Abstract
Synthesis of tRNA and 5S rRNA by RNA polymerase (pol) III is regulated by the mTOR pathway in mammalian cells. The mTOR kinase localizes to tRNA and 5S rRNA genes, providing an opportunity for direct control. Its presence at these sites can be explained by interaction with TFIIIC, a DNA-binding factor that recognizes the promoters of these genes. TFIIIC contains a TOR signaling motif that facilitates its association with mTOR. Maf1, a repressor that binds and inhibits pol III, is phosphorylated in a mTOR-dependent manner both in vitro and in vivo at serine 75, a site that contributes to its function as a transcriptional inhibitor. Proximity ligation assays confirm the interaction of mTOR with Maf1 and TFIIIC in nuclei. In contrast to Maf1 regulation in yeast, no evidence is found for nuclear export of Maf1 in response to mTOR signaling in HeLa cells. We conclude that mTOR associates with TFIIIC, is recruited to pol III–transcribed genes, and relieves their repression by Maf1.
Keywords: pol III transcription, rapamycin
Synthesis of ribosomal components can monopolize 90% of all transcription under conditions of rapid growth (1). It requires RNA polymerase (pol) I to produce the large rRNAs, pol II to produce the mRNAs encoding ribosomal proteins (RPs), and pol III to produce 5S rRNA (2). The metabolic demands made by this biosynthetic program necessitate tight coordination and the ability to respond to energy status and nutrient availability. The target of rapamycin (TOR) pathway performs this role in yeast and higher organisms (3, 4). For example, inactivation of TOR in Saccharomyces cerevisiae using the drug rapamycin causes a rapid and coordinated decrease in expression of virtually all genes involved in ribosome production. This involves the down-regulated activity of pols I and III, along with a more selective decrease in pol II transcription of RP genes (5, 6). Exposure of this yeast to rapamycin also triggers a decrease in expression of the Ribi (ribosome biogenesis) regulon, which encodes many nonribosomal proteins involved in the production and maturation of ribosomes (7). The Ribi regulon is the largest set of coregulated genes in S. cerevisiae, further illustrating the magnitude of the task of coordinating ribosome production.
Expression of S. cerevisiae RP genes is controlled by the transcription factors SFP1 and CRF1. TOR regulates the localization of these factors and thereby dictates whether they can access RP gene promoters in the nucleus (7, 8). This does not require that TOR enter the nucleus itself, because SFP1 and CRF1 both traffic into the cytoplasm (7, 8). The subcellular localization of the pol III transcriptional repressor Maf1 is also controlled through TOR signaling, which can cause its nuclear exclusion (9–11). Here we show that mammalian Maf1 is subject to rapamycin-sensitive phosphorylation as well, but that this has a minimal effect on its distribution between nuclei and cytoplasm, in contrast to the situation in S. cerevisiae. Nuclear Maf1 may be accessed directly by mTOR, which is detected at tRNA and 5S rRNA genes in vivo. Its presence can be explained by interaction with TFIIIC, a transcription factor that binds to pol III promoters. Localization of mTOR to these sites may allow tRNA and 5S rRNA expression to respond rapidly and directly to the availability of nutrients and growth factors.
Results
Human Maf1 Is Dephosphorylated in Response to Rapamycin.
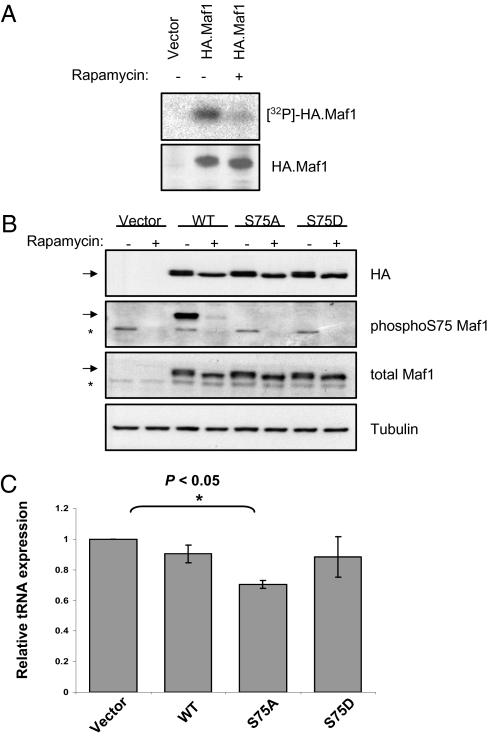
Maf1 is highly phosphorylated in actively growing yeast, but rapamycin triggers its dephosphorylation, which correlates with its activation as a repressor (12–18). To test for similar control in mammals, HeLa cells expressing HA-tagged Maf1 were labeled in vivo with [32P]orthophosphate. Anti-HA antibody immunoprecipitated phospholabeled Maf1 from these cells, but incorporation of [32P] into Maf1 was substantially decreased by rapamycin (Fig. 1A).
Fig. 1.
Serine 75 is a functionally significant site in Maf1 that is phosphorylated in a rapamycin-sensitive manner. (A) HeLa cells transfected with vector or HA-tagged Maf1 were labeled for 3 h with [32P] orthophosphate in the presence of vehicle or rapamycin, as indicated. Panels show autoradiography (Upper) and Western blot analysis (Lower) after immunoprecipitation with anti-HA antibody. (B) Cells were transfected with empty vector or with vector encoding WT, S75A, or S75D Maf1, as indicated, and treated with vehicle or rapamycin. Western blots are shown with antibodies against HA, S75-phosphorylated Maf1, total Maf1, and tubulin. The arrow indicates transfected HA-tagged Maf1; the asterisk indicates endogenous Maf1. (C) Expression of pre-tRNATyr was assessed by real-time qRT-PCR following transfection with empty vector or with vector encoding WT, S75A, or S75D Maf1. Data presented represent levels of pre-tRNATyr after normalization to TFIIB mRNA, with empty vector assigned a value of 1.0. n = 3.
A mass spectrometry screen identified serine 75 (S75) of human Maf1 as being phosphorylated in vivo (19). We raised a phosphospecific antibody that recognizes this site. The specificity of the antibody was confirmed by the finding that it failed to detect transfected HA-tagged Maf1 if S75 is substituted by alanine (S75A) or aspartate (S75D), although these mutants were expressed as efficiently as the wild type (Fig. 1B). This antibody revealed that rapamycin treatment severely diminishes S75 phosphorylation in both transfected WT Maf1 and endogenous Maf1, indicating that the mTORC1 pathway controls phosphorylation at this site in vivo. However, it is unlikely to be the only site phosphorylated in response to mTOR signaling, given that the S75 mutants show a clear electrophoretic mobility shift after rapamycin treatment. Multisite phosphorylation is also a feature of 4E-BPs and S6Ks, the best-characterized mTOR substrates (20).
To assess the functional significance of S75, we compared the expression of pre-tRNATyr after transfection of WT and mutant Maf1 (Fig. 1C). Under the conditions of our assay, WT Maf1 was not strongly repressive, decreasing pre-tRNATyr levels by only 10%. This might be because it is heavily phosphorylated in growing HeLa cells. Consistent with this, phosphomimetic S75D substitution had minimal effect on activity. In contrast, the phosphoresistant S75A mutant suppressed pre-tRNATyr expression significantly (P < 0.05), causing a 30% decrease in pre-tRNATyr. Similar results were obtained with tRNAiMet, which was 19% suppressed by WT Maf1 but 39% suppressed by the S75A mutant (Fig. S1). These data demonstrate that S75 contributes to Maf1 function. Substitution of this residue does not overcome sensitivity to rapamycin, consistent with our belief that additional phosphoacceptor sites are also involved in controlling Maf1. Nevertheless, we can conclude that S75 contributes to Maf1 function in vivo and is phosphorylated in response to mTOR.
Rapamycin Does Not Cause Nuclear Import of Maf1 in HeLa Cells.
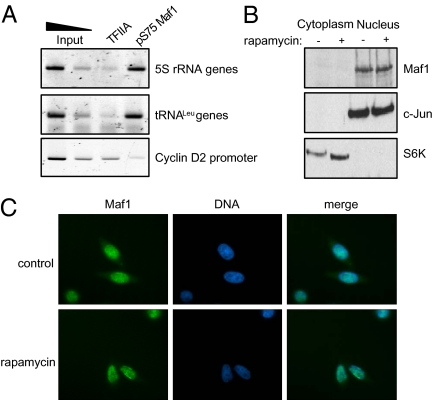
In S. cerevisiae, TOR-dependent phosphorylation of Maf1 can cause its dissociation from the pol III machinery and export from the nucleus (11–16). In contrast, ChIP assays with our phosphospecific antibody demonstrated that human Maf1 can be cross-linked to tRNA and 5S rRNA genes when it is phosphorylated at S75 (Fig. 2A). The pol II–specific transcription factor TFIIA served as a negative control. This finding suggests that phosphorylation of Maf1 might not cause its export in mammalian cells. Consequently, we used subcellular fractionation to examine the localization of Maf1 and to investigate whether this is affected by rapamycin. Maf1 was found predominantly in the nuclei irrespective of rapamycin (Fig. 2B). This was confirmed by immunofluorescence (Fig. 2C). As controls against the selective loss of cytoplasmic proteins, we confirmed the presence of S6 kinase in the subcellular fractions (Fig. 2B) and the detection of cytoplasmic actin by immunofluoresence (Fig. S2). We conclude that redistribution of Maf1 from the cytoplasm is not a feature of the response to rapamycin in HeLa cells. We extended our analysis to several other cell types, including human osteosarcoma and breast cancer lines, as well as murine fibroblasts. In each case, Maf1 immunofluorescence was predominantly nuclear and showed minimal response to rapamycin (Fig. S3).
Fig. 2.
Phosphorylated Maf1 is retained in the nucleus. (A) ChIP assay showing cross-linking in HeLa cells of S75-phosphorylated Maf1 at tRNA and 5S rRNA genes, but not the cyclin D2 promoter. TFIIA serves as negative control. (B) Western blots with equal amounts of protein from nuclear and cytoplasmic fractions of cells treated with vehicle (−) or rapamycin (+). c-Jun and S6 kinase serve as nuclear and cytoplasmic markers, respectively. (C) Indirect immunofluoresence with antibody against Maf1 in HeLa cells treated with vehicle or rapamycin.
Endogenous mTOR Can Be Cross-Linked to pol III-Transcribed Genes in Vivo.
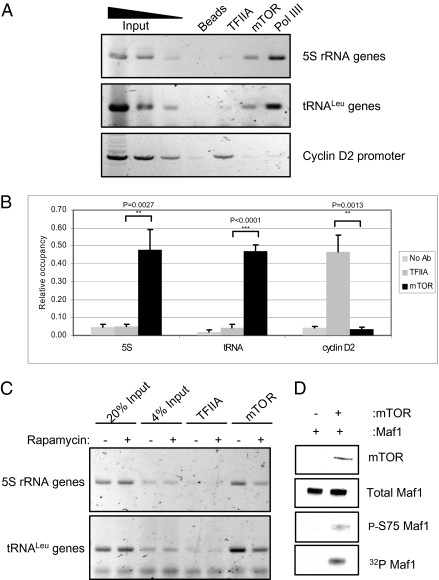
TOR1 was reported to ChIP on 5S rRNA genes in S. cerevisiae (18, 21). We tested whether mTOR can be cross-linked to such genes in mammalian cells. ChIP assays showed that this is indeed the case, in both HeLa cells (Fig. 3A) and fibroblasts (Fig. S4). The interaction appears to be specific, given that mTOR was not detected at the promoter of the pol II–transcribed cyclin D2 gene. This was not due to failure of the cyclin D2 ChIP, because TFIIA was detected specifically at this site. Endogenous mTOR not only was found at chromosomal 5S rRNA genes, but also was cross-linked to tRNA genes. This contrasts with what was reported for S. cerevisiae, where TOR1 was detected at 5S RNA genes but not at tRNA genes (18). Although 5S rRNA has been reported to respond more strongly than pre-tRNA following rapamycin treatment of S. cerevisiae (18), this was not the case in the mammalian cell lines that we examined (Fig. S5). Quantitation of multiple experiments confirmed that cross-linking of mTOR to 5S and tRNA genes is significantly above the background signal obtained with control antibody to TFIIA, with P < 0.0001 for tRNA genes in HeLa cells (Fig. 3B). The mTOR ChIP was rapamycin-sensitive, providing additional evidence of its authenticity (Fig. 3C). It was verified using an alternative mTOR antibody (Fig. S6).
Fig. 3.
mTOR is found at tRNA and 5S rRNA genes and may phosphorylate Maf1. (A) ChIP assays showing mTOR occupancy at tRNA and 5S rRNA genes in HeLa cells. Antibody to pol III was used as a positive control, and antibody to TFIIA and beads without antibody provide negative controls. (B) Quantification of A and two additional independent experiments. (C) ChIP assays showing mTOR occupancy at tRNA and 5S rRNA genes in HeLa cells treated with vehicle (−) or rapamycin (+). Antibody to TFIIA provided a negative control. (D) Recombinant mTOR fragment (residues 1362–2549) was used to phosphorylate recombinant Maf1 in vitro. Top three panels show Western blots using antibodies against mTOR, total Maf1, and phosphorylated S75 of Maf1, as indicated. (Bottom) Autoradiograph of 32P incorporation.
The presence of mTOR at pol III templates may allow it to regulate transcription of these genes directly. This possibility is supported by the fact that bacterially expressed recombinant Maf1 can be phosphorylated in vitro on S75 using immunoprecipitated mTOR (Fig. S7). Because an associated or contaminating kinase might have coimmunoprecipitated with mTOR and phosphorylate Maf1 in this assay, we repeated the experiment using a baculovirus-expressed recombinant C-terminal fragment of mTOR (22). Again we found phosphorylation of Maf1, as revealed by labeling with [32P], and blotting with the phosphospecific antibody confirmed S75 as a phosphoacceptor (Fig. 3D). These data provide evidence that S75 of Maf1 can serve as a substrate for mTOR in vitro and suggest that direct regulation also might occur in vivo.
TFIIIC Interacts with mTOR in the Nucleus.
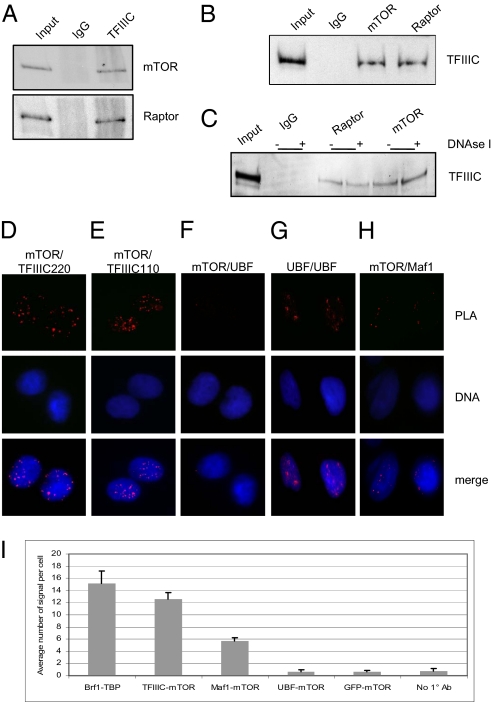
To explain the presence of mTOR at tRNA and 5S rRNA genes, we searched for interactions with the pol III transcription machinery. A stable association was detected with the promoter-binding factor TFIIIC. Thus, antibody against TFIIIC was found to coimmunoprecipitate endogenous mTOR (Fig. 4A) and vice versa (Fig. 4B). These interactions were confirmed with alternative antibodies (Fig. S8). In the same way, association was detected with endogenous raptor, which binds to mTOR in the mTORC1 complex (23, 24). The possibility that coimmunoprecipitation was mediated by contaminating DNA was excluded by treatment with DNase 1 (Fig. 4C). These data suggest that mTOR and raptor form a complex with the pol III–specific transcription factor TFIIIC that is stable to cell lysis. This complex can be detected without the need to overexpress any of its components.
Fig. 4.
TFIIIC associates with mTOR. (A) Western blot analysis using mTOR and raptor antibodies of input and immunoprecipitates produced with nonimmune IgG or antibody to TFIIIC110. (B) Western blot analysis using TFIIIC110 antibody of input and immunoprecipitates produced with nonimmune IgG and antibodies to raptor and mTOR. (C) Western blot analysis using TFIIIC110 antibody of immunoprecipitates produced in the presence (+) or absence (−) of DNase I with nonimmune IgG and antibodies to raptor and mTOR. (D) PLA using antibodies against mTOR and TFIIIC220. (E) PLA using antibodies against mTOR and TFIIIC110. (F) PLA using antibodies against mTOR and UBF. (G) PLA using two antibodies against UBF. (H) PLA using antibodies against mTOR and Maf1. (I) Quantification of PLA assays conducted using antibodies against the indicated pairs of proteins or without primary antibodies. The well-characterized interaction between TFIIIB subunits TBP and Brf1 was included as a positive control. GFP provided a negative control, because it is not expressed in these cells.
Proximity ligation assays (PLAs) provided additional evidence for association of mTOR and TFIIIC in vivo. This less-invasive technique allows detection of native complexes with minimal cellular disruption (25, 26). A signal is obtained only if oligonucleotides coupled to two separate antibodies are sufficiently close to allow enzymatic ligation; detection then relies on amplification from the ligated template, followed by hybridization with fluorescent probe (25, 26). A strong signal is detected using antibodies against mTOR and either of two TFIIIC subunits (Figs. 4 D and E). DAPI staining for DNA suggests that this in vivo interaction occurs predominantly in nuclei. In contrast to the results obtained with mTOR and TFIIIC, the low signal obtained with antibodies against mTOR and the abundant pol I–specific factor UBF is comparable to the background seen in the absence of primary antibody (Fig. 4F). Function of the UBF antibody in this assay was confirmed by detection of strong signals when it was combined with a second UBF antibody (Fig. 4G). However, nuclear association was detected between endogenous mTOR and Maf1, consistent with the latter being a substrate for the former (Fig. 4H). Quantitation of images of >100 cells from each of three independent experiments revealed that the mTOR/Maf1 signal was less pronounced than the mTOR/TFIIIC signals, but nevertheless well above the background obtained with a GFP antibody (Fig. 4I). These data suggest close proximity of endogenous mTOR with both Maf1 and TFIIIC in the nuclei of intact cells.
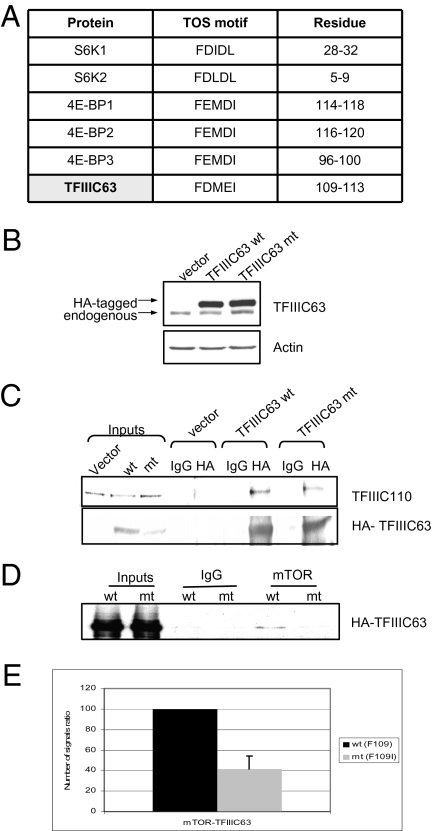
Targeting of S6K1 and 4E-BP1 involves a TOR signaling (TOS) motif in these substrates that is recognized by raptor (27–31). We identified a similar motif in TFIIIC63, a subunit of TFIIIC (Fig. 5A). To test its relevance, we substituted the phenylalanine at position 109 to isoleucine, because the equivalent substitution in the TOS motif of 4E-BP1 was shown to significantly compromise raptor binding (31). An HA-tagged version of this F109I TFIIIC63 mutant was expressed as efficiently as WT when transfected into cells (Fig. 5B). Its interaction with another TFIIIC subunit appeared to be normal, indicating that its conformation was not severely compromised by the substitution (Fig. 5C). However, endogenous mTOR coimmunoprecipitated less efficiently with the mutant than with WT TFIIIC63 (Fig. 5D). Evidence that the F109I substitution compromises interaction with mTOR also was provided by PLA (Fig. 5E). Residual association of the mutant with mTOR was more apparent on PLA than on coimmunoprecipitation, probably because the latter is more disruptive of weak interactions. Nevertheless, these data suggest that the TOS motif is involved in bringing mTOR into close proximity with TFIIIC in vivo. This could target mTORC1 to pol III–transcribed genes.
Fig. 5.
TFIIIC63 contains a TOS motif that contributes to the interaction between TFIIIC and mTOR. (A) Sequence and location of a putative TOS motif in TFIIIC63 and comparison with TOS motifs in S6K and 4E-BP proteins. (B) Western blot analysis with antibodies against TFIIIC63 and actin of lysates from cells transfected with empty vector, vector encoding HA-tagged WT TFIIIC63 (wt), or vector encoding HA-tagged mutant (mt) TFIIIC63 carrying a F109I substitution. (C) Western blot analysis for TFIIIC110 and HA-TFIIIC63 coimmunoprecipitated with HA antibody or nonimmune IgG from cells transfected with vectors encoding HA-tagged WT TFIIIC63 (wt) or F109I mutant TFIIIC63 (mt). (D) Western blot analysis with HA antibody for HA-TFIIIC63 coimmunoprecipitated with mTOR antibody or nonimmune IgG from cells transfected with vectors encoding HA-tagged WT TFIIIC63 (wt) or F109I mutant TFIIIC63 (mt). (E) Quantification of the interaction between mTOR and transfected WT or mutant (F109I) TFIIIC63 as determined by PLA in three independent experiments. The signals were normalized to cell number, and the WT signal was arbitrarily assigned a value of 100.
Discussion
We have established that endogenous mTOR associates with TFIIIC and can be cross-linked to tRNA and 5S rRNA genes in vivo. Such positioning may allow it to phosphorylate and regulate the pol III transcription apparatus directly. Maf1 appears to be a substrate, given that we have identified S75 as a phosphoacceptor site that is rapamycin-sensitive in vivo and can be phosphorylated directly in vitro by recombinant mTOR. Although other residues are phosphorylated as well, S75 clearly influences Maf1 activity, as indicated by the significant increase in its repressive effect on tRNA expression by S75A substitution. Thus, we postulate that TFIIIC recruits mTOR to promoters, which then stimulates pol III transcription by inactivating the repressor Maf1. This allows a cell to adapt pol III output to its energy status and the availability of nutrients and growth factors.
While this work was under review and revision, two overlapping studies were published. Tsang et al. (32) demonstrated that mTOR can be cross-linked to tRNA and 5S rRNA genes in mouse β-TC6 pancreatic cells, and Shor et al. (33) showed the same to be true in human MG63 osteosarcoma and HEK293 kidney cells. The latter study also found that raptor can be cross-linked to these loci (33). From these studies, along with our observations in HeLa cells and murine fibroblasts, the presence of mTOR at pol III–transcribed genes has been established in five distinct cell types of disparate origin, providing a strong indication that the phenomenon is widespread. It is also noteworthy that five different antibodies have been used among the three studies to ChIP mTOR at these sites. Further evidence against the possibility of fortuitous cross-reaction is the sensitivity to rapamycin and the absence of cross-linking to various control loci, including cyclin D2 (Fig. 3) and GAPDH (32). Thus, this appears to be a very robust observation.
The mTOR kinase domain is considered to have little inherent sequence specificity (34). Substrate preference instead appears to be conferred by targeting through mTOR-associated proteins, such as raptor, which recognizes the TOS motif (29–31). The presence of a functional TOS motif in TFIIIC63, which contributes to TORC1 binding, suggests that TFIIIC63 may be bound directly by raptor. Additional interactions are likely involved as well, given that diminished binding remains detectable by PLA with the F109I mutant of TFIIIC63.
When bound to TFIIIC, mTOR might be close enough to phosphorylate several components of the pol III machinery. Phosphorylation of Brf1 and Bdp1 has been shown to respond to PTEN (35), an effect that may be mediated by mTOR. We have found that rapamycin reduces phosphorylation of TFIIIC, although we have no evidence that this effect is functionally significant. Pol III output remains sensitive to rapamycin in cells transfected with the phosphomimetic S75D mutant of Maf1. Control may be complex and involve multiple targets. We chose to focus on Maf1 because of the genetic evidence that it is required for the response of pol III transcription to rapamycin in yeast (36). Strong evidence that Maf1 is a key target in higher organisms has come from the finding that pre-tRNA expression becomes insensitive to mTOR inhibitors when Maf1 is depleted from MG63 cells by RNAi (33). We found that human Maf1 is phosphorylated in vivo at serine 75 in a rapamycin-sensitive manner, although mobility shifts of an S75A mutant point to the existence of additional phosphoacceptor sites. Serine 75 was found to be relevant to transcriptional repression by human Maf1; however, it likely is only one component of a complex control system, given that pol III transcription remains responsive to rapamycin in cells transfected with the S75A mutant. In a global proteomic analysis in MDA361 breast cancer cells, Shor et al. (33) identified S75 of Maf1 as being phosphorylated in an mTOR-dependent manner. This phosphorylation decreased by >99% when the cells were treated with WYE-132, a potent and specific mTOR active site inhibitor with a different mode of action to rapamycin (33). These authors also showed that an S75A substitution enhances the ability of transfected Maf1 to suppress expression of pre-tRNAs (33). Despite using a different cell type, their findings are entirely consistent with our own data (Fig. 1C). Repression of pre-tRNA expression was further enhanced by combining the S75A mutation with alanine substitutions at three additional phosphoacceptor sites (33); however, whether these additional sites are responsive to mTOR signaling has yet to be confirmed.
We have shown that mTOR can phosphorylate Maf1 directly in vitro. Although this might be true in vivo as well, we would not rule out S6 kinase (S6K) as an intermediary. Woiwode et al. (35) found that a constitutively activated S6K1 mutant could reverse the repression of pol III activity seen when PTEN is overexpressed in glioblastoma cells, but that S6K1 did not promote pol III output in the absence of PTEN overexpression. Interpretation is complex, because a feedback loop allows S6K1 to influence the pathway regulated by PTEN (37–39). Perhaps S6K1 regulates pol III transcription in a manner controlled by mTOR, but this mechanism might show redundancy with mTORC1 acting directly when bound to TFIIIC. Such a situation might occur in S. cerevisiae; there is strong evidence that ScMaf1 is regulated by Sch9, a yeast kinase with functional similarities to mammalian S6Ks (16, 17, 40). However, ScMaf1 is subject to redundant control, with protein kinase A and Sch9 targeting the same phosphoacceptor sites (17). In addition, immunoprecipitated TOR1 has been found to phosphorylate recombinant ScMaf1 in vitro, although a phosphoacceptor has not been identified, and the possibility that PKA or Sch9 coimmunoprecipitate with TOR1 has not been excluded (18). The resemblance between Sch9 and S6Ks obviously strengthens the possibility that S6Ks might mediate an effect of mTOR on mammalian Maf1; however, all eight phosphoacceptor sites identified for Sch9 and PKA in ScMaf1 lie within domains that are absent from Maf1 in mammals, which is shorter than its yeast orthologs (Fig. S9). Thus, this aspect of the pathway might be fundamentally different in yeast and mammals. A serine is found in ScMaf1 (S166) at the equivalent position to S75 in humans when the sequences are aligned, but the surrounding residues are not conserved, again suggesting that regulation is distinct. TFIIIC also is poorly conserved through evolution (41), and the ortholog of TFIIIC63 in S. cerevisiae, TFC1p, does not contain the TOS motif, which appears to be important for mTOR interaction in humans.
Another key difference between yeast and mammals is that phosphorylation of ScMaf1 causes it to dissociate from pol III in yeast cells (13). In contrast, mammalian pol III, TFIIIB, and TFIIIC remain associated with Maf1 after its phosphorylation induced by serum stimulation (42). This can explain our finding that S75-phosphorylated Maf1 can be cross-linked to tRNA genes in HeLa cells (Fig. 2A).
Rapamycin treatment of budding yeast can cause Maf1 to become dephosphorylated and concentrated in the nucleus (12–17). However, pol III transcription remains responsive to rapamycin in mutant strains with constitutively nuclear Maf1 (12, 15, 18). This indicates that yeast TORC1 can regulate Maf1 without excluding it from the nucleus. Using microscopy and subcellular fractionation, we found that human Maf1 concentrates in the nuclei of HeLa cells irrespective of mTOR signaling. The same was true for each of the various other cell lines that we examined (MCF-7, U2OS, DU145, and A31 cells). Controls confirmed the efficacy of rapamycin treatment in each case. In contrast, Shor et al. (33) reported significant Maf1 staining in the cytoplasm of MG63 osteosarcoma cells, and noted that the nuclear content increased in response to WYE-132 or the rapamycin analog CCI-779. This discrepancy might reflect the use of different cell types and is reminiscent of the situation in yeast, where Maf1 is constitutively nuclear in one strain but not another (18). Our data demonstrate that this aspect of Maf1 regulation is dispensable for its function in some mammalian cell types, consistent with its inessential contribution in budding yeast.
Pol III is responsible for ∼10% of all nuclear RNA production, but its impact on biosynthetic capacity goes far beyond this. Translation is a primary determinant of the rate of cell growth, given that 80–90% of a cell's dry mass is protein (43). By providing tRNA and 5S rRNA, pol III has a major impact on translational capacity. Indeed, tRNA availability might be rate-limiting for protein synthesis under some conditions, as suggested by the increased translation observed in fibroblasts transfected with tRNAiMet genes (44). Direct control of pol III transcription by mTOR bound to TFIIIC might facilitate a rapid and efficient response to changes in growth conditions. This resembles the location of mTOR at genes involved in mitochondrial function through its interaction with the DNA-binding protein YY1 (45). Recruitment to key target genes may prove to be an important aspect of mTOR biology.
Materials and Methods
Cell Culture and Transfections.
Cell lines and culture and transfection conditions are described in SI Materials and Methods. Rapamycin was applied for 4 h at a final concentration of 100 nM.
Mutagenesis.
Mutations were introduced using the QuikChange II Site-Directed Mutagenesis Kit (Stratagene), as detailed in SI Materials and Methods
Cell Lysis and Fractionation.
SI Materials and Methods provides detailed descriptions of the preparation of cell extracts and subcellular fractions.
Phosphate Labeling and Kinase Assays.
Details of theses assays are provided in SI Materials and Methods.
Gene Expression.
RNA extraction, PCR, RT-qPCR, and Western blot analyses are described in SI Materials and Methods
ChIP and Coimmunoprecipitation.
ChIP assays were performed as described previously (46). Details of immunoprecipitation procedures, antibodies, and data analysis are provided in SI Materials and Methods.
Immunofluorescence and PLA.
Details of immunofluorescence analysis are provided in SI Materials and Methods PLA was performed using the Duolink In Situ PLA Kit (Olink Bioscience), as described in SI Materials and Methods
Supplementary Material
Acknowledgments
We thank Michael Hall, Eyal Gottlieb, Chris Proud, John Blenis, and David Sabatini for discussions regarding these data. This work was funded by Grant 081977/Z/07/Z from the Wellcome Trust, by a Glasgow University studentship, and by Cancer Research UK.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. S.H. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1005188107/-/DCSupplemental.
References
- 1.Warner JR. The economics of ribosome biosynthesis in yeast. Trends Biochem Sci. 1999;24:437–440. doi: 10.1016/s0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- 2.Mayer C, Grummt I. Ribosome biogenesis and cell growth: mTOR coordinates transcription by all three classes of nuclear RNA polymerases. Oncogene. 2006;25:6384–6391. doi: 10.1038/sj.onc.1209883. [DOI] [PubMed] [Google Scholar]
- 3.Mamane Y, Petroulakis E, LeBacquer O, Sonenberg N. mTOR, translation initiation and cancer. Oncogene. 2006;25:6416–6422. doi: 10.1038/sj.onc.1209888. [DOI] [PubMed] [Google Scholar]
- 4.Martin DE, Powers T, Hall MN. Regulation of ribosome biogenesis: Where is TOR? Cell Metab. 2006;4:259–260. doi: 10.1016/j.cmet.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 5.Zaragoza D, Ghavidel A, Heitman J, Schultz MC. Rapamycin induces the G0 program of transcriptional repression in yeast by interfering with the TOR signaling pathway. Mol Cell Biol. 1998;18:4463–4470. doi: 10.1128/mcb.18.8.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Powers T, Walter P. Regulation of ribosome biogenesis by the rapamycin-sensitive TOR-signaling pathway in Saccharomyces cerevisiae. Mol Biol Cell. 1999;10:987–1000. doi: 10.1091/mbc.10.4.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jorgensen P, et al. A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes Dev. 2004;18:2491–2505. doi: 10.1101/gad.1228804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martin DE, Soulard A, Hall MN. TOR regulates ribosomal protein gene expression via PKA and the Forkhead transcription factor FHL1. Cell. 2004;119:969–979. doi: 10.1016/j.cell.2004.11.047. [DOI] [PubMed] [Google Scholar]
- 9.Geiduschek EP, Kassavetis GA. Transcription: Adjusting to adversity by regulating RNA polymerase. Curr Biol. 2006;16:R849–R851. doi: 10.1016/j.cub.2006.08.071. [DOI] [PubMed] [Google Scholar]
- 10.Willis IM, Moir RD. Integration of nutritional and stress signaling pathways by Maf1. Trends Biochem Sci. 2007;32:51–53. doi: 10.1016/j.tibs.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 11.Cieśla M, Boguta M. Regulation of RNA polymerase III transcription by Maf1 protein. Acta Biochim Pol. 2008;55:215–225. [PubMed] [Google Scholar]
- 12.Moir RD, et al. Protein kinase A regulates RNA polymerase III transcription through the nuclear localization of Maf1. Proc Natl Acad Sci USA. 2006;103:15044–15049. doi: 10.1073/pnas.0607129103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oficjalska-Pham D, et al. General repression of RNA polymerase III transcription is triggered by protein phosphatase type 2A–mediated dephosphorylation of Maf1. Mol Cell. 2006;22:623–632. doi: 10.1016/j.molcel.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 14.Roberts DN, Wilson B, Huff JT, Stewart AJ, Cairns BR. Dephosphorylation and genome-wide association of Maf1 with Pol III–transcribed genes during repression. Mol Cell. 2006;22:633–644. doi: 10.1016/j.molcel.2006.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Towpik J, Graczyk D, Gajda A, Lefebvre O, Boguta M. Derepression of RNA polymerase III transcription by phosphorylation and nuclear export of its negative regulator, Maf1. J Biol Chem. 2008;283:17168–17174. doi: 10.1074/jbc.M709157200. [DOI] [PubMed] [Google Scholar]
- 16.Huber A, et al. Characterization of the rapamycin-sensitive phosphoproteome reveals that Sch9 is a central coordinator of protein synthesis. Genes Dev. 2009;23:1929–1943. doi: 10.1101/gad.532109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee J, Moir RD, Willis IM. Regulation of RNA polymerase III transcription involves SCH9-dependent and SCH9-independent branches of the target of rapamycin (TOR) pathway. J Biol Chem. 2009;284:12604–12608. doi: 10.1074/jbc.C900020200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wei Y, Tsang CK, Zheng XFS. Mechanisms of regulation of RNA polymerase III–dependent transcription by TORC1. EMBO J. 2009;28:2220–2230. doi: 10.1038/emboj.2009.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsuoka S, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 20.Proud CG. mTORC1 signalling and mRNA translation. Biochem Soc Trans. 2009;37:227–231. doi: 10.1042/BST0370227. [DOI] [PubMed] [Google Scholar]
- 21.Li H, Tsang CK, Watkins M, Bertram PG, Zheng XFS. Nutrient regulates Tor1 nuclear localization and association with rDNA promoter. Nature. 2006;442:1058–1061. doi: 10.1038/nature05020. [DOI] [PubMed] [Google Scholar]
- 22.Michlewski G, Sanford JR, Cáceres JF. The splicing factor SF2/ASF regulates translation initiation by enhancing phosphorylation of 4E-BP1. Mol Cell. 2008;30:179–189. doi: 10.1016/j.molcel.2008.03.013. [DOI] [PubMed] [Google Scholar]
- 23.Kim D-H, et al. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110:163–175. doi: 10.1016/s0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- 24.Hara K, et al. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell. 2002;110:177–189. doi: 10.1016/s0092-8674(02)00833-4. [DOI] [PubMed] [Google Scholar]
- 25.Fredriksson S, et al. Protein detection using proximity-dependent DNA ligation assays. Nat Biotechnol. 2002;20:473–477. doi: 10.1038/nbt0502-473. [DOI] [PubMed] [Google Scholar]
- 26.Söderberg O, et al. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat Methods. 2006;3:995–1000. doi: 10.1038/nmeth947. [DOI] [PubMed] [Google Scholar]
- 27.Schalm SS, Blenis J. Identification of a conserved motif required for mTOR signaling. Curr Biol. 2002;12:632–639. doi: 10.1016/s0960-9822(02)00762-5. [DOI] [PubMed] [Google Scholar]
- 28.Schalm SS, Fingar DC, Sabatini DM, Blenis J. TOS motif mediated raptor binding regulates 4E-BP1 multisite phosphorylation and function. Curr Biol. 2003;13:797–806. doi: 10.1016/s0960-9822(03)00329-4. [DOI] [PubMed] [Google Scholar]
- 29.Choi KM, McMahon LP, Lawrence JC., Jr. Two motifs in the translational repressor PHAS-I required for efficient phosphorylation by mammalian target of rapamycin and for recognition by raptor. J Biol Chem. 2003;278:19667–19673. doi: 10.1074/jbc.M301142200. [DOI] [PubMed] [Google Scholar]
- 30.Nojima H, et al. The mammalian target of rapamycin (mTOR) partner, raptor, binds the mTOR substrates p70 S6 kinase and 4E-BP1 through their TOR signaling (TOS) motif. J Biol Chem. 2003;278:15461–15464. doi: 10.1074/jbc.C200665200. [DOI] [PubMed] [Google Scholar]
- 31.Lee VHY, Healy T, Fonseca BD, Hayashi A, Proud CG. Analysis of the regulatory motifs in eukaryotic initiation factor 4E–binding protein 1. FEBS J. 2008;275:2185–2199. doi: 10.1111/j.1742-4658.2008.06372.x. [DOI] [PubMed] [Google Scholar]
- 32.Tsang CK, Liu H, Zheng XF. mTOR binds to the promoters of RNA polymerase I and III–transcribed genes. Cell Cycle. 2010;9:953–957. doi: 10.4161/cc.9.5.10876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shor B, et al. Requirement of the mTOR kinase for the regulation of Maf1 phosphorylation and control of RNA polymerase III dependent transcription in cancer cells. J Biol Chem. 2010;285:15380–15392. doi: 10.1074/jbc.M109.071639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sarbassov DD, Ali SM, Sabatini DM. Growing roles for the mTOR pathway. Curr Opin Cell Biol. 2005;17:596–603. doi: 10.1016/j.ceb.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 35.Woiwode A, et al. PTEN represses RNA polymerase III dependent transcription by targeting the TFIIIB complex. Mol Cell Biol. 2008;28:4204–4214. doi: 10.1128/MCB.01912-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Upadhya R, Lee J, Willis IM. Maf1 is an essential mediator of diverse signals that repress RNA polymerase III transcription. Mol Cell. 2002;10:1489–1494. doi: 10.1016/s1097-2765(02)00787-6. [DOI] [PubMed] [Google Scholar]
- 37.Manning BD. Balancing Akt with S6K: Implications for both metabolic diseases and tumorigenesis. J Cell Biol. 2004;167:399–403. doi: 10.1083/jcb.200408161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 39.Guertin DA, Sabatini DM. Defining the role of mTOR in cancer. Cancer Cell. 2007;12:9–22. doi: 10.1016/j.ccr.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 40.Wei Y, Zheng XFS. Sch9 partially mediates TORC1 signaling to control ribosomal RNA synthesis. Cell Cycle. 2009;8:4085–4090. doi: 10.4161/cc.8.24.10170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang Y, Maraia RJ. Comparison of the RNA polymerase III transcription machinery in S. pombe, S. cerevisiae and humans. Nucleic Acids Res. 2001;13:2675–2690. doi: 10.1093/nar/29.13.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goodfellow SJ, et al. Regulation of RNA polymerase III transcription by Maf1 in mammalian cells. J Mol Biol. 2008;378:481–491. doi: 10.1016/j.jmb.2008.02.060. [DOI] [PubMed] [Google Scholar]
- 43.Zetterberg A, Killander D. Quantitative cytophotometric and autoradiographic studies on the rate of protein synthesis during interphase in mouse fibroblasts in vitro. Exp Cell Res. 1965;40:1–11. doi: 10.1016/0014-4827(65)90284-3. [DOI] [PubMed] [Google Scholar]
- 44.Marshall L, Kenneth NS, White RJ. Elevated tRNAiMet synthesis can drive cell proliferation and oncogenic transformation. Cell. 2008;133:78–89. doi: 10.1016/j.cell.2008.02.035. [DOI] [PubMed] [Google Scholar]
- 45.Cunningham JT, et al. mTOR controls mitochondrial oxidative function through a YY1-PGC-1α transcriptional complex. Nature. 2007;450:736–740. doi: 10.1038/nature06322. [DOI] [PubMed] [Google Scholar]
- 46.Kantidakis T, White RJ. Dr1 (NC2) is present at tRNA genes and represses their transcription in human cells. Nucleic Acids Res. 2010;38:1228–1239. doi: 10.1093/nar/gkp1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.