Abstract
Microorganisms and zooplankton are both important components of aquatic food webs. Although both inhabit the same environment, they are often regarded as separate functional units that are indirectly connected through nutrient cycling and trophic cascade. However, research on pathogenic and nonpathogenic bacteria has shown that direct association with zooplankton has significant influences on the bacteria's physiology and ecology. We used stratified migration columns to study vertical dispersal of hitchhiking bacteria through migrating zooplankton across a density gradient that was otherwise impenetrable for bacteria in both upward and downward directions (conveyor-belt hypothesis). The strength of our experiments is to permit quantitative estimation of transport and release of associated bacteria: vertical migration of Daphnia magna yielded an average dispersal rate of 1.3 × 105·cells·Daphnia−1·migration cycle−1 for the lake bacterium Brevundimonas sp. Bidirectional vertical dispersal by migrating D. magna was also shown for two other bacterial species, albeit at lower rates. The prediction that diurnally migrating zooplankton acquire different attached bacterial communities from hypolimnion and epilimnion between day and night was subsequently confirmed in our field study. In mesotrophic Lake Nehmitz, D. hyalina showed pronounced diel vertical migration along with significant diurnal changes in attached bacterial community composition. These results confirm that hitchhiking on migrating animals can be an important mechanism for rapidly relocating microorganisms, including pathogens, allowing them to access otherwise inaccessible resources.
Keywords: bacterial dispersal, conveyer-belt hypothesis, migration
Microorganisms are the major component of the earth's biodiversity and indispensable in organic-matter cycling in aquatic systems (1). Understanding their distribution is crucial to elucidate evolutionary forces governing microbial community dynamics. The traditional view in microbiology that “everything is everywhere, but the environment selects” (2) assumes high dispersal rates of microorganisms, leading to their ubiquity. Supporting this view, the same species or lineages of pro- as well as eukaryotic microorganisms are found globally (3–5), although these findings are still debated (6) because of the difficulty in discriminating cryptic taxa using 18S and 16S rRNA marker genes. Studies in European lakes showed that bacterial dispersal is not limited, even at large spatial scales, and that fast bacterial growth rates facilitate species sorting along environmental gradients (7). Dispersal rates of bacteria in shallow and well-mixed lakes can be substantially increased by atmospheric transport (8, 9), and airborne dispersal is likely responsible for the global distribution of closely related thermophiles in hot springs (10). Aerial and terrestrial animals can also help disperse bacteria among different surface waters. For example, all lakes studied by Van der Gucht et al. (7) are located along the major European route of migrating birds, which fosters microbial exchange between these lakes, releasing the bacterial communities from local isolation.
Rapid species sorting along environmental gradients has also been shown using multiple molecular markers. For instance, Wu and Hahn (11) found clear differences in community composition of limnetic Polynucleobacter necessarius between different habitats but no large-scale geographic patterns within similar habitat types. The same is true for endosymbiotic P. necessarius strains that are host-specific and hence, follow the distribution of their ciliate host (12). In contrast, experiments using different inocula from several Swedish lakes found a major influence of the source community on establishment of bacterial populations under identical environmental conditions (13, 14). The latter suggests that low dispersal rates may prevent bacterial community homogenization, even within short geographical distances. This might also be true within large water bodies where stratification limits vertical mixing of bacteria. Within a stratified water column, exchanges between epipelagic bacteria and deep-water bacteria can be facilitated by sinking particles (15, 16), but this mechanism works primarily in the downward direction.
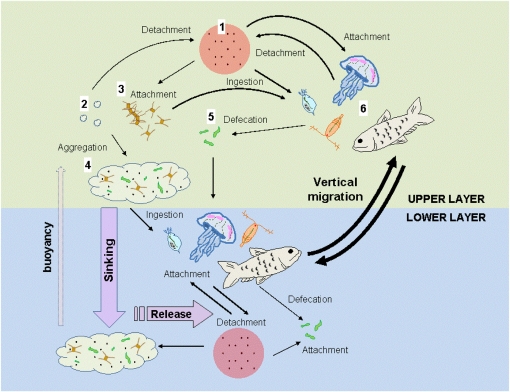
Because many zooplankton species harbor high numbers of bacteria on and inside their bodies (17–19) and perform diel vertical migration, they are expected to move attached bacteria through the water column on a daily basis. However, unless the zooplankton repeatedly acquire and release viable bacteria between the different water masses, zooplankton migration will have no effect on vertical dispersal and relocation of the bacterial populations. This is the essence of our conveyor-belt hypothesis (Fig. 1), which states that hitchhiking bacteria actively associate and dissociate from a migrating organism, leading to vertical dispersal of the bacterial population through otherwise impassable density discontinuities in both upward and downward directions. This dispersal mechanism can affect not only microbial dynamics in general but also the transmission of pathogens in the environment.
Fig. 1.
Transport and dispersal of bacteria in the water column. Free bacteria (1) may attach to suspended matters such as microgels (2) and particles (3) that form large sinking aggregates (4) and transport the bacteria to deeper water. Free bacteria may also be ingested by grazers and subsequently, released as part of the fecal matter (5), which also helps transport the bacteria to deeper water. These mechanisms work primarily in the downward direction. Alternatively, free bacteria may attach to large and motile organisms (6), which, through horizontal and vertical migration, transport the bacteria across aquatic boundary layers, such as thermo-, chemo-, and pycnoclines, that are otherwise impassable for the bacteria. Unlike passively sinking aggregates, large motile organisms can cover long distances in a short time and transport bacteria in both downward and upward directions. As such, migrating organisms function as a conveyor belt within the water column to facilitate the dispersal and exchanges of bacteria between isolated water masses.
Epipelagic and deeper waters are often characterized by large differences in environmental conditions. Vertical migration of higher organisms could allow hitchhiking bacteria to exploit favorable conditions in each water body, such as high concentrations of oxygen and algal-derived organic matter in the upper waters and high concentrations of inorganic nutrients in deeper waters. Hitchhiking on migrating organisms would also increase exchanges and interactions between different bacterial, viral, and grazer communities spatially separated by stratification (20).
To provide a quantitative estimate of bacteria dispersal through this conveyor-belt mechanism, we used migration columns with stable stratification separating two water bodies (slightly different in salinity) and measured the transport of bacteria by vertically migrating zooplankton across the density discontinuity in both upward and downward directions. Three bacterial species were isolated from oligotrophic Lake Stechlin (Brevundimonas sp., Pseudonocardia sp., and Pimelobacter sp.) and labeled with a GFP; GFP-labeled bacteria were added to either the upper water layer (downward transport experiments) or the lower water layer (upward transport experiments) of the migration columns, which contained different numbers of Daphnia magna. To stimulate zooplankton migration across the pycnocline, the upward and downward movements of the phototactic daphnids were guided by a focused light beam along the side of the migration columns. The abundance of GFP-labeled bacteria in the receiving end of the water column was counted through multiple migration cycles. Release of bacteria from precolonized daphnids into the surrounding water was further confirmed in a separate experiment. To complement our experimental observations, field samples were collected to determine diel differences in bacterial community composition (BCC) on migrating zooplankton.
Results and Discussion
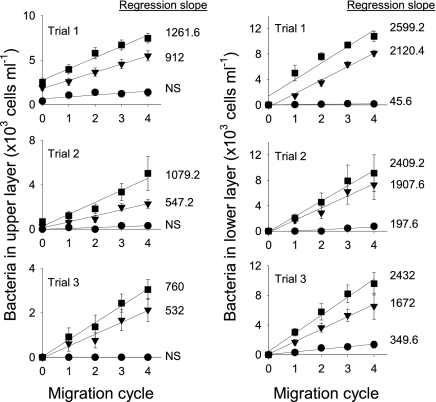
In all upward transport trials with Brevundimonas sp. (Fig. 2), no significant transport was detected in the control column (no D. magna) (Table S1), but when D. magna migrated, Brevundimonas sp. was effectively transported upward. Likewise, in the downward transport trials (Fig. 2), Brevundimonas sp. abundance in the lower layer increased much faster when D. magna was present, although there was a significant but small increase of Brevundimonas sp. in the lower layer of the control columns over time (Table S1). Comparison of all regression slopes (nonsignificant slopes were treated as zero) using two-way ANOVA yielded an overall significant model (P < 0.001). Post hoc pair-wise comparisons using Tukey's test showed a significant effect of the number of daphnids (80 daphnids > 20 daphnids > control; P < 0.005) and direction of transport (downward > upward; P < 0.001). The also significant interaction between number of daphnids and direction (P < 0.001) indicates a differential influence of the dispersal direction with a high or low number of daphnids. For identical numbers of D. magna, Brevundimonas sp. was dispersed 1.4–1.8 times faster in the downward direction than in the upward direction.
Fig. 2.
Upward (Left) and downward (Right) transport of Brevundimonas sp. in migration columns with different numbers of D. magna (● = 0; ▼ = 20; ■ = 80). D. magna completed one migration cycle (downward + upward) across the pycnocline every 2 h. Error bars represent SEs of 10 bacterial counts. Numbers next to lines are slopes of linear regressions (P < 0.05). NS, not significant. Full statistics are provided in Table S1.
Vertical transport of bacteria by migrating D. magna was also shown for two other bacterial species but at lower rates: both Pseudonocardia sp. and Pimelobacter sp. were effectively dispersed across the pycnocline in both upward and downward directions relative to controls (Table S2). Again, downward dispersal was 1.9 and 1.4 times faster than upward dispersal for Pseudonocardia sp. and Pimelobacter sp., respectively (comparison of two regression slopes by t test; P < 0.001 for Pseudonocardia and P < 0.05 for Pimelobacter). Differences in dispersal rates among the three bacterial species may reflect their different abilities to attach and detach, their likelihood to be ingested, their chance of surviving digestion, and their growth rates while attached to the host. We did observe that Pimelobacter sp. attached to the body surface of D. magna at a much lower density (mean ± SD; 1,992 ± 425 cells·mm−2) than Brevundimonas sp. (5,408 ± 582) and Pseudonocardia sp. (7,108 ± 1,181).
Data from the migration-column experiments showed that, although small and limited in their own motility, bacteria are able to travel and cross aquatic boundaries by hitchhiking on migrating organisms, which thus facilitates exchanges between separate microbial communities. Unlike slowly sinking aggregates and other detritus that transport bacteria primarily in the downward direction (15), motile and migrating hosts can cover long distances rapidly and disperse bacteria in all directions repeatedly and effectively. This dispersal mechanism would be particularly important for exchanging bacteria between layers in water bodies with permanent or seasonal stratification.
In our experiments, there are at least two means by which D. magna disperse bacteria bidirectionally: (i) bacteria actively attach to the exterior surfaces of the animal in one water body and then detach in another water body, and (ii) bacteria ingested by the animal in one water body survive digestion and are released through egestion into another water body. The latter mechanism requires that the bacteria survive digestion (21). Because our GFP-labeled bacteria cease to fluoresce shortly after death, our results represent the dispersal of mainly live bacteria. GFP-labeled bacteria were observed both on the carapace and inside the gut of D. magna (Fig. S1), which confirms that both dispersal mechanisms were acting simultaneously. Ingested bacteria may remain aggregated on defecation, evidenced by the presence of some microaggregates of bacteria in the columns with Daphnia; this gives them a higher sinking rate than individual cells, resulting in higher downward transport rate than upward transport rate.
Although specific rate measurements of the two dispersal mechanisms are not available, we estimate and compare the rates based on generalized models from the literature: the clearance rate of ambient bacteria caused by attachment can be estimated according to Kiørboe (22) as 4π × D × a, where D is the diffusion coefficient for motile bacteria (10−6–10−5 cm2·s−1) and a is the equivalent spherical radius for D. magna, which is approximated to be 0.15 cm. This yields a clearance rate of 0.2–1.6 mL·Daphnia−1·d−1 because of direct attachment. In comparison, clearance rate of ambient bacteria caused by filtration can be estimated from the daphnid's body length (L = 3 mm) as 12.9 × L1.545 (23), which is 70.5 mL·Daphnia−1·d−1. This calculation suggests that ingestion/defecation is potentially more efficient than attachment/detachment in dispersing bacteria.
To test how fast externally and internally attached bacteria were released into the surrounding water, we transferred D. magna precolonized with Brevundimonas sp. into bacteria-free water and measured the increase of bacteria in the surrounding water over time. The observed release rate can be described by the equation Y = 12,139 (1 − e−0.033X), where Y is number of cells released per animal and X is time in minutes (r2 = 0.93; P < 0.0001). This equation is consistent with the exponential decay function for bacterial detachment from marine snow particles (24) and suggests that a constant fraction of the attached bacterial population is released from the daphnid per unit of time. The exponential factor of 0.033 gives an average residence time of ca. 30 min for a bacterium on the daphnid, which is on par with the zooplankton's gut passage time at our experimental temperature (25) and the average searching time for highly motile bacteria to encounter a particle in the water column (24). Using this equation, we estimated a release rate of 1.2 × 104·cells·Daphnia−1·migration cycle−1 (120 min).
We independently calculated the release rate using the migration-column experimental data as slope of regression line (cells·mL−1·migration cycle−1) × volume (3,000 mL) ÷ number of D. magna, which yields an average release rate of 1.3 × 105·cells·Daphnia-1·migration cycle−1 for Brevundimonas sp. Because ≥10 times higher concentrations of D. magna were used in the release experiment than in the migration columns, the chance for the bacteria to reattach or be reingested was much higher. The larger surface-to-volume ratio of the container used in the release experiment relative to the migration columns would also lead to higher bacterial attachment rates to the container surfaces, resulting in the lower observed release rates.
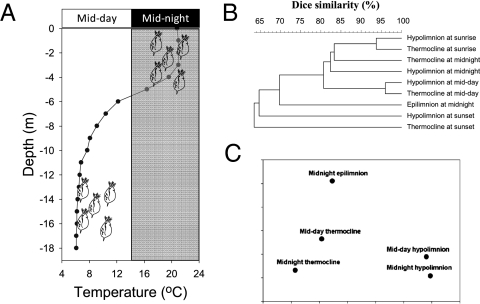
Both laboratory approaches independently showed that D. magna released a constant fraction of the attached bacteria per unit time. Together, these observations indicate that D. magna acquired (through direct attachment or ingestion) bacteria in one layer and released them into another layer at similar rates. The consequence would be an active exchange between the attached and the ambient bacterial communities, resulting in diurnal changes in bacterial community composition associated with a vertically migrating zooplankter. This prediction was subsequently confirmed in our field study in mesotrophic Lake Nehmitz. The water column of Lake Nehmitz was thermally stratified at the time of our study such that the water temperature was 21 °C in the epilimnion and <7 °C in the hypolimnion (Fig. 3A). D. hyalina showed a strong diel vertical migration: it was absent from the epilimnion at midday but appeared there at high abundance at midnight. Denaturing gradient gel electrophoreis (DGGE) analysis showed that the daphnids hosted very different bacterial communities between day and night (Analysis of Similarities [ANOSIM], R = 0.4; P = 0.001) and when they resided at different depths (ANOSIM, R = 0.174; P = 0.001) (Fig. 3 B and C).
Fig. 3.
(A) Water-temperature profile for Lake Nehmitz and relative vertical distribution of D. hyalina at midday and midnight. (B) Cluster analysis of DGGE banding patterns of bacterial communities on D. hyalina collected at different depths and times of the day. (C) ANOSIM analysis testing for differences in bacterial community composition on D. hyalina collected at different depths and time (details in Text).
Recently, Van der Gucht et al. (7) concluded that “…bacterial taxa need not to be everywhere at all times to yield the observed patterns: it is sufficient that low but sustained or regular dispersal is coupled with very efficient tracking of environmental conditions through local population dynamics.” Adding to their conclusion, our study provides a quantitative estimate of bacterial dispersal through migrating zooplankton and shows that bacteria may exploit resources in different water layers by hitchhiking on large migrating organisms. Daphnid's size, physiology, and abundance as well as environmental conditions and distances in lakes vary spatially and temporally, which may affect the rate functions of ambient and attached bacteria. Therefore, generalization of our experimental results should be done with care. In natural environments with diverse species of zooplankton and other motile organisms, dispersal of hitchhiking bacteria will not be uniform and can occur in different directions (vertical vs. horizontal), between different water bodies (littoral vs. pelagic zone; epipelagic vs. deeper waters), and on different temporal (diel vs. seasonal migration) and spatial scales (millimeter to kilometer scale).
The importance of zooplankton as carrier has been shown for pathogenic bacteria, most notably Vibrio spp. (26). However, the significance of zooplankton association goes beyond pathogenic bacteria, because zooplankton harbors very diverse bacterial phylotypes (19) in great abundances. For example, a single zooplankter can harbor up to 109 bacteria (27, 28), which is equivalent to or higher than ambient bacterial concentrations. With 10 zooplankters·L−1, not uncommon for lakes during the growth season (29), up to 1010 bacteria/L could be associated with the animals. The volume of Lake Stechlin, Germany, is 9.7 × 1010 L, with the epilimnion accounting for 40% of its volume in the summer. Assuming a population density of 10 zooplankter·L−1 (29) and a dispersal rate of 1.3 × 105·cells·zooplankter−1·migration cycle−1, dispersal by migrating zooplankton would replace about 1–2%·d−1 of the ambient bacteria in both epi- and hypolimnion. Although, based on our calculations, the redistribution of cells by migrating zooplankton seems to be small, it serves as a mechanism to inoculate a water body with bacteria from a remote source and allows bacteria to proliferate in an otherwise inaccessible environment.
In oceanic environments, many zooplankton species perform strong diel, seasonal, or ontogenetic vertical migration, sometimes up to thousands of meters (30, 31). They may, therefore, transport and disperse bacteria over a vast distance, affecting the ecology and physiology of even deep-sea microbes. There are other examples where bacteria use migrating organisms as vectors to efficiently exploit favorable growth conditions of even distant locations: the cave-dwelling amphipods of the genus Niphargus efficiently transport chemoautotrophic, sulfur-oxidizing bacteria of the genus Thiothrix all over the cave (32). The amphipods are even able to place the bacteria at the most favorable growth conditions along the chemical gradient on top of the anoxic bottom layer of the cave (32).
The life history of hitchhiking bacteria is inherently linked to that of their hosts, whereby bacteria growth and dispersal are directly influenced by the hosts’ physiology and behavior (18, 33). High dispersal rates of aquatic bacteria through hitchhiking would allow for rapid species sorting along environmental gradients (7). Long-range migration of hosts may also help spread pathogens across a broad geographical range, even among otherwise isolated water masses. Hence, dispersal rate plays a key role in determining community structure and function (e.g., productivity) over ecological as well as evolutionary time scales (34). Ecological and microbiological studies should take this important relationship into consideration to understand the full extent of microbial processes and evolutionary potential in aquatic and other ecosystems.
Materials and Methods
Migration Columns.
Experiments were conducted in transparent Plexiglas cylindrical columns 120 cm tall and 10 cm inner diameter. A sampling valve was affixed to the side 25 cm from the bottom of each column. To test that a stable stratification could be maintained in the migration columns, the columns were first filled to one-half with freshwater (0 psu; 15 °C). Thereafter, an equal amount of slightly saline water (2 psu; 5 °C) with food color (to aid visual observation) was slowly added through a siphon to the bottom to slowly displace the freshwater layer upward. After filling the columns, the siphon was slowly retrieved without breaking the stratification, as indicated by persistent color contrast at the pycnocline. The columns were allowed to equilibrate to the experimental temperature (21 °C), and both water layers remained well-separated for up to 1 wk, as indicated by the color contrast. This test confirmed that a stable density discontinuity could be maintained for an extended time, mimicking a stratified lake water column. Before each experiment, the columns were thoroughly cleaned with research-grade hypochlorite solution followed by several rinses with deionized water to remove all organic matter and bacteria.
Sampling of Water.
Experimental water was taken in early July 2009 from the epilimnion of Lake Stechlin, an oligotrophic lake in northeastern Germany. The water was sequentially filtered through a 22-μm nylon sieve, a 5-μm polycarbonate membrane filter, and a 0.2-μm Sterivex cartridge (Millipore) to remove ambient bacteria and particles. The water was then split into two portions; to one portion, NaCl was added to increase the salinity to 2 psu. The water was kept at 5 °C in the dark until use, usually within 24 h.
Zooplankton.
The freshwater cladoceran D. magna, originating from a monoclonal culture, was kept in M4 medium (35) and fed with a diet of yeast (36). We picked individuals of ca. 2.5–3 mm in size for the experiments, but occasionally, smaller individuals were included. Preliminary tests confirmed that D. magna tolerates a salinity of 2 psu without any noticeable adverse effects. The animals used in this study were positively phototactic such that we were able to control their vertical movement by placing a small and focused light source to the side of the migration column. We are aware that this behavior was opposite to the negatively phototactic migration pattern normally observed in nature (37), but for the purpose of this study, it was only necessary to manipulate the animals’ vertical movement, regardless of whether they responded positively or negatively to light. The animals were acclimated to filtered Lake Stechlin water (21 °C) for several hours before each experiment.
Bacteria Labeling with GFP.
Fresh zooplankton were collected from Lake Stechlin and washed with sterile water to remove loosely attached bacteria. The animals were then streaked out on agar plates (1.5%) with lactose broth (DEV) medium (10 g peptone·L−1, 10 g meat extract·L−1, 5 g NaCl·L−1). The resultant colonies were picked and streaked out on new DEV agar plates. Pure isolates were transferred and grown in liquid DEV medium. At exponential growth, bacteria were harvested by short centrifugation (8,000 × g for 6 min), heat shocked at 42 °C for 1 min, and inoculated with extracted GFP plasmids from Escherichia coli. Cells with a GFP plasmid were grown and selected in liquid DEV medium containing the antibiotics erythromycin, because a gene for erythromycin resistance was present on the GFP plasmid (38). After 1 d of growth, we checked the bacteria under an epifluorescence microscope for their fluorescent signal. With this procedure, we isolated three bacterial pure cultures from the zooplankton and successfully labeled them with the GFP. Sequencing of their 16S rRNA gene revealed a 99–100% similarity to Brevundimonas sp. (99% similarity to DQ825663), Pseudonocardia sp. (99% similarity to EU741109), and Pimelobacter sp. (100% similarity to AY509240).
Upward Transport of Brevundimonas sp.
All migration-column experiments were conducted in a dark room at 21 °C. Three columns were first filled with 3 L of the filtered lake water (15 °C), and 4 L of the saline water (5 °C) was slowly added through a siphon to the bottom. The columns were allowed to equilibrate to the room temperature, and concentrated aliquots of the bacterial culture, free of growth medium, were injected with syringes through the sampling valves to the lower layer for an initial concentration of 1.6 × 105 cells·mL−1. D. magna was added with a wide-mouth pipette to the upper layer of two of the columns (20 and 80 animals, respectively). The column without D. magna was used as control. A small light source was placed near the bottom of each column, including the control, to attract the animals to the lower layer. Initial samples (20 mL) were taken with pipettes from the top layer about 3 cm below the surface. After 1 h, the light source was moved to near the top of the columns to attract the animals to the upper layer. The light source was alternated between upper and lower positions every 1 h, and samples were taken from the upper layer every 2 h. The experiment lasted for 8 h (four migration cycles) and was repeated two times using new water, bacteria, and Daphnia. Water samples were filtered onto black 0.2-μm polycarbonate membrane filters (Whatman), and the GFP-labeled bacteria were counted using a Zeiss epifluorescence microscope with a blue filter set (475-nm excitation and 530-nm emission).
Downward Transport of Brevundimonas sp.
The setup was similar but with a few differences: each column was first filled with 4 L of the filtered lake water, and 3 L of the saline water was slowly added to the bottom. Aliquots of the bacterial culture were added to the upper layer (1.6 × 105 cells·mL−1; final concentration). D. magna was added to the upper layer of two of the columns (20 and 80 animals, respectively). Initial samples (20 mL) were taken from the bottom layer through the sampling valves (>10 cm below the pycnocline). The light source was alternated between upper and lower positions every 1 h, and samples were taken from the bottom layer every 2 h. The experiment was repeated two times using new water, bacteria, and D. magna.
Transport Experiments with Other Bacterial Species.
To verify that the observed vertical transport of Brevundimonas sp. by migrating daphnids was applicable to other bacterial species, we repeated the upward and downward transport experiments with Pseudonocardia sp. and Pimelobacter sp. Experimental designs were the same as described above with the exception that 40 D. magna were used in each treatment, and the experiments were conducted only one time.
Release Experiment with Brevundimonas sp.
To provide additional evidence that D. magna releases previously attached bacteria when moved to sterile filtered lake water, we took 40 animals that had been previously colonized by Brevundimonas sp. for 1 d and transferred them to 150 mL of sterile-filtered Lake Stechlin water in an Erlenmeyer flask. A 10-mL aliquot was drawn from the surrounding water at 0, 10, 30, 60, and 120 min for bacterial counts.
Field Sampling.
Field sampling was conducted in early July 2008 in Lake Nehmitz, a mesotrophic lake connected to Lake Stechlin through a narrow artificial channel. The sampling station was 19 m deep with a thermocline at ca. 6 m. The zooplankton community was dominated by D. hyalina at the time of the study. Zooplankton within the epilimnion (0–4 m) and hypolimnion (12–16 m) were sampled with an open-close net at midday, sunset, sunrise, and midnight. Temperature profiles were measured with a Wissenschaftlich Technische Werkstätten (WTW) probe to determine the exact position of the thermocline. Collected zooplankton were concentrated onto 200-μm meshes on board, rinsed with deionized water to remove loosely attached bacteria, and kept in Petri dishes on ice. D. hyalina was picked individually, briefly rinsed in sterile deionized water, and transferred to Eppendorf vials for DGGE (15 animals per vial). The vials were kept at −20 °C until DNA extraction. Extraction of DNA was done according to Zhou et al. (39), and parts of the 16S rDNA were amplified using the primers 341f-gc and 907r. Thereafter, DGGE was run for 20 h at 100 V with a 7% acrylamide gel and a denaturing gradient of 40–70%. After staining with SybrGold, gels were illuminated by UV and photographed with a digital camera.
Data Analysis.
For the upward and downward transport experiments with Brevundimonas sp., linear-regression functions were fitted to the data, and the significance level for the y intercept and slope was set at P = 0.05. The slopes of the regressions, representing the dispersal rates, were compared with two-way ANOVA, and number of daphnids and dispersal directions was independent factors. Tukey's test was used for posthoc comparisons. Because experiments with Pseudonocardia sp. and Pimelobacter sp. were done only one time with one abundance of D. magna, t test was used to compare the linear-regression slopes between upward and downward transport experiments. For bacteria-release experiment, an exponential function with maximum was fitted to the data at a significance level of P = 0.05.
For the field experiment, DGGE banding patterns of epilimnion and hypolimnion samples and different time points were compared with cluster analysis using Dice coefficient and the program GelComparII. Afterward, the similarity matrix was loaded into the software Primer6 (Primer-E Ltd.) with Analysis of Similarities (ANOSIM) for differences between sampling depth and time.
Supplementary Material
Acknowledgments
We thank Søren Molin from the Danmarks Tekniske Universitet (Copenhagen, Denmark) for the E. coli culture with a GFP plasmid and Andrea Paul from the Bundesanstalt für Materialforschung (Berlin, Germany) for the monoclonal culture of D. magna. We also thank Thomas Kiørboe, Roman Stocker, and Kirsten Pohlmann for helpful comments on the manuscript. Funding for H.P.G, C.D., and F.L. was provided by German Science Foundation Grants DFG-GR 1540/11-1 and PA 1655/1-1 and the Leibniz Foundation. K.W.T. was supported by US National Science Foundation Grant OCE-0814558 and a visiting scientist fellowship from Leibniz-Institute of Freshwater Ecology and Inland Fisheries, Berlin.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1000668107/-/DCSupplemental.
References
- 1.Azam F. Microbial control of oceanic carbon flux: The plot thickens. Science. 1998;280:694–696. [Google Scholar]
- 2.Baas-Becking LGM. In: Geobiological Introduction to Environmental Sciences. Van Stockum WP, Zoon NV, editors. The Hague: Diligentia Wetensch Series 18/19, Van Stockum's Gravenhange; 1934. pp. 249–254. [Google Scholar]
- 3.Finlay BJ, Clarke KJ. Ubiquitous dispersal of microbial species. Nature. 1999;400:828. [Google Scholar]
- 4.Finlay BJ. Global dispersal of free-living microbial eukaryote species. Science. 2002;296:1061–1063. doi: 10.1126/science.1070710. [DOI] [PubMed] [Google Scholar]
- 5.Fenchel T, Finlay BJ. Bacteria and island biogeography. Science. 2005;309:1997–1999. doi: 10.1126/science.309.5743.1997. [DOI] [PubMed] [Google Scholar]
- 6.Foissner W. Biogeography and dispersal of micro-organisms: A review emphasizing protists. Acta Protozool. 2006;45:111–136. [Google Scholar]
- 7.Van der Gucht K, et al. The power of species sorting: Local factors drive bacterial community composition over a wide range of spatial scales. Proc Natl Acad Sci USA. 2007;104:20404–20409. doi: 10.1073/pnas.0707200104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hervàs A, Camarero L, Reche I, Casamayor EO. Viability and potential for immigration of airborne bacteria from Africa that reach high mountain lakes in Europe. Environ Microbiol. 2009;11:1612–1623. doi: 10.1111/j.1462-2920.2009.01926.x. [DOI] [PubMed] [Google Scholar]
- 9.Hervàs A, Casamayor EO. High similarity between bacterioneuston and airborne bacterial community compositions in a high mountain lake area. FEMS Microbiol Ecol. 2009;67:219–228. doi: 10.1111/j.1574-6941.2008.00617.x. [DOI] [PubMed] [Google Scholar]
- 10.Bonheyo GT, Frias-Lopez J, Fouke BW. In: Geothermal Biology and Geochemistry in Yellowstone National Park. Inskeep WP, McDermott TR, editors. Bozeman, MT: Montana State University Publications; 2005. pp. 327–340. [Google Scholar]
- 11.Wu QL, Hahn MW. Differences in structure and dynamics of Polynucleobacter communities in a temperate and a subtropical lake, revealed at three phylogenetic levels. FEMS Microbiol Ecol. 2006;57:67–79. doi: 10.1111/j.1574-6941.2006.00105.x. [DOI] [PubMed] [Google Scholar]
- 12.Vannini C, et al. Endosymbiosis in statu nascendi: Close phylogenetic relationship between obligately endosymbiotic and obligately free-living Polynucleobacter strains (Betaproteobacteria) Environ Microbiol. 2007;9:347–359. doi: 10.1111/j.1462-2920.2006.01144.x. [DOI] [PubMed] [Google Scholar]
- 13.Langenheder S, Lindström ES, Tranvik LJ. Weak coupling between community composition and functioning of aquatic bacteria. Limnol Oceanogr. 2005;50:957–967. [Google Scholar]
- 14.Langenheder S, Ragnarsson H. The role of environmental and spatial factors for the composition of aquatic bacterial communities. Ecology. 2007;88:2154–2161. doi: 10.1890/06-2098.1. [DOI] [PubMed] [Google Scholar]
- 15.Turley CM, Mackie PJ. Bacterial and cyanobacterial flux to the deep. NE Atlantic on sedimenting particles. Deep Sea Res A. 1995;42:1453–1474. [Google Scholar]
- 16.Simon M, Grossart HP, Schweitzer B, Ploug H. Microbial ecology of organic aggregates in aquatic ecosystems. Aquat Microb Ecol. 2002;28:175–211. [Google Scholar]
- 17.Carman KR, Dobbs FC. Epibiotic microorganisms on copepods and other marine crustaceans. Microsc Res Tech. 1997;37:116–135. doi: 10.1002/(SICI)1097-0029(19970415)37:2<116::AID-JEMT2>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 18.Tang KW. Copepods as microbial hotspots in the ocean: Effects of host feeding activities on attached bacteria. Aquat Microb Ecol. 2005;38:31–40. [Google Scholar]
- 19.Grossart HP, Dziallas C, Tang KW. Bacterial diversity associated with freshwater zooplankton. Env Microbiol Reports. 2009;1:50–55. doi: 10.1111/j.1758-2229.2008.00003.x. [DOI] [PubMed] [Google Scholar]
- 20.Allgaier M, Grossart HP. Diversity and seasonal dynamics of Actinobacteria populations in four lakes in northeastern Germany. Appl Environ Microbiol. 2006;72:3489–3497. doi: 10.1128/AEM.72.5.3489-3497.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.King CH, Sanders RW, Shotts EB, Jr, Porter KG. Differential survival of bacteria ingested by zooplankton in an eutrophic lake. Limnol Oceanogr. 1991;36:829–845. [Google Scholar]
- 22.Kiørboe T. A Mechanistic Approach to Plankton Ecology. Princeton: Princeton University Press; 2008. p. 228. [Google Scholar]
- 23.Porter KG, Feig YS, Vetter EF. Morphology flow regimes and filtering rate of Daphnia, Ceriodaphnia and Bosmina fed natural bacteria. Oecologia. 1983;58:156–163. doi: 10.1007/BF00399211. [DOI] [PubMed] [Google Scholar]
- 24.Kiørboe T, Grossart HP, Ploug H, Tang K. Mechanisms and rates of bacterial colonization of sinking aggregates. Appl Environ Microbiol. 2002;68:3996–4006. doi: 10.1128/AEM.68.8.3996-4006.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dam HG, Peterson WT. The effect of temperature on the gut clearance rate constant of planktonic copepods. J Exp Mar Biol Ecol. 1988;123:1–14. [Google Scholar]
- 26.Rawlings TK, Ruiz GM, Colwell RR. Association of Vibrio cholerae O1 El Tor and O139 Bengal with the Copepods Acartia tonsa and Eurytemora affinis. Appl Environ Microbiol. 2007;73:7926–7933. doi: 10.1128/AEM.01238-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lawrence SG, Ahmad A, Azam F. Fate of particle bound bacteria ingested by Calanus pacificus. Mar Ecol Prog Ser. 1993;97:299–307. [Google Scholar]
- 28.Delille D, Razouls S. Community structures of heterotrophic bacteria of copepod fecal pellets. J Plankton Res. 1994;16:603–615. [Google Scholar]
- 29.Balcer MD, Korda NL, Dodson SI. Zooplankton of the Great Lakes: A Guide to the Identification and Ecology of the Common Crustacean Species. Madison WI: University of Wisconsin Press; 1984. p. 188. [Google Scholar]
- 30.Kobari T, Ikeda T. Vertical migration and life cycle of Neocalanus plumchrus (Crustacea: Copepoda) in the Oyashio region, with notes on regional variations in body size. J Plankton Res. 2001;23:287–302. [Google Scholar]
- 31.Visser AW, Jónasdóttir SH. Lipids, buoyancy and the seasonal vertical migration of Calanus finmarchicus. Fish Oceanogr. 2003;8:100–106. [Google Scholar]
- 32.Dattagupta S, et al. A novel symbiosis between chemoautotrophic bacteria and a freshwater cave amphipod. ISME J. 2009;3:935–943. doi: 10.1038/ismej.2009.34. [DOI] [PubMed] [Google Scholar]
- 33.Tang KW, Bickel SL, Dziallas C, Grossart HP. Microbial activities accompanying decomposition of cladoceran and copepod carcasses under different environmental conditions. Aquat Microb Ecol. 2009;57:89–100. [Google Scholar]
- 34.Venail PA, et al. Diversity and productivity peak at intermediate dispersal rate in evolving metacommunities. Nature. 2008;452:210–214. doi: 10.1038/nature06554. [DOI] [PubMed] [Google Scholar]
- 35.Elendt BP, Bias WR. Trace nutrient deficiency in Daphnia magna cultured in standard medium for toxicity testing: Effects of the optimization of culture conditions on life history parameters of D. magna. Water Res. 1990;24:1157–1167. [Google Scholar]
- 36.Herring PJ. The carotenoid pigments of Daphnia magna Straus. II. Aspects of pigmentary metabolism. Comp Biochem Physiol. 1968;24:205–221. doi: 10.1016/0010-406x(68)90968-7. [DOI] [PubMed] [Google Scholar]
- 37.Forward RB. Diel vertical migration: Zooplankton photobiology and behavior. Oceanogr Mar Biol Annu Rev. 1988;26:361–392. [Google Scholar]
- 38.Lambertsen L, Sternberg C, Molin S. Mini-Tn7 transposons for site-specific tagging of bacteria with fluorescent proteins. Environ Microbiol. 2004;6:726–732. doi: 10.1111/j.1462-2920.2004.00605.x. [DOI] [PubMed] [Google Scholar]
- 39.Zhou J, Bruns MA, Tiedje JM. DNA recovery from soils of diverse composition. Appl Environ Microbiol. 1996;62:316–322. doi: 10.1128/aem.62.2.316-322.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.