Abstract
Three classes of proteins are known to nucleate new filaments: the Arp2/3 complex, formins, and the third group of proteins that contain ca. 25 amino acid long actin-binding Wiskott-Aldrich syndrome protein homology 2 domains, called the WH2 repeats. Crystal structures of the complexes between the actin-binding WH2 repeats of the Spire protein and actin were determined for the Spire single WH2 domain D, the double (SpirCD), triple (SpirBCD), quadruple (SpirABCD) domains, and an artificial Spire WH2 construct comprising three identical D repeats (SpirDDD). SpirCD represents the minimal functional core of Spire that can nucleate actin filaments. Packing in the crystals of the actin complexes with SpirCD, SpirBCD, SpirABCD, and SpirDDD shows the presence of two types of assemblies, “side-to-side” and “straight-longitudinal,” which can serve as actin filament nuclei. The principal feature of these structures is their loose, open conformations, in which the sides of actins that normally constitute the inner interface core of a filament are flipped inside out. These Spire structures are distant from those seen in the filamentous nuclei of Arp2/3, formins, and in the F-actin filament.
Keywords: cytoskeleton, X-ray crystallography, fluorescence assay
The actin cytoskeleton is involved in many cellular processes, including cell motility, cell adhesion, endo/exocytosis, intracellular and membrane trafficking, and the maintenance of cell shape and polarity. These cellular functions require the dynamic remodeling of the actin cytoskeleton, which depends on the transition between monomeric actin (G-actin) and its filamentous state (F-actin) (1). The initiation of actin polymerization from free actin monomers requires nucleation factors that help to overcome the kinetic barrier for formation of actin dimers and trimers (2, 3). Three classes of actin-nucleation proteins have been identified until today: the Arp2/3 complex together with newly recognized nucleation-promoting factors such as WASH, WHAMM, and JMY (4–8), formins (9–12), and the third group of proteins that contain 17–27 amino acid long actin-binding motifs called the WH2 repeats—the name derived from the WASP (Wiskott–Aldrich syndrome protein) homology domain 2 (13–15). The third group consists of Spire (16), Cordon-bleu (17), and Leiomodin from muscle cells (18). The molecular mechanism of actin nucleation is well described for the Arp2/3 complex (19, 20) and formins (21), whereas little molecular details are known about the nucleation by Spire.
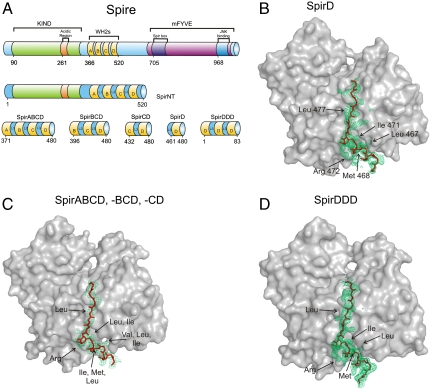
Spire was first identified as an actin-binding factor necessary for the correct establishment of polarity axes of the oocyte in Drosophila (22). It mediated actin-microtubule interactions and has important roles in membrane transport, although the upstream signaling pathways that regulate these functions have not been fully studied (23–25). Spire is a 1,020 amino acid long, multidomain protein (Fig. 1A); the most important domains, from the point of view of actin organization, are localized in the N-terminal part of the protein. For example, the N-terminal domain of Spire (SpirNT, residues 1–520 (Fig. 1A) was shown to interact with G-actin. The KIND domain (the kinase noncatalytic C-lobe domain, residues 90–328) is known to interact with another actin-nucleating factor, Cappuccino (fly formin) (24, 25). Spire has a cluster of four WH2 repeats. These WH2 domains, designated A, B, C, and D, are known to bind four actin monomers. Mutational studies indicated that, among the four WH2 domains of Spire, the C and D domains have the strongest nucleation potential and the C-D fragment is sufficient to initiate the polymerization process (16). In an actin assembly assay, the ABCD nucleating region of Spire nucleates actin at low Spire/actin ratios and sequesters actin at high Spire/actin ratios (8, 16, 26).
Fig. 1.
(A) Domain organization of the full-length Drosophila Spire and representation of the constructs used in this work. The WH2 domains are shown in yellow. KIND, the kinase noncatalytic C-lobe domain; mFYVE, a modified zinc finger motif found in four cysteine-rich proteins; JNKB, the c-Jun N-terminal kinase binding domain (16). (B–D) Comparison of the structures of WH2 domains of SpirD, SpirABCD, SpirBCD, SpirCD, and SpirDDD. Electron densities (omit maps) are colored green; the polyglycine model, represented as sticks, is in red; actins are shown as gray surface plots; conserved residues are marked with arrows. The side chains of SpirD and SpirDDD are visible and have well-defined electron densities. In the case of the averaged WH2 domains, conserved residues were recognizable because of an elongated electron density; however, the averaging precluded the assignment of these residues to specific WH2 domain. (B) The WH2 domain of SpirD. (C) The averaged WH2 domain of SpirABCD, SpirBCD, and SpirCD. (D) The WH2-D domain of SpirDDD.
WH2 motifs are known to be intrinsically disordered, adopting an α-helical structure only upon binding to actin (27). Alignments of WH2 domains indicate that the most conserved regions are the LKK motif and an α-helix at the N terminus (Fig. S1). Comparing NMR and crystal structures of different WH2 domains, this conserved α-helix is shown to be the principal structural actin-binding element that binds to actin in its hydrophobic pocket between actin’s subdomains 1 and 3 (28–32). The rest of the WH2, comprising the LKK motif, extends along the outer surface of the actin subdomain 1 up to subdomains 2 and 4.
Here we provide the structural insight into the interaction of Spire with actin. Together with the results of our actin assembly assays, the structures provide a mechanism by which Spire initiates actin nucleation into filaments.
Results
Interaction of Spire and Actin in Solution.
In order to physicochemically characterize the binding properties of Drosophila Spire, we performed NMR analyses (Fig. S2) and gel filtration mobility shift assays for several constructs of Spire. Both NMR and gel filtration data showed that quadruple (SpirABCD), triple (SpirBCD), three identical D repeats (SpirDDD), double (SpirCD), and single WH2 of SpirC and SpirD bind four, three, three, two, and one actin molecules, respectively, and create tight and stable complexes with actin (Fig. S3) (definition of the constructs is shown in Fig. 1A). In gel filtration, peaks of the complexes from Superdex 200 were sharp and clearly separated from the unbound actin, and the sizes of the complexes were determined by the number of WH2 domains in the constructs, which correlated with the number of bound actins. The elution profiles of the unbound monomeric AP-actin, the mutant that we used for crystallization of Spire/actin complexes (33), as well as wild-type rabbit muscle actin complexed with latrunculin B, were similar to the elution profile of ovalbumin (43 kDa, Stokes radius—30.5 Å). Because of the expected elongated shape of the complexes, the Stokes radii of complexes of SpirCD, SpirBCD, and SpirABCD were larger than for globular proteins of molecular weight 92, 136, 182 kDa, respectively. This, however, did not complicate the analysis because the binding of an additional actin per one WH2 domain more caused a significant shift in the peak position, well resolved from the species with one actin/WH2 unit less.
Our gel filtration data are, as expected, in agreement with the stoichiometry of Spire-actin binding determined by Bosch et al. (26), Rebowski et al. (34), and Quinlan et al. (16), who found that, for example, the NT-SpireABCD (SpirNT in Fig. 1A) “binds four actin monomers in a tight nonpolymerizable complex” (26). The minimal functional core of the protein that still can assemble actin filaments contains the C-terminal half of the Spire WH2 cluster (SpirCD), which precisely is composed of WH2-C, L-3, and WH2-D (residues 428–485) (Fig. 1A and Fig. S1; see also ref. 16).
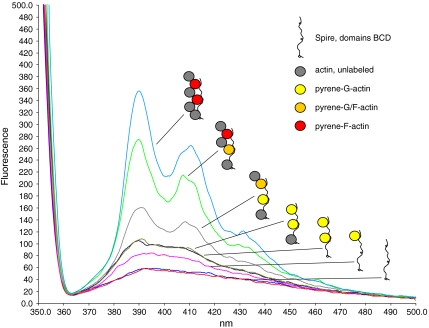
The interaction of SpirBCD with G-actin was studied by fluorescence spectroscopy (Fig. 2). The monomeric pyrene actin shows only a very low signal. Therefore, the Spire construct that contains 3 WH2 domains was primed with pyrene actin at a molar ratio of 2∶3 actin: WH2 domains. Under these conditions the actin remains in its G configuration. The stepwise titration of G-actin into a SpirBCD solution aims at the appearance of the first F-actin signal mediated by pyrenylated G-actin. We find the first F-actin signal only after all three WH2 domains of BCD have been filled with one actin monomer each. The addition of a fourth, even unlabeled G-actin caused a previously nonfluorescent pyrenylated G-actin in the BCD/3xactin complex to change into a fluorescent pyrenylated F-actin (this spectrum is shown in gray in Fig. 2). It is essential for this assay that the addition of unlabeled actin after the appearance of the first F-actin signal mediated by pyrenylated G-actin can lead to an increased fluorescence signal only if the preloaded pyrenylated G-actin is transformed into its F-actin conformation. This was the case when unlabeled actin was added beyond the 1∶1 ratio of actin: WH2 domains (Fig. 2). The data thus suggest a stepwise integration of the WH2-bound G-actins into an emerging F-actin nucleus.
Fig. 2.
Fluorescence emission spectra of SpirBCD titrated stepwise with a two molar excess of pyrene actin followed by the addition of unlabeled G-actin. SpirBCD, 0.2 μM (red, buffer only: blue) was incubated with 2 × 0.2 μM pyrene actin (purple, olive), which led to the expected small fluorescence increase of pyrenylated G-actin. Addition of 0.2 μM of unlabeled G-actin (dark blue) did not change this basic level of fluorescence. Only further addition of unlabeled actin in 0.2-μM aliquots (gray, green, light blue) raised the pyrene signal as it is typical for pyrenylated F-actin, which is completely integrated in a filament. The cartoons show the putative mechanism of the stepwise formation of an F-actin nucleus along Spire WH2 domains. The cartoons at the right do not imply a specific binding order of the actins; they just highlight the first and most-likely formation of an F-actin in the complex.
Crystal Structure of a Single WH2 Domain of Spire and Actin.
A high-resolution structure of the SpirD/actin-latrunculin B complex was determined at up to 1.6 Å in space group P212121 and a unit cell a = 52.96 Å, b = 71.81 Å, c = 100.72 Å by Patterson search molecular replacement using the model of rabbit actin (PDB ID code 1NWK) (35). The overall structure of the single WH2 domain is similar to those previously solved structures of WASP, WASP homologous proteins (PDB ID codes 2A32, 2A40, and 2A41) (30), N-WASP (PDB ID code 2VCP), and Ciboulot (PDB ID code 1SQK), (28) (Fig. 1B and Fig. S4). SpirD comprises residues Ser449-Glu484; Pro461 is the first residue defined by the electron density, which, at the C-terminal part, is truncated after Ile480. The structure of SpirD WH2 consists of a three-turn amphipatic α-helix, whose hydrophobic side includes the highly conserved residues Leu467, Met468, and Ile471. These residues are bound in the hydrophobic cleft formed between subdomains 1 and 3 at the barbed end of the actin monomer followed by the approximately 10 amino acids long C-terminal tail covering the surface of actin between subdomains 2 and 4.
Crystal Structures of the SpirCD/AP-Actin, SpirBCD/AP-Actin, SpirDDD/AP-Actin and SpirABCD/AP-Actin complexes.
Complexes of SpirCD/AP-actin, SpirBCD/AP-actin, SpireDDD/AP-actin, and SpirABCD/AP-actin, which diffracted to 2.6 Å, 2.0 Å, 2.1 Å, and 2.2 Å, respectively, crystallize in the space group P65, with one molecule in the asymmetric unit (Table S1). Processing the SpirBCD/AP-actin data in the subgroup of lower symmetry, P21, and three actin molecules in the asymmetric unit, gave insignificantly better integration statistics (Rmeas were 5.7% for P21 and 6.8% for P65). We carried out the analysis of the SpirBCD data in the lower symmetry to check for any differences in the electron densities among the three WH2 repeats and possibly to see electron density for the linker segments. This, however, was not materialized (see below).
In all complexes entire actin molecules are well-defined in the electron density maps, except for the first three amino acids and the region between residues 39–49, which corresponds to the Dnase I-binding loop in domain 2.
As expected from crystal symmetry and packing, the electron densities for the WH2 domains in all complexes correspond to one structure that resulted from averaging between the three B, C, and D domains in the SpirBCD/AP-actin complex, two C and D domains in the SpirCD/AP-actin complex and four A, B, C, and D domains in the SpirABCD/AP-actin, and three D domains in SpirDDD (Fig. 1C). Except for SpirDDD (see below), we thus could not fit specific amino acids of WH2-A, WH2-B, WH2-C, and WH2-D to the electron density (except for the conserved residues), and therefore a polyglycine model of SpirABCD, SpirBCD, and SpirCD was built in. The conserved residues of the three WH2 repeats marked in Fig. 1 B–D can be recognized, but have not well-defined electron densities (Fig. 1C and Fig. S4 A–C). The fact that there is no difference in the structures of the SpirBCD, SpirABCD, and SpirCD obtained from the P65 and lower symmetry groups argues for the equivalent averaging of all WH2 domains.
It is important to realize that the “averaging” could result from two scenarios. The first possibility is the crystallization of a mixture of complexes where one actin binds one WH2 domain of SpireCD, SpireBCD, or SpirABCD, with the rest of the WH2 domains having a random conformation. In this case the stoichiometry of the complex, for example, for SpirCD would be one actin molecule per one SpirCD. This scenario is not in agreement with the experimental data because we have always the binding of two actin molecules per one SpirCD and so on. In the second mechanism, the averaging in the crystals of SpirCD/AP-actin, SpirBCD/AP-actin, and SpirABCD/AP-actin results from the “shifted” binding of different WH2 domains to equivalent actins along the longitudinal arrangement of actin “columns” in the crystal. To unequivocally characterize the averaging seen in the crystals, we prepared an artificial Spire WH2 construct comprising three identical D repeats, SpirDDD. We could determine the structure of the complex SpirDDD/AP-actin (Table S1, Fig. 1D, and Fig. S4B), and this structure unequivocally points toward the second mechanism, as now the side chains of the Spire helix are well determined by the electron density (Fig. 1D and Fig. S4B). The fact that the electron density for the linkers between D-D modules is not seen in the maps suggests that these fragments may be flexible.
The structures show that the “averaged” WH2 domain of SpirABCD, SpirBCD, SpirCD, and the D domain of SpirDDD binds between subdomains 1 and 3 of actin, and starting at the N-terminal part, forms a short 11-residue helix that further extends to the top of actin between subdomains 2 and 4 (Figs. 1C and 3 B and C). The C-terminal tail of each of the WH2 domains is terminated around Asp25 of the actin molecule. In total, it gives 19 amino acids (glycines or alanines) that could be fitted into the electron density. Linkers L-2 and L-3, consisting of 11 and 12 amino acids, respectively, and L1 from SpirABCD containing 10 amino acids, have no electron density and are not visible in the structure (Fig. 1C).
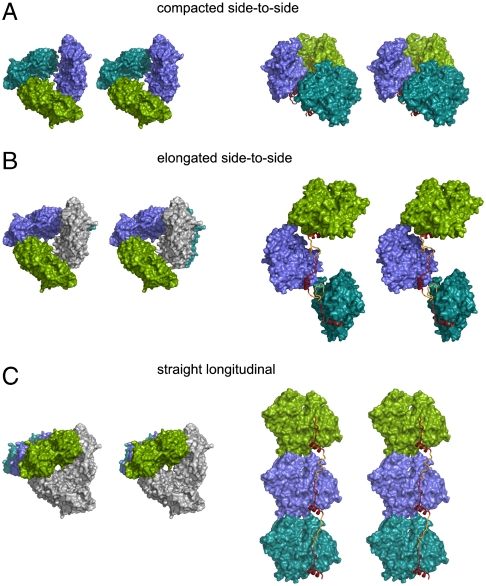
Fig. 3.
The two main architectures of packing in the SpirBCD/AP-actin crystal. Actin molecules are shown as surface plots, WH2 domains as ribbon plots in red/yellow. “Missing” linkers are depicted as ribbons and colored in yellow. (Left) Views along the axis of actin-nucleation seed. (Right) Side views ca. perpendicular to the long axis of the actin-nucleation seed. A and B show the side-to-side architectures. (A) The primary nucleus (colored green, blue, and cyan) is built by the Spire/AP-actin complex inside one asymmetric unit (compacted side-to-side). (B) The primary nucleus is built by the Spire/AP-actin complex, which belongs to two neighboring asymmetric units. Actin in gray belongs to the asymmetric unit that contains the two other actins in green and blue. (C) shows the straight-longitudinal architecture of packing in the SpirBCD/AP-actin crystal. In this model one molecule of actin (i.e., the molecule colored in green) and its two symmetry related ones (actins in blue and cyan) constitute an actin-nucleation seed. Actins in gray belong to the asymmetric unit that contains the actin shown in green.
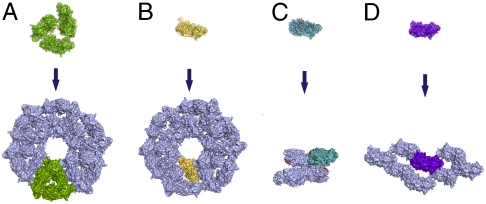
The arrangements of the complexes in the unit cell of SpirCD, SpirBCD, SpirDDD, and that of SpirABCD, are identical, but different from the organization of actin molecules in the crystals of the AP-actin alone (33) and that of our SpirD/actin-latrunculin B complex (Fig. 4). In the Spire/actin complexes the actin-actin interfaces are through the sides of the actin molecules on which the binding of Spire takes place (designated as the outer side of actin, Fig. S5 A and B), whereas in actin alone and in the SpirD/actin-latrunculin B, the actin-actin interfaces are mostly through their inner sides—the parts of actins that participate normally in the central core of a filament (Fig. S5 B and C). This suggests that the specific arrangement of Spire/actin complexes is determined by the cluster of WH2 domains (although the crystal packing for short-range interactions between subdomains 1–3 of one actin molecule and subdomains 2–4 of the neighboring actins are solely determined by contacts between the large actin molecules) (Fig. 4).
Fig. 4.
Crystal packing of different Spire/actin complexes and AP-actin alone. (A) SpirBCD/AP-actin, space group P21. (B) SpirCD/AP-actin, SpirABCD/AP-actin, and SpirDDD/AP-actin, space group P65. (C) SpirD/actin-latrunculin B, space group P212121. (D) AP-actin, space group C121 [PDB ID code 2HF4; (33)]. Crystal packing of SpirBCD/AP-actin and SpirCD/AP-actin, SpirABCD/AP-actin and SpirDDD/AP-actin reveals that, despite different space groups, the arrangement of molecules in both crystals is similar. This suggests the influence of double and triple WH2 domains (in red) on the organization of actin in forming filaments. For comparison, the SpirD/actin-latrunculin B and AP-actin alone do not reveal similarities to the SpirBCD/AP-actin, SpirABCD/AP-actin, SpirCD/AP-actin, and SpirDDD/AP-actin architectures.
Models for the Nucleating Arrangement Between Actin and Spire.
The structures of the SpirCD/AP-actin, SpirBCD/AP-actin, SpirDDD/AP-actin, and SpirABCD/AP-actin complexes suggest the molecular architecture of an actin/Spire nucleation seed. Because of the averaging of the structures of the WH2 domains, model building was based on the analysis of the distances between the C- and N-terminal parts of each single Spire repeat in the crystal arrangement. For this purpose, we searched for distances containing a proper number of amino acids of the linkers (not seen in the structures) that would be compatible with filling the gaps between the WH2 domains’ N and C termini. There are actually only two ways for connecting the averaged WH2 domains of Spire. The nucleating arrangement can be either “side-to-side” (Fig. 3 A and B) or “straight-longitudinal” (Fig. 3C). The first one can be “compacted” when formed inside the one asymmetric unit (Fig. 3A) or more “elongated” when connecting the neighboring asymmetric unit (Fig. 3B). Here we present the models derived from the structure of the SpirBCD/AP-actin complex, identical to those of the SpirCD/AP-actin, SpirABCD/AP-actin, and SpirDDD/AP-actin complexes.
The arrangement of actin molecules in the SpirBCD/AP-actin complex is such that one asymmetric unit contains three WH2 domains (Figs. 3 and 4A). In the “elongated side-to-side” model in Fig. 3B, two WH2 domains bound to the two actins colored in green and blue are linked to one WH2 domain bound to the actin located below this asymmetric unit. In Fig. 3B, the distance between the C terminus of the Spire molecule bound to one actin and the N terminus of Spire bound to neighboring actin is approximately 16 Å, which is sufficient for both linkers L-2 and L-3 to connect the corresponding WH2 domains.
For the “compacted side-to-side” architecture, the distances between the N and C termini of WH2 domains bound to actins inside one asymmetric unit are approximately 23 Å. The calculated area of the interface between two actin molecules for this configuration (compacted side-to-side arrangement) is ca. 250 Å2, and it is twice as large as the one calculated for the elongated side-to-side arrangement. Because there are three actins in the asymmetric unit, the total area of actin-actin interactions for SpirBCD/AP-actin is equal to 750 Å2, which in solution should produce a tight Spire/actin complex. It should be noted here that the compacted side-to side conformation for SpirABCD would have to include a fourth actin molecule (Fig. S6), which is located in the neighboring asymmetric unit; however, the actin-actin interface between the third and fourth actins is still 250 Å2.
In the straight-longitudinal model, Spire connects one actin with its two symmetry closest molecules (Fig. 3C). The distances between the N terminus of one WH2 domain and the C terminus of the next molecule are approximately 30 Å. This configuration of Spire/actin shows some intersubunit actin contacts that occur in one strand of the filament helix: i.e., the interface between subdomains 4 and 2 of one actin molecule (the bottom actin in Fig. 3C) and subdomains 3 and 1 of the next actin molecule (the middle actin molecule in Fig. 3C). The area of the actin-actin interface in the straight-longitudinal model is approximately 350 Å2.
Discussion
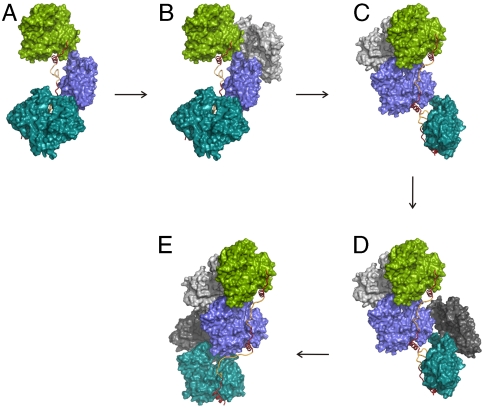
The spatial arrangements of actin molecules in the four SpirABCD, SpirBCD, SpirCD, and SpirDDD structures are of significance because they directly show the organization of a seed for actin nucleation, which represents a primary species that can evolve into a filament (Figs. 3, 5, Fig. S7, and Movie S1). A stretch of WH2 domains is assumed to promote filament nucleation by positioning actin subunits for an emerging filament. In Fig. 3, Spire assembles the nucleus by tethering three WH2-bound actin monomers either into the side-to-side conformations or into the straight-longitudinal arrangement. In the former ones, each actin molecule is at 60° to each other. The elongated side-to-side conformation is much looser than that seen in F-actin; there are short pitch actin-actin contacts between one actin molecule and its neighboring actins, but these are not the F-actin type interfaces (36, 37). This Spire-actin structure is different from the long-pitch twist seen in the filament assemblies (36, 37). The principal difference between this structure and the filament-like arrangement is that in the Spir(A)(B)CD/AP-actin complexes the actin sides that participate in the inner interface of a filament are flipped inside out, with Spire in the core of the side-to-side nucleus (cf. Fig. S5). This last feature of the elongated side-to-side conformation is also present in the compacted side-to-side arrangement. These arrangements might be the representation of a “nonpolimerizable” SA4 complex described by Bosch et al. (26). To “decorate” a full-fledged filament, the Spire has to be on the outside of the filament, and thus it must assume a less energetically favorable conformation, in which all loops (L-1, L-2, and L-3) are stretched into extended secondary structures (Fig. 5 and Fig. S7).
Fig. 5.
Model of actin nucleation by Spire. The stereo expanded version is presented in Fig. S7. (A) The crystal structure of a SpirBCD/AP-actin primary nucleus (elongated side-to-side configuration) (side view perpendicular to the long axis). (B and C) Model of G-actin recruitment and nucleus rearrangement. (D) Filament formation and elongation by addition of actin monomers to the barbed end of growing filament. (E) The filament has been formed.
In the straight-longitudinal model, the twist characteristics of the F-actin filament are not observed. There are some intersubunit actin contacts between adjacent actins. It is clear that both architectures constitute “open” conformations that are solvent permeable with the exposed ATP binding sites, the elongated side-to-side conformation being looser than the straight-longitudinal one.
In our actin polymerization assay, our models would predict that Spir/pyrene-actin complexes show comparable fluorescence emission spectra to pyrenyl-G-actin alone. This is indeed observed. Further, and crucially important, the binding of the unlabeled G-actin to the preformed purified complex of SpirBCD/pyrenyl-actin should be associated with a significant increase of fluorescence after each step of the titration until complete saturation of all lateral binding sites and the formation of longitudinal bonds between actin subunits in one strand of the filament has taken place, as seen in Fig. 2.
Studies of Bosch and co-workers (26) show that at concentrations of SpirNT/actin 0–0.1 μM/2.5 μM, respectively, Spire effectively nucleates actin (SpirNT, residues 1–520 (Fig. 1A). However, at these concentration ratios more than 99% of SpirNT is in the SpirNT/actin complex. At the concentration ratio of SpirNT/actin of 10 nM/2.5 μM, the polymerization is 2 orders of magnitude slower than expected if a total fraction of the SpirNT/actin complex was an actual nucleus. This suggests that only part of the Spire/actin complex forms a highly active nucleus that leads to rapid polymerization of actin. We propose that the compacted side-to-side conformation represents an initial step of binding of Spire to actin and constitutes a stable complex, which then isomerizes to the elongated side-to-side orientation and later to the straight-longitudinal configuration. The latter configuration might represent a direct nucleus (the actin-actin contacts are more extensive), which would be active just before the rotation to form the filamentously twisted structure. This suggests a simple, yet internally consistent, mechanism for Spire-mediated filament nucleation. The transition from the compacted side-to-side arrangement to the elongated side-to-side conformation of Fig. 5A and Fig. S7A is followed by recruitment of free actin monomers (Fig. 5B and Fig. S7B). In the side-to-side arrangement, all “inner” sides of actins (as defined in Fig. S5) are pointing away from the center of the complex. Binding of two free actins to these domains of the actins in the SpirD and SpirC positions (Fig. 5B and Fig. S7B) induces rotation of the SpirC/actin by ca. 120° with respect to the SpirD/actin (Fig. 5C and Fig. S7C) and so on (Fig. 5D and Fig. S7D). Alternatively, the recruitment of free actins might occur just after the isomerization of Spire/actin complexes from the side-to-side arrangements into the straight-longitudinal configuration. This zipper-like nucleation leads to the mature filament (Movie S1). A stable filamentous assembly is thus formed, which is sufficient for rapid filament elongation. In this interpretation, Spire would arrange for filament-like actin conformations (Fig. 5E and Fig. S7E) that are “tightening” into the “packed” full filaments after being processed by the ring-like structures of formins (14, 25, 26).
It was initially proposed that actin subunits in the Spire/actin complex interact via the longitudinal bonds that form one strand of the long-pitch helix in the filament, thus making a filament template or nucleus (16). There was, however, little experimental evidence to support such a model (which has different packing of actin molecules from that of our straight-longitudinal configuration). Recent X-ray scattering analysis of the complex of actin with an artificial tandem of WH2 domains could be interpreted as if a Spire-like protein promotes polymerization by aligning actin subunits in a single strand along the long-pitch helix of the actin filament (34). We, however, propose that the model described by Rebowski et al. (34), which would also be compatible with our data, shows a final stage of the nucleation rather than the initial step of the actin seed formation.
Our model of the nucleation by Spire is also supported by the nucleotide exchange studies of Bosch et al. (26), which show that the nucleotide exchange is only slightly slowed down in the SpirNT-actin complex, indicating an actin arrangement different to filamentous assembly. In SpirNT/actin, ATP dissociates from all four actins in the complex in a single exponential process, indicating that the nucleotide exchange rate is similar for all actin subunits within the complex (26). Thus, the ATP binding sites in all actin molecules have to be freely accessible for the ATP exchange. This is true for the loose elongated side-to-side structure. In F-actin, the ATP exchange is totally inhibited, as the ATP binding sites are blocked by adjacent actin molecules (26).
Our Spire nucleation model is also in agreement with the mutational data of Quinlan et al. (16). Spire promotes filament formation even when all three linker sequences are mutated to the flexible Gly Ser residues, as long as the lengths of the linkers are preserved. In our structures and side-to-side model the linkers are flexible, but their lengths may influence the specific arrangements of actin molecules. On the other hand, recent studies show that the L3 linker is important for actin assembly, and when it is introduced between the two rat N-WASP WH2 domains, L3 can convert rat N-WASP into the Arp2/3-independent nucleator (7). Although the linkers are not visible in our structure, their presence might be vital for free actin recruitment and further nucleation. It is worth mentioning here that the L3 linker is the longest amino acid sequence among all the linkers in Spire [12 residues long vs. 10 and 11 for the two other linkers (Fig. S1)] and longer than the linkers in N-WASP (8 residues, rat and human).
Spire-mediated actin nucleation can also involve direct interaction of the KIND domain of Spire with the FH2 domain of the Cappuccino formin (25). This interaction influences the actin-nucleation properties of both binding partners. Quinlan et al. (25) suggest that Spire initiates the nucleation of filaments, which are then elongated at the barbed end by FH2 domains of the Cappuccino formin. This mode of Spire action would be in agreement with the results presented here, which suggest that Spire stays associated with the pointed end of actin filaments, whereas formins processively move along with the actin barbed ends of the actin structures similar to those shown in Fig. 5.
Materials and Methods
Detailed protocols of protein purification, crystallization, and X-ray data analysis are available in SI Materials and Methods.
Data Collection and Structure Determination.
X-ray datasets of the SpirD/actin-latrunculin B (1.6 Å), SpirBCD/AP-actin (2.0 Å), SpirABCD/AP-actin (2.2 Å), SpirDDD/AP-actin (2.1 Å), and SpirCD/AP-actin (2.6 Å) were collected on the Swiss Light Source beamline PXII at the Paul Scherrer Institut. The datasets were integrated, scaled, and merged by XDS and XSCALE programs (38) (Table S1). The structures were determined by molecular replacement.
Fluorescence Measurements.
Fluorescence measurements were carried out on a Perkin Elmer fluorometer with SpirBCD and rabbit skeletal muscle actin. Pyrenylated and unlabeled actin was isolated according to routine purification procedures. Fluorescence emission was monitored under polymerizing conditions between 350 and 500 nm at an excitation of 342-nm and 8-nm slit widths. The actin was added stepwise to a SpirBCD solution in 800 μL F buffer (10 mM imidazole, pH 7.2, 2 mM MgCl2, 1 mM ATP, 0.2 mM CaCl2), the sample was mixed and incubated for 3 min at room temperature before starting the scan. Under the conditions used (actin concentrations below 1 μM, short time schedule of the measurements), actin alone did not polymerize and the obtained changes in fluorescence emission required the presence of the Spire construct.
Supplementary Material
Acknowledgments.
We thank Daniela Rieger for excellent technical assistance. The work was supported by funds from the Deutsche Forschungsgemeinschaft (to A.A.N. and M.S.) and from the National Institutes of Health (Grant HL38113 to K.M.T.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The crystallography, atomic coordinates, and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3MMV, 3MN5, 3MN6, 3MN7, and 3MN9).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1005347107/-/DCSupplemental.
References
- 1.Pollard TD, Cooper JA. Actin, a central player in cell shape and movement. Science. 2009;27:1208–1212. doi: 10.1126/science.1175862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sept D, McCammon JA. Thermodynamics and kinetics of actin filament nucleation. Biophys J. 2001;81:667–674. doi: 10.1016/S0006-3495(01)75731-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chesarone MA, Goode BL. Actin nucleation and elongation factors: Mechanisms and interplay. Curr Opin Cell Biol. 2009;21:28–37. doi: 10.1016/j.ceb.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mullins RD, Pollard TD. Rho-family GTPases require the Arp2/3 complex to stimulate actin polymerization in Acanthamoeba extracts. Curr Biol. 1999;9:405–415. doi: 10.1016/s0960-9822(99)80187-0. [DOI] [PubMed] [Google Scholar]
- 5.Campellone KG, Webb NJ, Znameroski EA, Welch MD. WHAMM is an Arp2/3 complex activator that binds microtubules and functions in ER to Golgi transport. Cell. 2008;134:148–161. doi: 10.1016/j.cell.2008.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu R, et al. Wash functions downstream of Rho and links linear and branched actin nucleation factors. Development. 2009;136:2849–2860. doi: 10.1242/dev.035246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zuchero JB, Coutts AS, Quinlan ME, Thangue NB, Mullins RD. p53-cofactor JMY is a multifunctional actin nucleation factor. Nat Cell Biol. 2009;11:451–459. doi: 10.1038/ncb1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campellone KG, Welch MD. A nucleator arms race: Cellular control of actin assembly. Nat Rev Mol Cell Biol. 2010;11:237–251. doi: 10.1038/nrm2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Evangelista M, Pruyne D, Amberg DC, Boone C, Bretscher A. Formins direct Arp2/3-independent actin filament assembly to polarize cell growth in yeast. Nat Cell Biol. 2002;4:32–41. doi: 10.1038/ncb718. [DOI] [PubMed] [Google Scholar]
- 10.Evangelista M, Zigmond S, Boone C. Formins: signaling effectors for assembly and polarization of actin filaments. J Cell Sci. 2003;116:2603–2611. doi: 10.1242/jcs.00611. [DOI] [PubMed] [Google Scholar]
- 11.Sagot I, Rodal AA, Moseley J, Goode BL, Pellman D. An actin nucleation mechanism mediated by Bni1 and profilin. Nat Cell Biol. 2002;4:626–631. doi: 10.1038/ncb834. [DOI] [PubMed] [Google Scholar]
- 12.Xu JB, et al. Crystal structures of a formin homology-2 domain reveal a tethered dimer architecture. Cell. 2004;116:711–723. doi: 10.1016/s0092-8674(04)00210-7. [DOI] [PubMed] [Google Scholar]
- 13.Symons M, et al. Wiskott-Aldrich syndrome protein, a novel effector for the GTPase CDC42Hs, is implicated in actin polymerization. Cell. 1996;84:723–734. doi: 10.1016/s0092-8674(00)81050-8. [DOI] [PubMed] [Google Scholar]
- 14.Qualmann B, Kessels MM. New players in actin polymerization-WH2-domain-containing actin nucleators. Trends Cell Biol. 2009;19:276–285. doi: 10.1016/j.tcb.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 15.Pechlivanis M, Samol A, Kerkhoff E. Identification of a short Spir interaction sequence at the C-terminal end of formin subgroup proteins. J Biol Chem. 2009;284:25324–25333. doi: 10.1074/jbc.M109.030320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Quinlan ME, Heuser JE, Kerkhoff E, Mullins RD. Drosophila Spire is an actin nucleation factor. Nature. 2005;433:382–388. doi: 10.1038/nature03241. [DOI] [PubMed] [Google Scholar]
- 17.Ahuja R, et al. Cordon-Bleu is an actin nucleation factor and controls neuronal morphology. Cell. 2007;131:337–350. doi: 10.1016/j.cell.2007.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chereau D, et al. Leiomodin is an actin filament nucleator in muscle cells. Science. 2008;320:239–243. doi: 10.1126/science.1155313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- 20.Robinson RC, et al. Crystal structure of Arp2/3 complex. Science. 2001;294:1679–1684. doi: 10.1126/science.1066333. [DOI] [PubMed] [Google Scholar]
- 21.Otomo T, et al. Structural basis of actin filament nucleation and processive capping by a formin homology 2 domain. Nature. 2005;433:488–494. doi: 10.1038/nature03251. [DOI] [PubMed] [Google Scholar]
- 22.Manseau LJ, Schupbach T. Cappuccino and spire: two unique maternal-effect loci required for both the anteroposterior and dorsoventral patterns of the Drosophila embryo. Genes Dev. 1989;3:1437–1452. doi: 10.1101/gad.3.9.1437. [DOI] [PubMed] [Google Scholar]
- 23.Kerkhoff E, et al. The Spir actin organizers are involved in vesicle transport processes. Curr Biol. 2001;11:1963–1968. doi: 10.1016/s0960-9822(01)00602-9. [DOI] [PubMed] [Google Scholar]
- 24.Rosales-Nieves AE, et al. Coordination of microtubule and microfilament dynamics by Drosophila Rho1, Spire and Cappuccino. Nat Cell Biol. 2006;8:367–376. doi: 10.1038/ncb1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quinlan ME, Hilgert S, Bedrossian A, Mullins RD, Kerkhoff E. Regulatory interactions between two actin nucleators, Spire and Cappuccino. J Cell Biol. 2007;179:117–128. doi: 10.1083/jcb.200706196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bosch M, et al. Analysis of the function of spire in actin assembly and its synergy with formin and profilin. Mol Cell. 2007;28:555–568. doi: 10.1016/j.molcel.2007.09.018. [DOI] [PubMed] [Google Scholar]
- 27.Czisch M, Schleicher M, Horger S, Voelter W, Holak TA. Conformation of thymosin beta 4 in water determined by NMR spectroscopy. Eur J Biochem. 1993;218:335–344. doi: 10.1111/j.1432-1033.1993.tb18382.x. [DOI] [PubMed] [Google Scholar]
- 28.Hertzog M, et al. The β-thymosin/WH2 domain; structural basis for the switch from inhibition to promotion of actin assembly. Cell. 2004;117:611–623. doi: 10.1016/s0092-8674(04)00403-9. [DOI] [PubMed] [Google Scholar]
- 29.Irobi E, et al. Structural basis of actin sequestration by thymosin β4: Implications for WH2 proteins. EMBO J. 2004;23:3599–3608. doi: 10.1038/sj.emboj.7600372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chereau D, et al. Actin-bound structures of Wiskott-Aldrich syndrome protein (WASP) homology domain 2 and the implications for filament assembly. Proc Natl Acad Sci USA. 2005;102:16644–16649. doi: 10.1073/pnas.0507021102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aguda AH, Xue B, Irobi E, Preat T, Robinson RC. The structural basis of actin interaction with multiple WH2/betathymosin motif-containing proteins. Structure. 2006;14:469–476. doi: 10.1016/j.str.2005.12.011. [DOI] [PubMed] [Google Scholar]
- 32.Lee SH, et al. Structural basis for the actin-binding function of missing-in-metastasis. Structure. 2007;15:145–155. doi: 10.1016/j.str.2006.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rould MA, Wan Q, Joel PB, Lowey S, Trybus KM. Crystal structures of expressed non-polymerizable monomeric actin in the ADP and ATP states. J Biol Chem. 2006;281:31909–31919. doi: 10.1074/jbc.M601973200. [DOI] [PubMed] [Google Scholar]
- 34.Rebowski G, et al. X-ray scattering study of actin polymerization nuclei assembled by tandem W domains. Proc Natl Acad Sci USA. 2008;105:10785–10790. doi: 10.1073/pnas.0801650105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Graceffa P, Dominguez R. Crystal structure of monomeric actin in the ATP state. Structural basis of nucleotide-dependent actin dynamics. J Biol Chem. 2003;278:34172–34180. doi: 10.1074/jbc.M303689200. [DOI] [PubMed] [Google Scholar]
- 36.Holmes KC, Popp D, Gebhard W, Kabsch W. Atomic model of the actin filament. Nature. 1990;347:44–49. doi: 10.1038/347044a0. [DOI] [PubMed] [Google Scholar]
- 37.Oda T, Iwasa M, Aihara T, Maeda Y, Narita A. The nature of the globular- to fibrous-actin transition. Nature. 2009;457:441–445. doi: 10.1038/nature07685. [DOI] [PubMed] [Google Scholar]
- 38.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Crystallogr. 1993;26:795–800. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.