer,23 and interactions of the TMDs are involved in dimerization.24 Upon Epo binding the receptor is activated and the activation process involves reorientation of the extracellular domains,22 which might translate to structural changes in the TMD. In the TMD sequence of the EpoR two polar residues are located one residue apart from each other. Since individual polar residues are known to stabilize TM helix-helix interactions,25,26 it appears to be a possibility that one residue is involved in stabilizing a signaling competent active state, whereas the other stabilizes an inactive state. An Epo induced conformational change in the extracellular domain could then lead to a shift from one TM helix structure to another. This could then subsequently result in reorientation of the intracellular associated Janus kinase in a way that permits receptor phosphorylation (rotational or pivot movement, Fig. 1). Noteworthy, signal transduction via the EpoR can be disturbed by the viral TM protein Gp55.27 The gp55-P protein of the spleen focus forming virus (SFFV) is able to activate the murine EpoR via its TMD but not the human counterpart unless a single leucine residue (Leu238) is mutated to a serine.28 Gp55 directly interacts with the TMD of the murine EpoR and thereby deregulates receptor function, resulting in erythroleukemia. This observation suggests that TM helix-helix interactions are critically involved in signal transduction via the EpoR, as described previously for cadherins. Also in the case of the EpoR interactions of soluble domains appear to sterically hinder defined interactions between the TMDs and only upon ligand binding the TM α-helices can form a TM helix dimer structure, which promotes signal transduction across the membrane. As discussed in the following paragraph, this basic principle, which is similar for cadherins and the EpoR, appears to also control functioning of human receptor tyrosine kinases, such as the ErbB receptors.
Receptor Tyrosine Kinases (RTKs)
While this review focuses on the family of human ErbB RTKs, we briefly introduce the entire superfamily of human RTKs. More details about this family are discussed in the complementary review article by K. Histrova and coworkers in this issue. It has to be noted that generalizing descriptions are often only valid for the majority of the family members since human RTKs comprise a very large and diverse family.
In the human genome 58 different RTKs are encoded which are classified into 20 subfamilies.29 RTKs generally consist of an N-terminal extracellular ligand binding domain, a single TM helix and an intracellular C-terminally located kinase domain. Upon ligand binding to the extracellular domain most RTKs are believed to dimerize, which positions the intracellular kinase domains in a way that allows transphosphorylation of defined tyrosine residues. Many RTKs form specific homo- and heterodimers, which highly diversifies downstream signaling events.30,31 But how is RTK dimerization achieved?
Different domains appear to be involved in dimerization of RTKs. In the insulin growth factor 1-receptor (IGF1R) the dimer is permanently stabilized by the formation of disulfide bridges in between two adjacent extracellular domains,32 and thus this receptor is always a dimer. Therefore, at least in case of the human IGF1R the ligand does not directly contribute to the formation of dimers by connecting two monomers. While one ligand binds to a IGF1R monomer and the ligand binding domains of two monomers are well separated in the IGF1R dimer, ligand binding to the extracellular domain was reported to be involved in dimerization of the fibroblast growth factor receptor (FGFR).33 However, since this study concentrates on the isolated ligand binding domain, the contributions of individual domains to the overall dimer stability cannot simply be deduced.
Noteworthy, while the IGF1R is a dimer, which is disulfide bridged in the extracellular domain, the isolated IGF1R TMDs additionally display a very strong interaction propensity and seem to have an important regulatory function in signal transduction.34,35 Therefore, defined interactions of the TMDs are most likely critically involved in IGF1R signaling, as it appears to be common for all members of the insulin receptor family.36–39 In a recent in vivo study the interaction propensities of the individual TMDs of all human RTKs have been determined.35 Importantly, all TMDs of human RTKs form stable TM oligomers, and this observation provides a framework for estimating potential contributions of RTK TMDs to dimer formation and stability. Defined interactions between RTK TMDs could generally be involved in RTK signaling. The involvement of the TM domains in signaling and dimerization of human epidermal growth factor receptors has been studied by various techniques in the recent years and will be discussed further below. Nevertheless, the individual studies on the interaction propensity of isolated RTK domains have to be complemented by interaction studies of full length receptors. Only studies in a full receptor will allow to properly describe the role of interactions between individual domains to the functional mechanism of the entire receptor. An approach to produce an active, membrane-spanning form of ErbB1 for subsequent structural and functional analyzes has recently been described.40
After signal transduction by the TMDs to the intracellular RTK domains, defined tyrosine residues are trans-phosphorylated by the intracellular kinase domains. Once phosphorylated, proteins containing Src homology region 2 (SH2) or phosphotyrosine binding (PTB) domains specifically recognize the phosphotyrosine residues of the intracellular RTK domain and mediate diverse downstream signaling events.41 Proteins containing a SH2 or PTB domain and interacting with phosphorylated RTKs belong to various protein classes, such as enzymes, docking proteins, transcription factors or regulators.
ErbB Receptors
One of the 20 families of receptor tyrosine kinases,29 which has been investigated in greater detail in the recent years, is the family of epidermal growth factor receptors (EGFRs), also known as HERs or ErbBs. In humans, this family contains the four proteins: ErbB1 (a.k.a. EGFR or HER1),42 ErbB2 (a.k.a. HER2), ErbB3 (a.k.a. HER3) and ErbB4 (a.k.a. HER4).43
The overall structure of the ErbB family members is similar to the structures of other RTK families. ErbB receptors are bitopic membrane proteins that contain a short single spanning TMD and water soluble extra- and intracellular domains (Fig. 2). The structures of the extra- and intracellular soluble domains of ErbB proteins have been solved by X-ray crystallography.44–49 Ligand binding to the extracellular domain results in structural alterations in the extracellular domain, as well as in the TMD and in the intracellular domain. Different ligands can be involved in ErbB receptor activation, such as the epidermal growth factor (EGF), the transforming growth factor alpha (TGFα), the heparin-binding EGF-like growth factor (HB-EGF), amphiregulin, betacellulin, epigen, epiregulin (to ErbB1), as well as four different neuregulins, which bind to ErbB3 and/or ErbB4 (reviewed in ref. 50). Noteworthy, no ligand binding to ErbB2 has been identified so far. Ligands can bind to preformed ErbB1 dimers51,52 and also for all other ErbB receptors preformed homo- and heterodimers have been observed in vivo.53 In general, heterooligomerization of individual ErbB receptors greatly diversifies signaling events of ErbB receptors.
Figure 2.
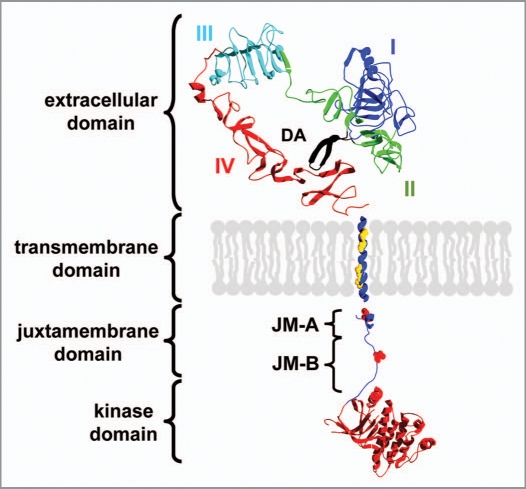
Domain structure of ErbB receptors. The extracellular ligand binding domain is composed of four subdomains (I–IV) and the dimerization arm (DA). Upon ligand binding to domain I and III, the tether between domain II and IV is released leading to exposure of the dimerization arm. The extracellular domain of ErbB1 is shown (pdb code: 1nql). The structure of the ErbB2 TMD was used and the position of two GxxxG-like motifs is highlighted (pdb code: 2jwa). The intracellular domain consists of the juxtamembrane domain and regulatory tyrosine residues are highlighted. The kinase domain is shown in red (pdb code: 3gop). The unstructured C-terminal domain is not shown.
While all ErbB receptors are able to form homooligomers, for ErbB2, homooligomerization is rather weak.54 Since no ligand appears to exist for ErbB2, this protein is the favored partner for ErbB heterodimer formation.55,56 ErbB3 lacks an active intracellular kinase domain,57 and thus signaling in a homooligomeric assembly would not be possible, which predestines also ErbB3 to participate in heterooligomeric assemblies.55
After ligand binding to the extracellular domain, structural changes in the intracellular domain, which contains the active site of the kinase, subsequently lead to phosphorylation of tyrosine residues in the juxtamembrane part and the C-terminal part of one ErbB monomer by the intracellular kinase domains of the adjacent monomer.58 Phosphorylation enables binding of defined adaptor proteins that bind specifically to phosphotyrosine residues and the signal is further relayed.41 These events will not be further discussed here and are described in more detail in several recent review articles (reviewed in refs. 31, 59 and 60).
In the next paragraphs we will follow signal progression from the ligand activated extracellular domain via the TMD to the intracellular domain. Which structural changes in the receptor domains transmit the information of ligand binding? How is the ErbB receptor activity regulated? How is the signaling process deregulated in ErbB associated diseases? And what is the specific role of the TMD in signal transduction by human ErbB receptors?
Signal Progression: The Extracellular Ligand Binding Domain
The X-ray crystallographic structures of the isolated extracellular domains have been solved for all human ErbB family members.44,46,48,49 It has been shown that the full-length ErbB1 receptor can exist as an inactive preformed dimer,52 and it was initially assumed that the other family members form a stable dimeric structure upon ligand binding,61 except ErbB2, for which no ligand has been identified to date.55 However, for all human ErbBs, preformed dimers have now been described in vivo.53 Indeed, in the ErbB1 receptor two ligand molecules bind independently to two receptors, and thus do not directly participate in dimer formation, which is seen in the crystal structure of the extracellular ErbB1 domain.44,62 While homodimerization of individual ErbB receptor extracellular domains has been shown, heterodimerization of extracellular ErbB domains is only weak or even undetectable in vitro.63 These findings suggest that other receptor domains, such as the TMD, could be crucial for heterodimerization.
Structurally, the extracellular domain can be divided into four subdomains (see Fig. 2), and the structural changes that occur in the extracellular domain upon ligand binding have been elucidated by comparing the structures of the ErbB1 extracellular domain with and without a bound ligand.44,64 Domains I and III are mainly involved in ligand binding, whereas domains II and IV form interactions in the absence of a ligand, resulting in a tethered conformation that separates the ligand binding domains of the inactive receptor.49 Although this interaction may only have a minor contribution to formation of the tethered conformation, this tether also exists in solution.65 However, a merely untethered state does not necessarily activate the receptor.66 In order to obtain a mechanistic understanding of extracellular domain movements upon ligand binding, it will be necessary to identify all restraints, which keep the extracellular domain in such an auto-inhibited tethered state.
Upon ligand binding to the ErbB1 extracellular domains of a preformed dimer, different patches on the ligand surface interact with domain I and III.44 The ligand thereby forms a bridge between these two domains and stabilizes a conformation, where the two domains are both in close proximity to each other and bound to the ligand. The involved structural changes disrupt interactions between domains II and IV resulting in formation of an extended or untethered receptor conformation. A region known as the dimerization arm44 is exposed after this structural rearrangement (Fig. 2 and DA), and dimer formation is strengthened by interaction of two such dimerization arms from two adjacent activated ErbB1 receptors. Whereas dimerization of ErbB receptors is facilitated by the dimerization arm in the extracellular domain, in case of ErbB3 and ErbB4 it was initially assumed that the ligand binds to both subunits of the dimer, resulting in defined interactions of the extracellular domains.62,67 However, recent data suggest a ligand binding model similar to ErbB1.46,49,65,68
No ligand appears to exist for the ErbB2 receptor, and the soluble extracellular domain of ErbB2 always resides in the extended state.48 However, since, for example, the ErbB2 extracellular domain is always present in an untethered conformation with little intracellular kinase activity, even an untethered extracellular ErbB domain appears to be insufficient for signal transduction to the intracellular part. The extracellular domain of an invertebrate ErbB from the fruit fly Drosophila melanogaster (dEGFR) is, like ErbB2, also present in an untethered extended conformation in the absence of ligands, although, unlike for ErbB2, numerous ligands are described to activate this receptor.66 Thus, solely the formation of the untethered extracellular domain and exposure of the dimerization arm is insufficient to drive effective kinase signaling in the intracellular part. Auto-inhibition appears to be facilitated by another mechanism in dEGFR when compared to human ErbB receptors. A rearrangement of the juxtamembrane part of domain IV in the ErbB extracellular domain was also suggested to participate in signal transduction, but the exact mechanism remains unclear.69
Finally, a signal received at the extracellular surface of the membrane induces a complex signaling cascade within a cell and this cascade is triggered by an activation of the intracellular kinase domains.
Signal Progression: The Intracellular Domain
At the cytoplasmic side of the plasma membrane a juxtamembrane domain of about 40 amino acids follows the ErbB TMD. This region was only recently identified as an important factor in the kinase activation process.45,70 In contrast to other RTKs, the ErbB juxtamembrane region is involved in activation of the intracellular kinase domain and does not inhibit its activity.71 Furthermore, this region contains charged residues that are supposed to be attached to the charged lipid head groups of the cell membrane.45,72 The small juxtamembrane domain is divided into approximately 20 residue N-terminal JM-A and C-terminal JM-B subdomains (Fig. 2). The JM-A subdomains of two adjacent ErbB receptors form an antiparallel coiled-coil structure and thereby this subdomain also participates in subunit interactions and receptor dimerization.73,74 The JM-B region contains two tyrosine residues that can be phosphorylated and appear to have a regulatory function75 (highlighted in Fig. 2).
The juxtamembrane region is followed by the kinase domain. While initially a symmetric orientation of the active kinase domains was assumed, recent structural and biochemical studies suggest an asymmetric head to tail orientation of the kinase domains in the activated dimer.45,70,73 Due to this asymmetry, the two kinase domains in a dimer can have different functions: the kinase domain of one monomer is termed the receiver kinase and is activated by the activator kinase of the other monomer. The kinases are supposed to switch between the two positions in order to activate each other via transphosphorylation.73 The unstructured juxtamembrane JM-B subdomain from the receiver kinase was also shown to form interactions with the C-terminus of the kinase domain of the activator kinase.47
In ErbB1, the C-terminal tail succeeding the kinase domain is about 225 amino acids long. This domain contains tyrosine residues, which become phosphorylated upon receptor activation and serve as docking sites for phosphotyrosine binding proteins. Upon activation, the intracellular ErbB kinase domains use the γ phosphate group from an ATP molecule for transphosphorylation of the receiver kinase. Upon transphosphorylation of the activated receptors, signaling factors that contain SH2 or PTB domains bind to these phosphorylated sites and further relay the signal.41 SH2 domain containing proteins occur in enzymes, adaptors, scaffold proteins, signal regulators and transcription factors. PTB containing proteins, on the other hand, serve as a platform for the binding of many downstream signaling molecules. The functions of all these downstream effectors are very diverse like proliferation, cell migration and adhesion. These events are described in great detail in other reviews (reviewed in refs. 41 and 58) and will not be further discussed here.
Signal Progression: A Potential Role of the TMD
Initially, the TMD of ErbB receptors were regarded as a short protein domain that only anchors the receptor in the cell membrane, and this domain was often omitted at all from early considerations regarding receptor function. The individual TM α-helices were considered to connect the extracellular domains of ErbB receptors with the intracellular kinase domains and activation was suggested to involve a translational movement of individual, monomeric RTKs without a direct involvement of the TMDs. However, this simple model has already been challenged by the observation that the IGF1R forms a disulfide linked preformed dimer. Reports indicating the presence of preformed inactive ErbB1 dimers52 and of chimeric variants of ErbB2 that form strong dimers but are inactive in vivo76 favor a more flexible mechanism, which involves an interplay between the soluble domains and the TMD in receptor activation. Thus, rearrangements of preformed, dimeric ErbB structures could be involved in ErbB signaling. Furthermore, a potential role of the ErbB TM domains has been emphasized when it was demonstrated that the TMDs of ErbB receptors are vital for the receptor function and can even be involved in receptor dysfunctions leading to diseases (as further discussed below). Interestingly, truncation of the extracellular domain of human ErbB receptors often leads to constitutively activated ErbB dimers, even in the absence of a ligand.77,78 This indicates that the extracellular domain exerts an auto-inhibitory function, as described above for cadherins and the EpoR, and emphasizes a critical function of the TMDs in receptor activation and signal transduction. In addition, introduction of a flexible linker between the extracellular domain and the TMD activates the receptor,69 which suggests that the autoinhibitory effect of the extracellular domain is thereby uncoupled from the remaining receptor domains.45 Ligand binding has been shown to induce homodimerization of the ErbB1 extracellular domains in detergent, although dimerization was far more efficient in the additional presence of the ErbB1 TMD,79 which further suggests an important role of the TMDs in ErbB receptor function. The isolated TMDs of human ErbBs can form homodimers as well as heterodimers,80–82 and it has been suggested that defined TM helix-helix interactions are involved in ErbB signaling. In a recent in vitro analysis of ErbB1-4 TM peptide homo- and heterooligomerization a dimerization hierarchy for ErbB receptor homo- and heterooligomerization was proposed, solely based on the observed interaction tendencies of the isolated TMDs.82 However, it has to be mentioned that due to technical reasons some amino acids of the peptides were mutated when compared to the original human TMD sequence. Furthermore, since synthetic peptides were analyzed in detergent micelles, potential contributions of different interaction motifs within the ErbB TMD for dimerization were not analyzed or defined (as further discussed below). Peptides corresponding to the ErbB1 and ErbB2 TMDs, respectively, are able to interrupt signaling of the full length ErbB receptor in living cells,34,83,84 most likely by interfering with the ErbB TMD of the full length receptor.
Taken together, these observations strongly suggest that defined interactions of the TM domains are involved in ErbB signaling in vivo.
TM Helix-Helix Interactions Can Involve Defined Interaction Motifs
In recent years the TMDs of membrane proteins have received increasing attention since mutations in TMDs are often related to diseases and are involved in many functional aspects of membrane proteins.1,85
A simple folding pathway of membrane proteins has been suggested about two decades ago to involve two steps.86 According to this model, individual TM helices integrate in a first stage independently into a membrane and subsequently interact with each other to form higher-ordered oligomeric structures in a second, independent step. Based on this simplifying model, interactions between TM α-helices are a key aspect in folding of integral membrane proteins, and interactions of individual transmembrane helices in a membrane environment are uncoupled from their membrane integration. However, in recent years many structures of α-helical membrane proteins have been solved and it became obvious that this simplistic model can not describe folding of more complex membrane proteins, which contain reentry loops or half-spanning TM helices.87 Nevertheless, the two-stage model still provides a useful framework for understanding interactions of individual single TM helices within membranes, as in the case of ErbB receptors.
In recent years the TM helix of human glycophorin A (GpA) became a paradigm for studying the second step of the two-stage model. The single GpA TM helix forms a rather stable, non-covalently associated homodimer, and dimerization is largely mediated by the seven residue motif L75 IxxGVxxGVxxT.87–89 Subsequent studies have shown that especially the GxxxG motif is important for dimerization of the GpA TM helices.89,90 While this motif has initially been identified to mediate dimerization of the GpA TM helix, it was later shown that the motif can in general create a framework for TM helix dimerization, and this motif was found to be highly overrepresented in TM α-helices.91,92 The two glycine side chains are roughly positioned on the same side of a helix, and thus the GxxxG motif leads to a void in the space occupied by the TM α-helix side chains. Two helices that contain such a motif can come in close proximity to each other, pack tightly and strong helix packing promotes further interactions between other residues of the two helices. Besides glycine, other small amino acids can also mediate helix-helix interactions in a more general (small)-xxx-(small) or GxxxG-like motif.80,93–95 However, the presence of a GxxxG-like motif does not necessarily stand for an interaction since the sequence context around a GxxxG(-like) motif is also highly important and can determine the actual strength of a given TM helix-helix interaction mediated by such a motif.96,97 Moreover, individual polar amino acids have been shown to drive TM helix-helix interactions and other amino acid motifs have been identified in recent years to mediate specific interactions of individual TM helices (reviewed in ref. 98). Nevertheless, the most prominent interaction motif still is the GxxxG motif, and GxxxG-like motifs have been identified in several cases in recent years to be involved in mediating and stabilizing defined TM helix-helix interactions. These studies as well as many others have shown that defined interactions in TM helices are structurally important.
The progresses of the recent years, which have advanced our global understanding of membrane protein folding as well as of the principles governing TM helix-helix interactions, are also significantly promoting our understanding of TM receptor functioning. While it is often only rudimentarily understood how ligand binding at the extracellular domain is transmitted via the TMD of RTKs, specific and promiscuous interactions of the TMDs are most likely critically involved in ErbB signaling.
Interaction Motifs in the TMDs are Involved in ErbB Signaling
While many observations had indicated that interactions of the ErbB TMDs are critically involved in ErbB signaling, introducing the strongly dimerizing GpA interaction motif into the ErbB2 TMD sequence resulted in receptor dimerization but not in receptor activation.76 Since, as discussed above, TM helix-helix interactions appear to be crucial for ErbB signaling, this result indicates that the TMDs do not have to simply interact somehow, but specific interactions of the TMDs might have to induce or stabilize a defined structure of the TM helix dimer. Replacement of a series of individual amino acids in the ErbB2 juxtamembrane region with individual cysteine residues has shown that only a subset of the cysteine mutants showed transforming activity by stabilizing an active ErbB structure due to the formation of a disulfide bond.99 Based on these data it was suggested that defined interactions in the α-helical juxtamenbrane region are needed to activate the receptor dimer. Ligand binding to the extracellular domains was suggested to induce a rotation or twist in the juxtamembrane as well as in the TM region, which subsequently results in receptor activation.52 When a dimerization inducing motif with two glutamate residues was shifted periodically across a simple TMD, which replaced the native TMD of the rat homolog of human ErbB2, different protein conformations were promoted in the absence of a ligand. The kinase activity of this modified RTK was activated only when the kinase domains of two monomers were positioned in a specific rotational conformation.100 Thus, only certain orientations of the TMDs allow intracellular kinase activation, and rotations of the TM helices are coupled to the intracellular kinase activity.
Almost 20 years ago it was noted that the single TM domains of several RTKs contain a characteristic GxxxG-like motif, which is also know as Sternberg-Gullick motif, and a function of this motif in TM signaling has been proposed.101 Each TMD of the individual human ErbB receptors contains two distinct GxxxG-like motifs, one in the N-terminal (towards the extracellular space) and one in the C-terminal part (towards the intracellular space) (Fig. 3). Only the human ErbB3 protein contains just a single GxxxG-like motif in its N-terminal TMD segment and interactions of the C-terminus seem to be facilitated by an unknown interaction motif.102 Since members of the ErbB family form various homo- and heterodimers, the N-terminal GxxxG-like motif could be critical for heterodimer formation whereas the C-terminal motif mediates homodimerization.103 Recently, the propensity of the human ErbB1-4 TMDs to homooligomerize within the E. coli inner membrane has been followed with a genetic system.80 While this study has demonstrated that all the ErbB TM domains form homooligomers within a membrane, the role of the conserved GxxxG-like motifs for heterodimerization was not addressed and also the involvement of the GxxxG-like motifs in homodimer formation and stabilization was not conclusively explained. For ErbB1 and ErbB3 the motif within the N-terminal helix part did not appear to be involved in homodimerization of the TM helix, whereas the corresponding motifs in the ErbB2 and ErbB4 TMDs were. Similarly, the GxxxG-like motifs within the C-terminal helix parts of ErbB1 and ErbB2 appeared to mediate and to stabilize a TM helix dimer, whereas the C-terminal motif of ErbB4 did not. Surprisingly, the single GxxxG-like motif in the ErbB3 TM helix was suggested to not be critical for stabilizing the helix dimer, although the TM helix strongly dimerized. Thus, while this study has clearly shown a homodimerization propensity for all ErbB TMDs, it could not conclusively answer the role of the individual ErbB TM helix parts and of the two GxxxG-like motifs for dimer formation.
Figure 3.
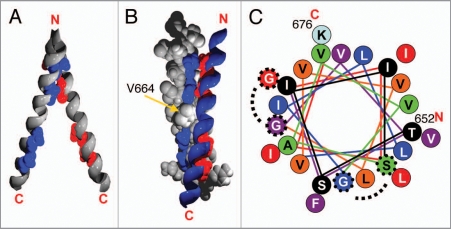
NMR structure of the ErbB2 TMD in detergent (PDB-ID: 2jwa). (A) Side view of two ErbB2 TMD helices with highlighted GxxxG-like motifs and top view with highlighted orientation of the interaction motifs. (B) Ribbon and space filling representation of the two interacting helices with highlighted interaction motifs and the position of Val664 is indicated as well. (C) Helical-wheel representation of the ErbB2 TMD. Colors change every four residues for clarity. The SxxxG and the GxxxG motifs are highlighted. The angle between the two interaction motifs corresponds to approximately 120°.
To elucidate the potential role of two conserved GxxxG-like motifs for mediating and/or stabilizing homo- and hetero-oligomeric interactions of the human ErbB TMDs, the interaction propensities of the N- or C-terminal GxxxG-like motifs of all human ErbB family members and the potential to mediate ErbB TM homo- and heterooligomerization were subsequently analyzed.102 This study has clearly demonstrated that the TMDs of all ErbB receptors form specific and stable homo- and hetero-oligomers in a biological membrane, although effects of the other ErbB domains of the dimerization propensity were also not considered at all. Nevertheless, the results clearly indicate that not only the extracellular or intracellular domains are involved in oligomerization of ErbB receptors but also the TMD can contribute to the specificity of oligomerization in vivo. The C- and N-terminal parts of the ErbB TMDs were found to differ in their tendency to form oligomers. The N-terminal motif of the ErbB TMDs was found to mediate and stabilize a slightly stronger interaction than the C-terminally located motif. A suggested more stable TM structure mediated by the N-terminal GxxxG-like motif is supported by the recently solved solid-state NMR structure of the ErbB2 receptor TMD dimer. Here, the N-terminal SxxxG motif is located at the dimer interface and is mainly involved in the interactions between the two helices. The TM dimer structure stabilized by this N-terminal GxxxG-like motif is supposed to represent the active state of the receptor.104 Thus, the isolated receptor TMDs form a TM helix dimer stabilized by the N-terminal GxxxG-like motif, which is in line with the afore mentioned observed higher interaction propensity of the N-terminal part of the ErbB receptor TMD.
Computational mapping studies of the ErbB2 TMD structure have identified two local energy minima, which could correspond to an active and inactive TMD structure. In the two energy minima the TM dimer is stabilized by the two respective GxxxG-like motifs.105 The two interaction motifs in the ErbB2 TMD are distinctly oriented on the TM α-helix (Fig. 3), so that only one motif (N- or C-terminal) can interact with the respective motif on the adjacent helix at a time. Since the calculated energy barrier between these two states appears to be relatively low, the TM structure can probably switch between these two stages, most likely induced by small structural changes in the extracellular ligand binding domain. Noteworthy, the structure of the ErbB2 TMD does not show a defined persistent groove connecting the two motifs with each other. Such a groove would be essential if the two TMDs of a dimer smoothly move along the helix axes from one TM conformation into the other in a screw-like fashion. Thus, the two helices probably have to bounce from one structure into the other, and this switch could involve a transient separation of the TM helices. However, the TMDs could separate and re-associate. Since the extra- and intracellular domains can also stabilize homotypic interactions (see above) the TMDs would remain in close spatial proximity.
It has been suggested that one of the TMD motifs is mainly responsible for ErbB homooligomerization whereas the other one may be responsible for heterooligomerization.103 ErbB heterooligomerization is not significantly facilitated by the extracellular ligand binding domains,63 and deletion of the intracellular domain still allows homo- and heterooligomerization of ErbB receptors.54 Thus, the TMDs could be critically involved in defined oligomerization, and one TMD interaction motif could further strengthen homooligomerization whereas the other strengthens heterooligomerization. However, both GxxxG-like motifs of the human ErbB proteins can be involved in the formation and/or stabilization of both ErbB TM homo- and heterodimers,102 and it is likely that specific interactions in the soluble domains mainly determine formation of defined dimers. Additional structures of different ErbB heterooligomers could answer the question whether the N- and C-terminal motifs are further responsible for homo- and heterooligomeric contacts, respectively. However, generating a population of pure heterodimers between different ErbB TMDs will be challenging since all ErbB TM helices form homodimers as well as heterodimers, which results in formation of different homo- and heterodimeric structures.
GxxxG-like interaction motifs appear to be conserved in ErbB TM helices from mammals and other vertebrates (see Fig. 4). In ErbB1 and ErbB4 homologous proteins the two motifs in the two helix segments appear to be conserved, whereas in several cases only a single motif is present, such as in the TM helix of the ErbB2 and ErbB3 homologs as well as in the homolog from Caenorhabditis elegans. In a recent study a cryptic, not yet identified interaction motif in the TMD of the ErbB homolog from Drosophila melanogaster has been proposed.66 In the case of the human ErbB3 TMD other residues can form interactions in the C-terminal part of the TMD and these interactions are mediated by a yet uncharacterized cryptic TMD interaction motif.102 Interestingly, when two GxxxG-like motifs are present in a TMD (Fig. 4), the distance between these two motifs is always conserved. Therefore, not only the existence of two motifs appears to be critical and conserved but also the distance in between these two motifs. This observation further supports a model where activation of the kinase domain is controlled by a pivot movement of the TM helices, which also includes a rotation of the two helices with respect to each other.
Figure 4.
Alignment of ErbB receptor TMD sequences from different species. The sequences were derived from the ExPASy proteomics server (http://www.expasy.org) and the TMD was identified by the TMHMM server (http://www.cbs.dtu.dk/services/TMHMM-2.0). GxxxG(-like) motifs are highlighted.
It has to be mentioned that some ErbB receptors form additional higher ordered oligomers. Upon activation of ErbB1, the formation of tetrameric assemblies was reported,106 and a side-by-side interaction of two dimers was suggested. Nevertheless, currently no concept has emerged about how these higher ordered oligomers could form and which domains are involved in this additional oligomerization step.
A Model Emerges: TMD Interactions Control ErbB Receptor Activation
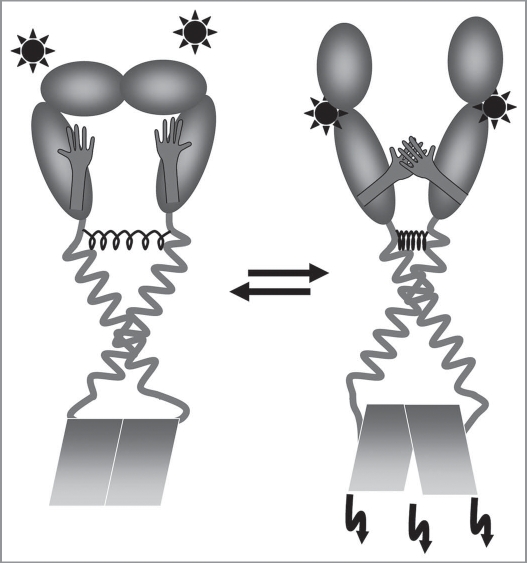
The above described structural features of the individual ErbB domains as well as the observed structural rearrangements indicate a mechanism of ErbB activation and signaling. As previously described, the general mode of action of ErbB receptors appears to be comparable to cadherins and to the EpoR. The ErbB receptors are most likely present as preformed dimers in the cell membrane. Receptor dimerization is (at least partly) mediated by the C-terminal half of the ErbB TM helix, although interaction of two helices via the N-terminal motif is energetically favored. This energetically favored TM helix interaction is hindered by the structure of the extracellular ligand binding domain in its ligand free form, and thus the TM helix dimer is in an energetically less favored state, comparable to a strained spring. Ligand binding induces structural changes in the extracellular domain, and upon ligand binding the autoinhibitory tether in the extracellular domain is removed and two adjacent dimerization arms can interact (Fig. 5). A structural rearrangement of the dimeric structure of the ligand binding domain induces or allows a shift of the inactive TM structure, which is stabilized by the C-terminal interaction motif (that orients the kinase domains in an inactive state) to the energetically favored N-terminal interaction motif (that orients the kinase domains in an active state). This screw-like rotation of approximately 120° corresponds to a transition from a preformed, inactive dimer to an active dimer (Fig. 3A and C). The rearrangement of the TM dimer structure results in a subsequent reorganization of the intracellular kinase domain structure and in kinase activation. Interestingly, in the inactive state, where the TM helix dimer is stabilized by the C-terminal GxxxG-like motif, the ends of the two TM helices are in closer proximity than in the active state, which is stabilized by the N-terminal motif. Thus, the N-termini of the kinase domains are also in closer proximity in the receptor inactive state, and not only the orientation of the kinase domains with respect to each other but also the distance in between these domains could be critical for kinase function.
Figure 5.
A model of ErbB TM signaling. The preformed ErbB dimer is stabilized in the TM region by the C-terminal GxxxG-like motif. The energetically preferred TM structure stabilized by the N-terminal motif is hindered because of a steric barrier defined by the soluble extracellular domain. Ligand binding induces structural rearrangements in the ligand binding domain and allows the “strained spring” to relax, and the TM structure switches into the structure stabilized by the N-terminal GxxxG-like motif of the TMD as well as by interactions of the dimerization arms. This structural rearrangement places the two intracellular kinase domains into a different position resulting in kinase activation and in downstream signaling.
The suggested model, which explains how ligand binding to the extracellular ErbB domain can be communicated across the membrane by the TMD, is in line with many of the above mentioned observations and highlights the very important role of the human ErbB TMDs in receptor signaling. Very likely, other interactions, such as interactions of the juxtamembrane region with each other and with lipids or interactions of the extracellular domain, are also involved in receptor activation. These interactions are not directly considered and the proposed (simplifying) model focuses on the important function of the ErbB TMD in receptor activation and signaling.
ErbB Receptor TMDs in Pathological Dysfunctions
ErbB receptors have received much attention in the recent years because of their involvement in cancer progression, and many forms of cancer are caused by ErbB receptor dysfunctions (reviewed in refs. 85, 107 and 108). ErbB2, especially, is able to transform cells upon overexpression, and an ErbB2 dysfunction is responsible for many types of cancers.108 Treatments of these cancer types include the use of monoclonal antibodies, such as Trastuzumab109 (a.k.a. Herceptin, from Roche, Switzerland), which binds to the extracellular domain of ErbB2. The monoclonal antibody Pertuzumab directly interferes with the dimerization interface in the extracellular domain of ErbB2 and thereby sterically blocks proper interactions of the receptor.110
The above presented observations indicate an important role of the TMD in ErbB signaling. Mutations resulting in amino acid exchanges in the TMD of different RTKs (including members of the ErbB family) have indeed been observed to lead to various forms of cancer.85,111 Although the ErbB2 receptor has no known ligand and appears to serve exclusively as a co-receptor in hetero-oligomers with other ErbB family members, the rat ErbB2 homologue, which is termed the Neu protooncogene, is constitutively activated if the amino acid valine 664 is mutated to glutamate (termed as Neu*).112,113
Continuous receptor activation leads to development of cancer. Many studies have indicated that the introduced glutamate residue constitutively activates the intracellular Neu* kinase domains by orienting them in a signaling-competent conformation that permits transphosphorylation.113–115 Subsequent studies have indicated that the mutation in the Neu* TMD is responsible for a change in helix-helix interaction and that the helices and the kinase domain in this variant are also rotationally coupled.100,114,115 This further supports the above suggested role of the TMD in signal progression and the notion that distinct interaction motifs are associated with the active or inactive state of the receptor. A corresponding amino acid exchange in the human ErbB2 receptor also leads to constitutive receptor activation.111 Interestingly, Val664 is located directly in between the two interaction motifs on one side of the helix (see Fig. 3B and C). The interaction tendency of the mutated human ErbB2 TMD is lowered,80 and thus, in line with a rotational coupling mechanism, an induced change in the TM structure probably stabilizes or creates an interaction that promotes kinase activation. It has been suggested that the Val664 → Glu mutation favors a TM dimer structure which is stabilized by a different amino acid motif involving amino acids Ile659, Ala661, Val663 and G665 from the Neu* TMD.116
Furthermore, an amino acid exchange from isoleucine at position 655 to valine in the TMD of human ErbB2,117 was shown to increase the risk of breast cancer.118 The residue Ile655 is located directly in front of the serine residue of the N-terminal SxxxG motif of the ErbB2 TMD, which stabilizes the active receptor conformation. Thus, residues close to the SxxxG motif appear to be of key importance for the activity of the receptor. This finding again points towards an important role of the TMD in signal progression.
Interestingly, the avian erythroblastosis virus (AEV) encodes a truncated version of an ErbB homologue.78 This viral version of an ErbB receptor (v-ErbB) is truncated in the extracellular domain and essentially only contains the TMD and the intracellular kinase domain. v-ErbB is able to transform murine cells that have been infected by a murine retrovirus encoding for v-ErbB.78,119 Interestingly, the sequence of the v-ErbB TMD (Fig. 4) contains an N-terminal as well as a C-terminal GxxxG motif, which have been shown to be involved in stabilization of defined TM helix structures (see above). Since constitutively active v-ErbB does not contain the large extracellular domain, the dimeric v-ErbB structure is most likely stabilized by the N-terminal GxxxG motif, in line with the above proposed model that the extracellular domain is responsible for the auto-inhibition of the receptor and the observations that ErbB variants truncated in the extracellular domain are constitutively active.74,120 However, while this concept explains the function of the N-terminal GxxxG motif of the v-ErbB TMD, the function of the C-terminal GxxxG motif remains open. In the human ErbB proteins this motif was suggested to stabilize an inactive receptor dimer, which forms due to restrictions defined by the extracellular ErbB domains. The v-ErbB protein does not contain this large domain, and thus the tether is absent. Therefore, the presence of a C-terminal located interaction motif might indicate that also the v-ErbB can rest in an inactive dimeric conformation, which is stabilized by the C-terminal GxxxG motif. Since the activity of v-ErbB involves defined interactions of the TMD it is tempting to design drugs that modulate TM interactions involved in signaling,121 and it could be possible to block the activity of v-ErbB by a synthetic peptide that interacts with the TMD. Synthetic peptides that bind to the ErbB2 TMD have been shown to be able to inhibit interactions in truncated and full length receptor constructs and to prevent transformation of cells.83,103 Such intervening peptides emerge more and more as potent pharmaceuticals and may help to overcome resistance of cancers towards conventional treatments.121,122
The Next Level of Complexity: ErbB Receptor Organization in Lipid Microdomains
While the above described features of ErbB receptors allow deducing structure-function relationships, one has to keep in mind that the expression pattern of the individual ErbB receptors varies between different cell types and tissues. It is equally important to consider the spatial organization of ErbB receptors in a membrane of a given cell.
ErbB receptors have been identified on the cell surface to be located in specific heterooligomer clusters and cluster formation depended on the added ligand.123 Clusters of ErbB1 and ErbB2 formed upon addition of EGF and clusters of ErbB2 and ErbB3 formed upon addition of heregulin (a.k.a. neuregulin type 1). These observations indicate that defined ErbB clusters can form and that ligands can induce such clusters. Interestingly, incubation of ErbB1 clusters with the kinase inhibitor AG1478 leads to the dissociation of these clusters,124 and a structural change in the receptor dimer could be involved.125 Other ErbB receptors are also known to form large clusters.126,127
It has been shown that ErbB1 is organized in caveolae, which represent cholesterol-rich invaginations in the plasma membrane caused by the caveolin-1 protein.128–130 Initial controversial results about the ErbB1 organization in such caveolae131 were later resolved by showing that a part of the ErbB1 population is located in caveolae and the other part seems to be organized in other lipid microdomains within the plasma membrane. Temperature and the presence of ligands influence the relative distribution of the receptors in defined microdomains.132 Upon activation of ErbB1 the receptors move out of the caveolae and are internalized via clathrin dependent endocytosis (reviewed in ref. 133) and are eventually degraded in lysosomes.134 These observations indicate that microdomain organization can potentially control receptor activation and function.
Lipid microdomains are islands within a membrane that contain specific lipids, selectively incorporate or exclude membrane integral or anchored proteins and offer a microenvironment that allows a high local concentration of certain membrane proteins.134 The local composition of defined lipids in such microdomains can significantly differ from that of the bulk part of the plasma membrane.135 It is possible that cell specific expression levels of the individual ErbB proteins combined with preferred localization of certain receptors in lipid microdomains promotes formation of specific homo- and heterodimers. Microdomains containing clustered ErbB receptors may represent signaling patches that are able to amplify receptor activation and increase the signaling output by transphosphorylation of many adjacent intracellular kinase domains. Due to the higher local protein concentrations in lipid microdomains, higher ordered oligomeric structures appear to be likely, although both higher oligomeric states and receptors with a lower oligomeric state that are non-cluster-forming may coexist in a dynamic equilibrium.136,137
But what could cause specific organization of ErbB receptors (and other TM proteins) in such microdomains? The TMDs of the individual ErbB receptors could be crucial for the subcellular distribution of individual ErbBs in defined membrane environments, and the membrane domain could serve as a marker for microdomain localization. TMDs often tend to localize in membranes with a hydrophobic core thickness, which matches about the length of the hydrophobic region of the individual TMDs. It has been noted already many years ago that certain TM helices of the Golgi membrane have on average a shorter TM sequence than TM α-helices residing in the plasma membrane,136,137 and it has been suggested that the length of the hydrophobic region defines subcellular sorting of a protein in the exocytotic pathway. Thus, the length of the hydrophobic region of a TM α-helix can influence and determine the membrane localization of a TM protein. Certain microdomains are rich in cholesterol and the high cholesterol content results in a significant increase in the local membrane thickness.138 How much, however, the thickness of a membrane can stabilize or destabilize a defined ErbB TM helix dimer has not been shown yet. During receptor activation the TM structure of the ErbB TM α-helices changes and structural differences in the TM region could be involved in sorting of ErbBs between defined microdomains and the bulk lipid fraction.
In recent years it became evident that protein-lipid interactions can control the structure and function of TM proteins and certain membrane proteins only function in a defined lipid environment, and e.g., certain gangliosides appear to inhibit the kinase activity of ErbB1.139 Gangliosides are lipids which can also be involved in formation of membrane microdomains and in the localization of ErbB1 and ErbB2 into these domains,140,141 which further indicates a complex network of factors controlling ErbB receptor microdomain organization. It has also been suggested that lipid microdomains form since ErbB receptors attract acidic lipids, such as phosphatidylinositol 4,5-bisphosphate (PIP2), which results in subsequent microdomain formation.72 All these observations indicate that protein-lipid interactions can also be involved in controlling ErbB functions in vivo, and changes in the structure of the TMD and the juxtamembrane regions could influence such protein-lipid interactions. Since in lipid microdomains the lipid composition differs from the bulk of the membrane, the physical properties of the membrane differ as well in such domains. Interestingly, an influence of membrane curvature on oligomerization of the ErbB2 TMD has been demonstrated in vitro,142 which suggests that changes in the local membrane properties may stabilize or destabilize the oligomeric state of the receptor.
Taken together, the presence of charged lipids, the presence of ligands, the kinase activity and the ErbB oligomerization can influence the distribution of ErbB receptors between different lipid environments, their function and their degradation. The individual TMDs of the ErbB RTKs could be involved in the organization of individual receptors in lipid microdomains and in binding of specific lipids. Furthermore, a structural rearrangement of the TM helix dimer could also be critical for ErbB signaling in defined lipid environments. An organization of ErbB receptors in lipid microdomains makes it even more difficult to understand the function and regulation of ErbB receptors but could offer new opportunities in the design of new pharmaceuticals.
Acknowledgements
Financial support from the Deutsche Forschungsgemeinschaft (SCHN 690/2-3 and GRK 1478), the Ministry of Science, Research and Arts of Baden-Württemberg and from the German-Israeli Foundation is gratefully acknowledged. We thank Carmen Finger for critically reading of the manuscript and for helpful suggestions.
Abbreviations
- TM
transmembrane
- TMD
transmembrane domain
- RTK
receptor-tyrosine kinase
- epo
erythropoietin
- EpoR
erythropoietin receptor
Footnotes
Previously published online: www.landesbioscience.com/journals/celladhesion/article/11191
References
- 1.Sanders CR, Myers JK. Disease-related misassembly of membrane proteins. Annu Rev Biophys Biomol Struct. 2004;33:25–51. doi: 10.1146/annurev.biophys.33.110502.140348. [DOI] [PubMed] [Google Scholar]
- 2.Wallin E, von Heijne G. Genome-wide analysis of integral membrane proteins from eubacterial, archaean and eukaryotic organisms. Protein Sci. 1998;7:1029–1038. doi: 10.1002/pro.5560070420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hopkins AL, Groom CR. The druggable genome. Nat Rev Drug Discov. 2002;1:727–730. doi: 10.1038/nrd892. [DOI] [PubMed] [Google Scholar]
- 4.Alberts B. Molecular biology of the cell. New York: Garland Science; 2008. [Google Scholar]
- 5.Langosch D, Arkin IT. Interaction and conformational dynamics of membrane-spanning protein helices. Protein Sci. 2009;18:1343–1358. doi: 10.1002/pro.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matthews EE, Zoonens M, Engelman DM. Dynamic helix interactions in transmembrane signaling. Cell. 2006;127:447–450. doi: 10.1016/j.cell.2006.10.016. [DOI] [PubMed] [Google Scholar]
- 7.Levchenko A. Dynamical and integrative cell signaling: challenges for the new biology. Biotechnol Bioeng. 2003;84:773–782. doi: 10.1002/bit.10854. [DOI] [PubMed] [Google Scholar]
- 8.von Heijne G. The membrane protein universe: what’s out there and why bother? J Intern Med. 2007;261:543–557. doi: 10.1111/j.1365-2796.2007.01792.x. [DOI] [PubMed] [Google Scholar]
- 9.Pokutta S, Weis WI. Structure and mechanism of cadherins and catenins in cell-cell contacts. Annu Rev Cell Dev Biol. 2007;23:237–261. doi: 10.1146/annurev.cellbio.22.010305.104241. [DOI] [PubMed] [Google Scholar]
- 10.Stemmler MP. Cadherins in development and cancer. Mol Biosyst. 2008;4:835–850. doi: 10.1039/b719215k. [DOI] [PubMed] [Google Scholar]
- 11.Goodwin M, Yap AS. Classical cadherin adhesion molecules: coordinating cell adhesion, signaling and the cytoskeleton. J Mol Histol. 2004;35:839–844. doi: 10.1007/s10735-004-1833-2. [DOI] [PubMed] [Google Scholar]
- 12.Ozawa M, Baribault H, Kemler R. The cytoplasmic domain of the cell adhesion molecule uvomorulin associates with three independent proteins structurally related in different species. EMBO J. 1989;8:1711–1717. doi: 10.1002/j.1460-2075.1989.tb03563.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tomschy A, Fauser C, Landwehr R, Engel J. Homophilic adhesion of E-cadherin occurs by a co-operative two-step interaction of N-terminal domains. EMBO J. 1996;15:3507–3514. [PMC free article] [PubMed] [Google Scholar]
- 14.Ozawa M, Kemler R. The membrane-proximal region of the E-cadherin cytoplasmic domain prevents dimerization and negatively regulates adhesion activity. J Cell Biol. 1998;142:1605–1613. doi: 10.1083/jcb.142.6.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brieher WM, Yap AS, Gumbiner BM. Lateral dimerization is required for the homophilic binding activity of C-cadherin. J Cell Biol. 1996;135:487–496. doi: 10.1083/jcb.135.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huber O, Kemler R, Langosch D. Mutations affecting transmembrane segment interactions impair adhesiveness of E-cadherin. J Cell Sci. 1999;112:4415–4423. doi: 10.1242/jcs.112.23.4415. [DOI] [PubMed] [Google Scholar]
- 17.Ozawa M. Lateral dimerization of the E-cadherin extracellular domain is necessary but not sufficient for adhesive activity. J Biol Chem. 2002;277:19600–19608. doi: 10.1074/jbc.M202029200. [DOI] [PubMed] [Google Scholar]
- 18.Yonekura S, Ting CY, Neves G, Hung K, Hsu SN, Chiba A, et al. The variable transmembrane domain of Drosophila N-cadherin regulates adhesive activity. Mol Cell Biol. 2006;26:6598–6608. doi: 10.1128/MCB.00241-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jelkmann W, Bohlius J, Hallek M, Sytkowski AJ. The erythropoietin receptor in normal and cancer tissues. Crit Rev Oncol Hematol. 2008;67:39–61. doi: 10.1016/j.critrevonc.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 20.Wilson IA, Jolliffe LK. The structure, organization, activation and plasticity of the erythropoietin receptor. Curr Opin Struct Biol. 1999;9:696–704. doi: 10.1016/s0959-440x(99)00032-9. [DOI] [PubMed] [Google Scholar]
- 21.Darnell JE, Jr, Kerr IM, Stark GR. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 22.Syed RS, Reid SW, Li C, Cheetham JC, Aoki KH, Liu B, et al. Efficiency of signalling through cytokine receptors depends critically on receptor orientation. Nature. 1998;395:511–516. doi: 10.1038/26773. [DOI] [PubMed] [Google Scholar]
- 23.Livnah O, Stura EA, Middleton SA, Johnson DL, Jolliffe LK, Wilson IA. Crystallographic evidence for preformed dimers of erythropoietin receptor before ligand activation. Science. 1999;283:987–990. doi: 10.1126/science.283.5404.987. [DOI] [PubMed] [Google Scholar]
- 24.Kubatzky KF, Ruan W, Gurezka R, Cohen J, Ketteler R, Watowich SS, et al. Self assembly of the transmembrane domain promotes signal transduction through the erythropoietin receptor. Curr Biol. 2001;11:110–115. doi: 10.1016/s0960-9822(01)00018-5. [DOI] [PubMed] [Google Scholar]
- 25.Zhou FX, Merianos HJ, Brunger AT, Engelman DM. Polar residues drive association of polyleucine transmembrane helices. Proc Natl Acad Sci USA. 2001;98:2250–2255. doi: 10.1073/pnas.041593698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gratkowski H, Lear JD, DeGrado WF. Polar side chains drive the association of model transmembrane peptides. Proc Natl Acad Sci USA. 2001;98:880–885. doi: 10.1073/pnas.98.3.880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li JP, D’Andrea AD, Lodish HF, Baltimore D. Activation of cell growth by binding of Friend spleen focus-forming virus gp55 glycoprotein to the erythropoietin receptor. Nature. 1990;343:762–764. doi: 10.1038/343762a0. [DOI] [PubMed] [Google Scholar]
- 28.Constantinescu SN, Liu X, Beyer W, Fallon A, Shekar S, Henis YI, et al. Activation of the erythropoietin receptor by the gp55-P viral envelope protein is determined by a single amino acid in its transmembrane domain. EMBO J. 1999;18:3334–3347. doi: 10.1093/emboj/18.12.3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robinson DR, Wu YM, Lin SF. The protein tyrosine kinase family of the human genome. Oncogene. 2000;19:5548–5557. doi: 10.1038/sj.onc.1203957. [DOI] [PubMed] [Google Scholar]
- 30.Hubbard SR, Miller WT. Receptor tyrosine kinases: mechanisms of activation and signaling. Curr Opin Cell Biol. 2007;19:117–123. doi: 10.1016/j.ceb.2007.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol. 2001;2:127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- 32.McKern NM, Lawrence MC, Streltsov VA, Lou MZ, Adams TE, Lovrecz GO, et al. Structure of the insulin receptor ectodomain reveals a folded-over conformation. Nature. 2006;443:218–221. doi: 10.1038/nature05106. [DOI] [PubMed] [Google Scholar]
- 33.Plotnikov AN, Schlessinger J, Hubbard SR, Mohammadi M. Structural basis for FGF receptor dimerization and activation. Cell. 1999;98:641–650. doi: 10.1016/s0092-8674(00)80051-3. [DOI] [PubMed] [Google Scholar]
- 34.Bennasroune A, Gardin A, Auzan C, Clauser E, Dirrig-Grosch S, Meira M, et al. Inhibition by transmembrane peptides of chimeric insulin receptors. Cell Mol Life Sci. 2005;62:2124–2131. doi: 10.1007/s00018-005-5226-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Finger C, Escher C, Schneider D. The single transmembrane domains of human receptor tyrosine kinases encode self-interactions. Sci Signal. 2009;2:56. doi: 10.1126/scisignal.2000547. [DOI] [PubMed] [Google Scholar]
- 36.Takahashi K, Yonezawa K, Nishimoto I. Insulin-like Growth Factor I Receptor Activated by a Transmembrane Mutation 1074/jbc.270.32.19041. Journal of Biological Chemistry. 1995;270:19041–19045. doi: 10.1074/jbc.270.32.19041. [DOI] [PubMed] [Google Scholar]
- 37.Cheatham B, Shoelson SE, Yamada K, Goncalves E, Kahn CR. Substitution of the erbB-2 oncoprotein transmembrane domain activates the insulin receptor and modulates the action of insulin and insulin-receptor substrate 1. Proc Natl Acad Sci USA. 1993;90:7336–7340. doi: 10.1073/pnas.90.15.7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Flörke RR, Schnaith K, Passlack W, Wichert M, Kuehn L, Fabry M, et al. Hormone-triggered conformational changes within the insulin-receptor ectodomain: requirement for transmembrane anchors. Biochem J. 2001;360:189–198. doi: 10.1042/0264-6021:3600189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamada K, Goncalves E, Kahn CR, Shoelson SE. Substitution of the insulin receptor transmembrane domain with the c-neu/erbB2 transmembrane domain constitutively activates the insulin receptor kinase in vitro. J Biol Chem. 1992;267:12452–12461. [PubMed] [Google Scholar]
- 40.Qiu C, Tarrant MK, Boronina T, Longo PA, Kavran JM, Cole RN, et al. In vitro enzymatic characterization of near full length EGFR in activated and inhibited states. Biochemistry. 2009;48:6624–6632. doi: 10.1021/bi900755n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schlessinger J, Lemmon MA. SH2 and PTB domains in tyrosine kinase signaling. Sci STKE. 2003;2003:12. doi: 10.1126/stke.2003.191.re12. [DOI] [PubMed] [Google Scholar]
- 42.Carpenter G, Cohen S. Human epidermal growth factor: binding of the polypeptide to human fibroblasts and stimulation of cell proliferation. Natl Cancer Inst Monogr. 1978:149–156. [PubMed] [Google Scholar]
- 43.Hynes NE, Horsch K, Olayioye MA, Badache A. The ErbB receptor tyrosine family as signal integrators. Endocr Relat Cancer. 2001;8:151–159. doi: 10.1677/erc.0.0080151. [DOI] [PubMed] [Google Scholar]
- 44.Ogiso H, Ishitani R, Nureki O, Fukai S, Yamanaka M, Kim JH, et al. Crystal structure of the complex of human epidermal growth factor and receptor extracellular domains. Cell. 2002;110:775–787. doi: 10.1016/s0092-8674(02)00963-7. [DOI] [PubMed] [Google Scholar]
- 45.Jura N, Endres NF, Engel K, Deindl S, Das R, Lamers MH, et al. Mechanism for activation of the EGF receptor catalytic domain by the juxtamembrane segment. Cell. 2009;137:1293–12307. doi: 10.1016/j.cell.2009.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bouyain S, Longo PA, Li S, Ferguson KM, Leahy DJ. The extracellular region of ErbB4 adopts a tethered conformation in the absence of ligand. Proc Natl Acad Sci USA. 2005;102:15024–15029. doi: 10.1073/pnas.0507591102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wood ER, Shewchuk LM, Ellis B, Brignola P, Brashear RL, Caferro TR, et al. 6-Ethynylthieno[3,2-d]- and 6-ethynylthieno[2,3-d]pyrimidin-4-anilines as tunable covalent modifiers of ErbB kinases. Proc Natl Acad Sci USA. 2008;105:2773–2778. doi: 10.1073/pnas.0708281105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cho HS, Mason K, Ramyar KX, Stanley AM, Gabelli SB, Denney DW, Jr, et al. Structure of the extracellular region of HER2 alone and in complex with the Herceptin Fab. Nature. 2003;421:756–760. doi: 10.1038/nature01392. [DOI] [PubMed] [Google Scholar]
- 49.Cho HS, Leahy DJ. Structure of the extracellular region of HER3 reveals an interdomain tether. Science. 2002;297:1330–1333. doi: 10.1126/science.1074611. [DOI] [PubMed] [Google Scholar]
- 50.Linggi B, Carpenter G. ErbB receptors: new insights on mechanisms and biology. Trends Cell Biol. 2006;16:649–656. doi: 10.1016/j.tcb.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 51.Chu CT, Everiss KD, Wikstrand CJ, Batra SK, Kung HJ, Bigner DD. Receptor dimerization is not a factor in the signalling activity of a transforming variant epidermal growth factor receptor (EGFRvIII) Biochem J. 1997;324:855–861. doi: 10.1042/bj3240855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moriki T, Maruyama H, Maruyama IN. Activation of preformed EGF receptor dimers by ligand-induced rotation of the transmembrane domain. J Mol Biol. 2001;311:1011–1026. doi: 10.1006/jmbi.2001.4923. [DOI] [PubMed] [Google Scholar]
- 53.Tao RH, Maruyama IN. All EGF(ErbB) receptors have preformed homo- and heterodimeric structures in living cells. J Cell Sci. 2008;121:3207–3217. doi: 10.1242/jcs.033399. [DOI] [PubMed] [Google Scholar]
- 54.Wehrman TS, Raab WJ, Casipit CL, Doyonnas R, Pomerantz JH, Blau HM. A system for quantifying dynamic protein interactions defines a role for Herceptin in modulating ErbB2 interactions. Proc Natl Acad Sci USA. 2006;103:19063–19068. doi: 10.1073/pnas.0605218103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Citri A, Skaria KB, Yarden Y. The deaf and the dumb: the biology of ErbB-2 and ErbB-3. Exp Cell Res. 2003;284:54–65. doi: 10.1016/s0014-4827(02)00101-5. [DOI] [PubMed] [Google Scholar]
- 56.Klapper LN, Glathe S, Vaisman N, Hynes NE, Andrews GC, Sela M, et al. The ErbB-2/HER2 oncoprotein of human carcinomas may function solely as a shared coreceptor for multiple stroma-derived growth factors. Proc Natl Acad Sci USA. 1999;96:4995–5000. doi: 10.1073/pnas.96.9.4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guy PM, Platko JV, Cantley LC, Cerione RA, Carraway KL., 3rd Insect cell-expressed p180erbB3 possesses an impaired tyrosine kinase activity. Proc Natl Acad Sci USA. 1994;91:8132–8136. doi: 10.1073/pnas.91.17.8132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2000;103:211–225. doi: 10.1016/s0092-8674(00)00114-8. [DOI] [PubMed] [Google Scholar]
- 59.Fuller SJ, Sivarajah K, Sugden PH. ErbB receptors, their ligands, and the consequences of their activation and inhibition in the myocardium. J Mol Cell Cardiol. 2008;44:831–854. doi: 10.1016/j.yjmcc.2008.02.278. [DOI] [PubMed] [Google Scholar]
- 60.Marmor MD, Skaria KB, Yarden Y. Signal transduction and oncogenesis by ErbB/HER receptors. Int J Radiat Oncol Biol Phys. 2004;58:903–913. doi: 10.1016/j.ijrobp.2003.06.002. [DOI] [PubMed] [Google Scholar]
- 61.Lemmon MA. Ligand-induced ErbB receptor dimerization. Exp Cell Res. 2009;315:638–648. doi: 10.1016/j.yexcr.2008.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lemmon MA, Bu Z, Ladbury JE, Zhou M, Pinchasi D, Lax I, et al. Two EGF molecules contribute additively to stabilization of the EGFR dimer. EMBO J. 1997;16:281–294. doi: 10.1093/emboj/16.2.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ferguson KM, Darling PJ, Mohan MJ, Macatee TL, Lemmon MA. Extracellular domains drive homo- but not hetero-dimerization of erbB receptors. EMBO J. 2000;19:4632–4643. doi: 10.1093/emboj/19.17.4632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ferguson KM, Berger MB, Mendrola JM, Cho HS, Leahy DJ, Lemmon MA. EGF activates its receptor by removing interactions that autoinhibit ectodomain dimerization. Mol Cell. 2003;11:507–517. doi: 10.1016/s1097-2765(03)00047-9. [DOI] [PubMed] [Google Scholar]
- 65.Dawson JP, Bu Z, Lemmon MA. Ligand-induced structural transitions in ErbB receptor extracellular domains. Structure. 2007;15:942–954. doi: 10.1016/j.str.2007.06.013. [DOI] [PubMed] [Google Scholar]
- 66.Alvarado D, Klein DE, Lemmon MA. ErbB2 resembles an autoinhibited invertebrate epidermal growth factor receptor. Nature. 2009;461:287–291. doi: 10.1038/nature08297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gullick WJ. A new model for the interaction of EGF-like ligands with their receptors: the new one-two. Eur J Cancer. 1994;30:2186. doi: 10.1016/0959-8049(94)00365-c. [DOI] [PubMed] [Google Scholar]
- 68.Dawson JP, Berger MB, Lin CC, Schlessinger J, Lemmon MA, Ferguson KM. Epidermal growth factor receptor dimerization and activation require ligand-induced conformational changes in the dimer interface. Mol Cell Biol. 2005;25:7734–7742. doi: 10.1128/MCB.25.17.7734-7742.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sorokin A. Activation of the EGF receptor by insertional mutations in its juxtamembrane regions. Oncogene. 1995;11:1531–1540. [PubMed] [Google Scholar]
- 70.Red Brewer M, Choi SH, Alvarado D, Moravcevic K, Pozzi A, Lemmon MA, et al. The juxtamembrane region of the EGF receptor functions as an activation domain. Mol Cell. 2009;34:641–651. doi: 10.1016/j.molcel.2009.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Thiel KW, Carpenter G. Epidermal growth factor receptor juxtamembrane region regulates allosteric tyrosine kinase activation. Proc Natl Acad Sci USA. 2007;104:19238–19243. doi: 10.1073/pnas.0703854104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.McLaughlin S, Smith SO, Hayman MJ, Murray D. An electrostatic engine model for autoinhibition and activation of the epidermal growth factor receptor (EGFR/ErbB) family. J Gen Physiol. 2005;126:41–53. doi: 10.1085/jgp.200509274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang X, Gureasko J, Shen K, Cole PA, Kuriyan J. An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell. 2006;125:1137–1149. doi: 10.1016/j.cell.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 74.Chantry A. The kinase domain and membrane localization determine intracellular interactions between epidermal growth factor receptors. J Biol Chem. 1995;270:3068–3073. [PubMed] [Google Scholar]
- 75.Heisermann GJ, Wiley HS, Walsh BJ, Ingraham HA, Fiol CJ, Gill GN. Mutational removal of the Thr669 and Ser671 phosphorylation sites alters substrate specificity and ligand-induced internalization of the epidermal growth factor receptor. J Biol Chem. 1990;265:12820–12827. [PubMed] [Google Scholar]
- 76.Burke CL, Lemmon MA, Coren BA, Engelman DM, Stern DF. Dimerization of the p185neu transmembrane domain is necessary but not sufficient for transformation. Oncogene. 1997;14:687–696. doi: 10.1038/sj.onc.1200873. [DOI] [PubMed] [Google Scholar]
- 77.Yarden Y, Schlessinger J. Self-phosphorylation of epidermal growth factor receptor: evidence for a model of intermolecular allosteric activation. Biochemistry. 1987;26:1434–1442. doi: 10.1021/bi00379a034. [DOI] [PubMed] [Google Scholar]
- 78.Downward J, Yarden Y, Mayes E, Scrace G, Totty N, Stockwell P, et al. Close similarity of epidermal growth factor receptor and v-erb-B oncogene protein sequences. Nature. 1984;307:521–527. doi: 10.1038/307521a0. [DOI] [PubMed] [Google Scholar]
- 79.Tzahar E, Pinkas-Kramarski R, Moyer JD, Klapper LN, Alroy I, Levkowitz G, et al. Bivalence of EGF-like ligands drives the ErbB signaling network. EMBO J. 1997;16:4938–4950. doi: 10.1093/emboj/16.16.4938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mendrola JM, Berger MB, King MC, Lemmon MA. The single transmembrane domains of ErbB receptors self-associate in cell membranes. J Biol Chem. 2002;277:4704–4712. doi: 10.1074/jbc.M108681200. [DOI] [PubMed] [Google Scholar]
- 81.Sharpe S, Barber KR, Grant CW. Evidence of a tendency to self-association of the transmembrane domain of ErbB-2 in fluid phospholipid bilayers. Biochemistry. 2002;41:2341–2352. doi: 10.1021/bi011340f. [DOI] [PubMed] [Google Scholar]
- 82.Duneau JP, Vegh AP, Sturgis JN. A dimerization hierarchy in the transmembrane domains of the HER receptor family. Biochemistry. 2007;46:2010–2019. doi: 10.1021/bi061436f. [DOI] [PubMed] [Google Scholar]
- 83.Lofts FJ, Hurst HC, Sternberg MJ, Gullick WJ. Specific short transmembrane sequences can inhibit transformation by the mutant neu growth factor receptor in vitro and in vivo. Oncogene. 1993;8:2813–2820. [PubMed] [Google Scholar]
- 84.Bennasroune A, Fickova M, Gardin A, Dirrig-Grosch S, Aunis D, Cremel G, et al. Transmembrane peptides as inhibitors of ErbB receptor signaling. Mol Biol Cell. 2004;15:3464–3474. doi: 10.1091/mbc.E03-10-0753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li E, Hristova K. Role of receptor tyrosine kinase transmembrane domains in cell signaling and human pathologies. Biochemistry. 2006;45:6241–6251. doi: 10.1021/bi060609y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Popot JL, Engelman DM. Membrane protein folding and oligomerization: the two-stage model. Biochemistry. 1990;29:4031–4037. doi: 10.1021/bi00469a001. [DOI] [PubMed] [Google Scholar]
- 87.Engelman DM, Chen Y, Chin C-N, Curran AR, Dixon AM, Dupuy AD, et al. Membrane protein folding: beyond the two stage model. FEBS Lett. 2003;555:122–125. doi: 10.1016/s0014-5793(03)01106-2. [DOI] [PubMed] [Google Scholar]
- 88.Lemmon MA, Flanagan JM, Treutlein HR, Zhang J, Engelman DM. Sequence specificity in the dimerization of transmembrane alpha-helices. Biochemistry. 1992;31:12719–12725. doi: 10.1021/bi00166a002. [DOI] [PubMed] [Google Scholar]
- 89.MacKenzie KR, Prestegard JH, Engelman DM. A transmembrane helix dimer: structure and implications. Science. 1997;276:131–133. doi: 10.1126/science.276.5309.131. [DOI] [PubMed] [Google Scholar]
- 90.Brosig B, Langosch D. The dimerization motif of the glycophorin A transmembrane segment in membranes: importance of glycine residues. Protein Sci. 1998;7:1052–1056. doi: 10.1002/pro.5560070423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Senes A, Gerstein M, Engelman DM. Statistical analysis of amino acid patterns in transmembrane helices: the GxxxG motif occurs frequently and in association with beta-branched residues at neighboring positions. J Mol Biol. 2000;296:921–936. doi: 10.1006/jmbi.1999.3488. [DOI] [PubMed] [Google Scholar]
- 92.Russ WP, Engelman DM. The GxxxG motif: a framework for transmembrane helix-helix association. J Mol Biol. 2000;296:911–919. doi: 10.1006/jmbi.1999.3489. [DOI] [PubMed] [Google Scholar]
- 93.Kim S, Jeon TJ, Oberai A, Yang D, Schmidt JJ, Bowie JU. Transmembrane glycine zippers: physiological and pathological roles in membrane proteins. Proc Natl Acad Sci USA. 2005;102:14278–14283. doi: 10.1073/pnas.0501234102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Schneider D, Engelman DM. Involvement of transmembrane domain interactions in signal transduction by alpha/beta integrins. J Biol Chem. 2004;279:9840–9846. doi: 10.1074/jbc.M312749200. [DOI] [PubMed] [Google Scholar]
- 95.Schneider D, Engelman DM. Motifs of two small residues can assist but are not sufficient to mediate transmembrane helix interactions. J Mol Biol. 2004;343:799–804. doi: 10.1016/j.jmb.2004.08.083. [DOI] [PubMed] [Google Scholar]
- 96.Unterreitmeier S, Fuchs A, Schaffler T, Heym RG, Frishman D, Langosch D. Phenylalanine promotes interaction of transmembrane domains via GxxxG motifs. J Mol Biol. 2007;374:705–718. doi: 10.1016/j.jmb.2007.09.056. [DOI] [PubMed] [Google Scholar]
- 97.Melnyk RA, Kim S, Curran AR, Engelman DM, Bowie JU, Deber CM. The affinity of GXXXG motifs in transmembrane helix-helix interactions is modulated by long-range communication. J Biol Chem. 2004;279:16591–16597. doi: 10.1074/jbc.M313936200. [DOI] [PubMed] [Google Scholar]
- 98.Senes A, Engel DE, DeGrado WF. Folding of helical membrane proteins: the role of polar, GxxxG-like and proline motifs. Curr Opin Struct Biol. 2004;14:465–479. doi: 10.1016/j.sbi.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 99.Burke CL, Stern DF. Activation of Neu (ErbB-2) mediated by disulfide bond-induced dimerization reveals a receptor tyrosine kinase dimer interface. Mol Cell Biol. 1998;18:5371–5379. doi: 10.1128/mcb.18.9.5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bell CA, Tynan JA, Hart KC, Meyer AN, Robertson SC, Donoghue DJ. Rotational coupling of the transmembrane and kinase domains of the Neu receptor tyrosine kinase. Mol Biol Cell. 2000;11:3589–3599. doi: 10.1091/mbc.11.10.3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sternberg MJ, Gullick WJ. A sequence motif in the transmembrane region of growth factor receptors with tyrosine kinase activity mediates dimerization. Protein Eng. 1990;3:245–248. doi: 10.1093/protein/3.4.245. [DOI] [PubMed] [Google Scholar]
- 102.Escher C, Cymer F, Schneider D. Two GxxxG-like motifs facilitate promiscuous interactions of the human ErbB transmembrane domains. J Mol Biol. 2009;389:10–16. doi: 10.1016/j.jmb.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 103.Gerber D, Sal-Man N, Shai Y. Two motifs within a transmembrane domain, one for homodimerization and the other for heterodimerization. J Biol Chem. 2004;279:21177–21182. doi: 10.1074/jbc.M400847200. [DOI] [PubMed] [Google Scholar]
- 104.Bocharov EV, Mineev KS, Volynsky PE, Ermolyuk YS, Tkach EN, Sobol AG, et al. Spatial structure of the dimeric transmembrane domain of the growth factor receptor ErbB2 presumably corresponding to the receptor active state. J Biol Chem. 2008;283:6950–6956. doi: 10.1074/jbc.M709202200. [DOI] [PubMed] [Google Scholar]
- 105.Fleishman SJ, Schlessinger J, Ben-Tal N. A putative molecular-activation switch in the transmembrane domain of erbB2. Proc Natl Acad Sci USA. 2002;99:15937–15940. doi: 10.1073/pnas.252640799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Clayton AH, Walker F, Orchard SG, Henderson C, Fuchs D, Rothacker J, et al. Ligand-induced dimer-tetramer transition during the activation of the cell surface epidermal growth factor receptor-A multidimensional microscopy analysis. J Biol Chem. 2005;280:30392–30399. doi: 10.1074/jbc.M504770200. [DOI] [PubMed] [Google Scholar]
- 107.Khazaie K, Schirrmacher V, Lichtner RB. EGF receptor in neoplasia and metastasis. Cancer Metastasis Rev. 1993;12:255–274. doi: 10.1007/BF00665957. [DOI] [PubMed] [Google Scholar]
- 108.Holbro T, Hynes NE. ErbB receptors: directing key signaling networks throughout life. Annu Rev Pharmacol Toxicol. 2004;44:195–217. doi: 10.1146/annurev.pharmtox.44.101802.121440. [DOI] [PubMed] [Google Scholar]
- 109.Hudziak RM, Lewis GD, Winget M, Fendly BM, Shepard HM, Ullrich A. p185HER2 monoclonal antibody has antiproliferative effects in vitro and sensitizes human breast tumor cells to tumor necrosis factor. Mol Cell Biol. 1989;9:1165–1172. doi: 10.1128/mcb.9.3.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Franklin MC, Carey KD, Vajdos FF, Leahy DJ, de Vos AM, Sliwkowski MX. Insights into ErbB signaling from the structure of the ErbB2-pertuzumab complex. Cancer Cell. 2004;5:317–328. doi: 10.1016/s1535-6108(04)00083-2. [DOI] [PubMed] [Google Scholar]
- 111.Segatto O, King CR, Pierce JH, Di Fiore PP, Aaronson SA. Different structural alterations upregulate in vitro tyrosine kinase activity and transforming potency of the erbB-2 gene. Mol Cell Biol. 1988;8:5570–5574. doi: 10.1128/mcb.8.12.5570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bargmann CI, Hung MC, Weinberg RA. Multiple independent activations of the neu oncogene by a point mutation altering the transmembrane domain of p185. Cell. 1986;45:649–657. doi: 10.1016/0092-8674(86)90779-8. [DOI] [PubMed] [Google Scholar]
- 113.Smith SO, Smith CS, Bormann BJ. Strong hydrogen bonding interactions involving a buried glutamic acid in the transmembrane sequence of the neu/erbB-2 receptor. Nat Struct Biol. 1996;3:252–258. doi: 10.1038/nsb0396-252. [DOI] [PubMed] [Google Scholar]
- 114.Smith SO, Smith C, Shekar S, Peersen O, Ziliox M, Aimoto S. Transmembrane interactions in the activation of the Neu receptor tyrosine kinase. Biochemistry. 2002;41:9321–9332. doi: 10.1021/bi012117l. [DOI] [PubMed] [Google Scholar]
- 115.Cao H, Bangalore L, Dompe C, Bormann BJ, Stern DF. An extra cysteine proximal to the transmembrane domain induces differential cross-linking of p185neu and p185neu. J Biol Chem. 1992;267:20489–20492. [PubMed] [Google Scholar]
- 116.Beevers AJ, Kukol A. The transmembrane domain of the oncogenic mutant ErbB-2 receptor: a structure obtained from site-specific infrared dichroism and molecular dynamics. J Mol Biol. 2006;361:945–953. doi: 10.1016/j.jmb.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 117.Papewalis J, Nikitin A, Rajewsky MF. G to A polymorphism at amino acid codon 655 of the human erbB-2/HER2 gene. Nucleic Acids Res. 1991;19:5452. doi: 10.1093/nar/19.19.5452-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Xie D, Shu XO, Deng Z, Wen WQ, Creek KE, Dai Q, et al. Population-based, case-control study of HER2 genetic polymorphism and breast cancer risk. J Natl Cancer Inst. 2000;92:412–417. doi: 10.1093/jnci/92.5.412. [DOI] [PubMed] [Google Scholar]
- 119.Miller M, Kennewell A, Takayama Y, Bruskin A, Bishop JM, Johnson G, et al. Transformation of early erythroid precursor cells (BFU-E) by a recombinant murine retrovirus containing v-erb-B. Oncogene. 1990;5:1125–1131. [PubMed] [Google Scholar]
- 120.Zhu HJ, Iaria J, Orchard S, Walker F, Burgess AW. Epidermal growth factor receptor: association of extracellular domain negatively regulates intracellular kinase activation in the absence of ligand. Growth Factors. 2003;21:15–30. doi: 10.1080/0897719031000096424. [DOI] [PubMed] [Google Scholar]
- 121.Cymer F, Schneider D. Lessons from viruses: controlling the function of transmembrane proteins by interfering transmembrane helices. Curr Med Chem. 2008;15:779–785. doi: 10.2174/092986708783955545. [DOI] [PubMed] [Google Scholar]
- 122.Yin H, Slusky JS, Berger BW, Walters RS, Vilaire G, Litvinov RI, et al. Computational design of peptides that target transmembrane helices. Science. 2007;315:1817–1822. doi: 10.1126/science.1136782. [DOI] [PubMed] [Google Scholar]
- 123.Szabo A, Horvath G, Szollosi J, Nagy P. Quantitative characterization of the large-scale association of ErbB1 and ErbB2 by flow cytometric homo-FRET measurements. Biophys J. 2008;95:2086–2096. doi: 10.1529/biophysj.108.133371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Clayton AH, Tavarnesi ML, Johns TG. Unligated epidermal growth factor receptor forms higher order oligomers within microclusters on A431 cells that are sensitive to tyrosine kinase inhibitor binding. Biochemistry. 2007;46:4589–4597. doi: 10.1021/bi700002b. [DOI] [PubMed] [Google Scholar]
- 125.Gan HK, Walker F, Burgess AW, Rigopoulos A, Scott AM, Johns TG. The epidermal growth factor receptor (EGFR) tyrosine kinase inhibitor AG1478 increases the formation of inactive untethered EGFR dimers. Implications for combination therapy with monoclonal antibody 806. J Biol Chem. 2007;282:2840–2850. doi: 10.1074/jbc.M605136200. [DOI] [PubMed] [Google Scholar]
- 126.Nagy P, Jenei A, Kirsch AK, Szollosi J, Damjanovich S, Jovin TM. Activation-dependent clustering of the erbB2 receptor tyrosine kinase detected by scanning near-field optical microscopy. J Cell Sci. 1999;112:1733–1741. doi: 10.1242/jcs.112.11.1733. [DOI] [PubMed] [Google Scholar]
- 127.Kani K, Warren CM, Kaddis CS, Loo JA, Landgraf R. Oligomers of ERBB3 have two distinct interfaces that differ in their sensitivity to disruption by heregulin. J Biol Chem. 2005;280:8238–8247. doi: 10.1074/jbc.M410944200. [DOI] [PubMed] [Google Scholar]
- 128.Sigismund S, Woelk T, Puri C, Maspero E, Tacchetti C, Transidico P, et al. Clathrin-independent endocytosis of ubiquitinated cargos. Proc Natl Acad Sci USA. 2005;102:2760–2765. doi: 10.1073/pnas.0409817102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Couet J, Sargiacomo M, Lisanti MP. Interaction of a receptor tyrosine kinase, EGF-R, with caveolins. Caveolin binding negatively regulates tyrosine and serine/threonine kinase activities. J Biol Chem. 1997;272:30429–30438. doi: 10.1074/jbc.272.48.30429. [DOI] [PubMed] [Google Scholar]
- 130.Ushio-Fukai M, Hilenski L, Santanam N, Becker PL, Ma Y, Griendling KK, et al. Cholesterol depletion inhibits epidermal growth factor receptor transactivation by angiotensin II in vascular smooth muscle cells: role of cholesterol-rich microdomains and focal adhesions in angiotensin II signaling. J Biol Chem. 2001;276:48269–48275. doi: 10.1074/jbc.M105901200. [DOI] [PubMed] [Google Scholar]
- 131.Ringerike T, Blystad FD, Levy FO, Madshus IH, Stang E. Cholesterol is important in control of EGF receptor kinase activity but EGF receptors are not concentrated in caveolae. J Cell Sci. 2002;115:1331–1340. doi: 10.1242/jcs.115.6.1331. [DOI] [PubMed] [Google Scholar]
- 132.Keating E, Nohe A, Petersen NO. Studies of distribution, location and dynamic properties of EGFR on the cell surface measured by image correlation spectroscopy. Eur Biophys J. 2008;37:469–481. doi: 10.1007/s00249-007-0239-y. [DOI] [PubMed] [Google Scholar]
- 133.Waterman H, Yarden Y. Molecular mechanisms underlying endocytosis and sorting of ErbB receptor tyrosine kinases. FEBS Lett. 2001;490:142–152. doi: 10.1016/s0014-5793(01)02117-2. [DOI] [PubMed] [Google Scholar]
- 134.Fantini J, Garmy N, Mahfoud R, Yahi N. Lipid rafts: structure, function and role in HIV, Alzheimer’s and prion diseases. Expert Rev Mol Med. 2002;4:1–22. doi: 10.1017/S1462399402005392. [DOI] [PubMed] [Google Scholar]
- 135.Hanzal-Bayer MF, Hancock JF. Lipid rafts and membrane traffic. FEBS Lett. 2007;581:2098–2104. doi: 10.1016/j.febslet.2007.03.019. [DOI] [PubMed] [Google Scholar]
- 136.Webb RJ, East JM, Sharma RP, Lee AG. Hydrophobic mismatch and the incorporation of peptides into lipid bilayers: a possible mechanism for retention in the Golgi. Biochemistry. 1998;37:673–679. doi: 10.1021/bi972441+. [DOI] [PubMed] [Google Scholar]
- 137.Bretscher MS, Munro S. Cholesterol and the Golgi apparatus. Science. 1993;261:1280–1281. doi: 10.1126/science.8362242. [DOI] [PubMed] [Google Scholar]
- 138.Nezil FA, Bloom M. Combined influence of cholesterol and synthetic amphiphillic peptides upon bilayer thickness in model membranes. Biophys J. 1992;61:1176–1183. doi: 10.1016/S0006-3495(92)81926-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Lopez PH, Schnaar RL. Gangliosides in cell recognition and membrane protein regulation. Curr Opin Struct Biol. 2009;19:549–557. doi: 10.1016/j.sbi.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Roepstorff K, Thomsen P, Sandvig K, van Deurs B. Sequestration of epidermal growth factor receptors in non-caveolar lipid rafts inhibits ligand binding. J Biol Chem. 2002;277:18954–18960. doi: 10.1074/jbc.M201422200. [DOI] [PubMed] [Google Scholar]
- 141.Sottocornola E, Misasi R, Mattei V, Ciarlo L, Gradini R, Garofalo T, et al. Role of gangliosides in the association of ErbB2 with lipid rafts in mammary epithelial HC11 cells. Febs J. 2006;273:1821–1830. doi: 10.1111/j.1742-4658.2006.05203.x. [DOI] [PubMed] [Google Scholar]
- 142.Khemtemourian L, Buchoux S, Aussenac F, Dufourc EJ. Dimerization of Neu/Erb2 transmembrane domain is controlled by membrane curvature. Eur Biophys J. 2007;36:107–112. doi: 10.1007/s00249-006-0111-5. [DOI] [PubMed] [Google Scholar]