Abstract
p53 is a central player in responses to cellular stresses and a major tumor suppressor. The identification of unique molecules within the p53 signaling network can reveal functions of this important transcription factor. Here, we show that brain-expressed RING finger protein (BERP) is a gene whose expression is up-regulated in a p53-dependent manner in human cells and in mice. We generated BERP-deficient mice by gene targeting and demonstrated that they exhibit increased resistance to pentylenetetrazol-induced seizures. Electrophysiological and biochemical studies of cultured cortical neurons of BERP-deficient mice showed a decrease in the amplitude of GABAA receptor (GABAAR)-mediated miniature inhibitory postsynaptic currents as well as reduced surface protein expression of GABAARs containing the γ2-subunit. However, BERP deficiency did not decrease GABAARγ2 mRNA levels, raising the possibility that BERP may act at a posttranscriptional level to regulate the intracellular trafficking of GABAARs. Our results indicate that BERP is a unique p53-regulated gene and suggest a role for p53 within the central nervous system.
Keywords: brat, neurotransmitter, brain, RNF22, TRIM3
Normal function of the transcription factor p53 maintains genomic integrity in the face of DNA damage and other cellular stresses. The importance of p53 stems from its roles in controlling cell cycle arrest, senescence, apoptosis, DNA repair, and angiogenesis. The ability of p53 to prevent the propagation of damaged or potentially transformed cells is key to its function as a tumor suppressor (1, 2). Although many molecules in the p53 signaling network have been identified, including its transcriptional targets p21 (cell cycle arrest), GADD45 (DNA repair), and Bax (apoptosis) as well as its major negative regulator MDM2, our understanding of this complex intracellular web is incomplete (1, 3–6). As more molecules within the p53 network are identified and characterized, the apparent reach of this transcription factor is extended (7). We hypothesized that the identification of unique molecules in the p53 network might provide clues to p53’s involvement in previously unknown functions of this vital regulator.
Our laboratory previously performed an in vivo genetic modifier screen in Drosophila melanogaster using the UAS-GAL4 system (8) to identify unique molecules interacting with p53. One of the molecules emerging from this screen was D. melanogaster “brain tumor protein” (brat). Brat is a D. melanogaster tumor suppressor that represses the translation of hunchback (9) and negatively regulates ribosomal RNA synthesis (10). Brat also inhibits the self-renewal of Drosophila neuroblasts (11). Structurally, brat belongs to a family of proteins that contain an N-terminal zinc-finger and coiled-coil domains and a C-terminal NHL (ncl-1, HT2A and Lin-41) domain (12). The mammalian homologs of brat are members of the tripartite motif (TRIM)-NHL group of proteins and include brain-expressed RING finger protein (BERP), neural activity-related RING finger protein (NARF), and HT2A (13).
To identify the mammalian homolog closest to brat, we employed the best reciprocal hit (BRH) strategy (14, 15) utilizing the National Center for Biotechnology Information's basic local alignment search tool for proteins (BLASTP). However, brat had no BLASTP BRH partner in mammals. The best unidirectional BLASTP hit for brat was BERP, which was first cloned and characterized in the rat by El-Husseini et al. (16, 17). In rat cells, BERP binds to α-actinin-4 and myosin V, two proteins of the actin cytoskeleton that have been implicated in cell motility, organelle trafficking, and cancer (18–22). In the rat neuronal cell line PC12, overexpression of a BERP truncation mutant prevented nerve growth factor-induced differentiation and neurite outgrowth (17). In mice, BERP is strongly expressed in brain and is up-regulated following chemically induced seizures (23). In humans, BERP (as TRIM3) has been identified as the candidate tumor suppressor gene at 11p15.5 associated with loss of heterozygosity in human gliomas (24). In addition, a recent shotgun proteomics analysis has indicated that BERP is up-regulated in the brains of schizophrenic patients (25). These findings suggest that BERP may play an important role in the central nervous system (CNS).
GABA is the major inhibitory neurotransmitter in the mammalian CNS. GABA exerts its fast inhibitory effect on synaptic transmission by binding to postsynaptic GABAA receptors (GABAARs) (26). Within the mammalian brain, the most common composition of GABAARs is a combination of the α1-, β2-, and γ2-GABAAR subunit proteins (26). Recent studies have indicated that the trafficking of such GABAARs to and from the neuronal surface is crucial for posttranslational signaling mechanisms that modulate the stability, density, and function of these channels (27). In particular, the GABAARγ2-subunit is required for GABAAR trafficking to the postsynaptic membrane (27).
In this study, we investigate the physiological function of BERP and its relationship to both GABAARs and p53. No link between GABAARs and p53 has been reported to date, but we present several lines of evidence indicating that BERP is a unique p53-regulated gene that modulates seizure susceptibility and GABAAR cell surface expression.
Results
Human BERP Is a Unique p53 Target Gene with Functional p53 Response Elements.
p53 regulates transcription by binding to p53 response elements (p53REs) within or near its target genes. The prototypical p53RE consists of two 10-bp half-sites separated by a spacer that can range from 0 to 13 nucleotides (28). Using sequence analysis, we determined that a 14.8-kb genomic fragment upstream of the BERP translational start site contained four potential p53REs (Fig. 1 A and B). Of these, sites E, F, and G were located within BERP intron 1, whereas site B was located upstream of exon 1.
Fig. 1.
p53 binding and transcriptional competence of predicted BERP p53REs. (A) Human BERP p53REs. Schematic localization of four putative p53REs (gray ovals labeled B, E, F, and G) in the human genomic BERP locus, as indicated. ATG, BERP translation start site; Chrom., chromosome. (B) Nucleotide sequences of consensus p53RE of the four putative human BERP (B, E, F, and G) p53REs, and the putative mouse BERP p53RE. Pu = A/G, Py = C/T, n = A/G/C/T; uppercase, half-site nucleotides; lowercase, spacer nucleotides. *Mismatches from p53RE consensus sequence. Residue positions are expressed relative to the “A” of ATG (+1). Numbers of residues matching the p53RE consensus sequence are indicated. (C) Binding of p53 to putative human BERP p53REs. ChIP assays were performed in HCT116 p53+/+ and HCT116 p53−/− cells to detect binding of p53 to BERP B and EFG sites (anti-p53 lanes). The p21 p53RE was used as a positive control (p21). Cells were treated with DMSO (“untreated” control) or 5FU to induce p53. IgG, nonspecific mouse Ig (negative control); Input, 1% of total sonicated chromatin was reserved before IP; H2O, water control. Results shown are representative of three trials. (D) BERP E is a functional p53RE. HCT116 p53−/− cells were cotransfected with a vector expressing either p53wt or p53mut plus a luciferase reporter construct containing putative BERP p53REs (pGL3-BerpB, pGL3-BerpE, pGL3-BerpF, or pGL3-BerpG). pGL3-p21, positive control; pGL3-TATA, empty vector. Luciferase activity was normalized to β-gal activity and expressed relative to pGL3-TATA levels. Results shown are mean fold change in luciferase activity ± SEM of four independent experiments performed in triplicate. (E) Mutation of BERP E abrogates transactivation. HCT116 p53−/− cells were cotransfected with a vector expressing p53wt or p53mut as in D plus a luciferase construct containing either WT BERP E (pGL3-BerpE) or BERP E with a C→A point mutation (pGL3BerpE* = GGGAAGGCCCtGGGCTTGTTC). Results shown are the mean fold change in luciferase activity ± SEM of three independent experiments performed in quadruplicate. (F) Berp M is a functional p53RE, and mutation of Berp M abrogates transactivation. The functionality of Berp M was analyzed as in D. pGL3-BerpM, murine site Berp M; pGL3BerpM*, Berp M with a C→A point mutation (AGGAAGGCCCtGTGCATGTTC); pGL3-p21, positive control; pGL3-TATA, empty vector. Results shown are mean fold change in luciferase activity ± SEM of three independent experiments performed in triplicate.
To determine whether p53 could, in fact, bind to the putative p53REs in the human BERP promoter region, we treated isogenic HCT116 p53+/+ and HCT116 p53−/− cells with 5-fluorouracil (5FU) to activate p53. The cells were harvested 24 h later, and ChIP assays were performed using anti-p53 antibody and PCR primers specific for either BERP site B or site EFG (which were too close to separate). We found that p53 could indeed bind to BERP sites B and EFG in human cells (Fig. 1C). To determine whether this binding of p53 to the putative BERP p53REs could regulate transcription, oligonucleotides containing the sequences for BERP sites B, E, F, and G were cloned into the pGL3-TATA construct upstream of a luciferase reporter gene. These constructs were transfected into HCT116 p53−/− cells along with a vector expressing either WT p53 (p53wt) or a p53 protein with a mutation in its DNA-binding domain (p53mut). The p53RE sequence of the known p53 target gene p21 was used as a positive control (pGL3-p21). Of the four BERP sites tested, only cotransfection with BERP E resulted in an increase in luciferase activity in the presence of p53wt; no increase was seen in the presence of p53mut (Fig. 1D). Furthermore, a C→A point mutation at position 4 of the BERP E site abrogated this increase in luciferase activity (Fig. 1E). A similar response was observed for the murine Berp site M (Fig. 1F). These findings suggest that BERP is a bona fide p53 target gene, that the putative p53RE at BERP site E can bind p53, and that this binding regulates transcription in a p53-dependent manner.
To examine the dependency of BERP expression on p53 at the mRNA level, we treated HCT116 p53+/+ and HCT116 p53−/− cells with increasing doses of 5FU for 48 h and measured levels of BERP mRNA using real-time RT-PCR. Compared with untreated controls, BERP mRNA was up-regulated 4- to 5-fold by treatment with 600 μM 5FU, and this increase depended on the presence of functional p53 (Fig. 2A). A similar p53-dependent up-regulation of BERP occurred after 24 h in HCT116 p53+/+ cells (Fig. 2B) and in the glioblastoma cell lines A172 and U118 (Fig. S1). In contrast, the mRNAs for the BERP homologs NARF and HT2A were not up-regulated on 5FU treatment regardless of p53 status (Fig. 2B). Thus, in response to a DNA-damaging stimulus, BERP gene expression is specifically up-regulated in a p53-dependent manner.
Fig. 2.
BERP is specifically up-regulated by p53 in human cells. (A) Induction. HCT116 p53+/+ and HCT116 p53−/− cells were treated for 48 h with the indicated doses of 5FU to activate p53, and relative expression levels of BERP and p21 (control) mRNAs were analyzed by real-time RT-PCR. Values were normalized to hypoxanthine phosphoribosyltransferase 1 (HPRT1) and expressed relative to untreated controls. Results shown are the mean fold change in mRNA ± SEM of three independent experiments performed in triplicate. (B) Specificity. HCT116 p53+/+ and HCT116 p53−/− cells were treated for 24 h with the indicated doses of 5FU, and relative expression levels of BERP, NARF, HT2A, and p21 (control) mRNAs were analyzed by real-time RT-PCR as for A. Results shown are the mean fold change ± SEM of three independent experiments performed in triplicate.
Murine BERP Expression Is p53-Dependent in Vivo.
Our study of human cell lines suggested that BERP E was a functional p53RE, prompting us to undertake a sequence analysis of the murine BERP genomic locus. Multiple putative p53REs were identified, although no site identical to human BERP E was found. However, a murine Berp site M (Fig. 1B) was similar in sequence to human BERP E and located at a comparable relative position within murine BERP intron 1. Moreover, Berp site M was capable of regulating transcription in the presence of p53wt but not p53mut, and a C→A point mutation at position 4 of the Berp site M site abrogated this response (Fig. 1F). To determine whether BERP expression was p53-dependent in mice, we injected p53+/+ and p53−/− male mice (8–12 wk old) with pentylenetetrazol (PTZ), a compound previously shown to up-regulate murine BERP expression (23). The brains of these animals were harvested after injection, and histological sections were subjected to in situ hybridization using a 33P-labeled BERP riboprobe (Fig. 3). At 6 h post-PTZ administration, only base levels of endogenous BERP were detected in the cerebellum and hippocampus of p53−/− mice (Fig. 3 A and C). In contrast, strong up-regulation of BERP mRNA was seen in the brains of PTZ-treated p53+/+ mice (Fig. 3 B and D). The brains of sham-injected p53−/− and p53+/+ mice showed only base levels of BERP mRNA (Fig. 3 E and F). These results confirm that BERP gene expression is up-regulated in a p53-dependent manner in response to PTZ treatment in vivo.
Fig. 3.
PTZ up-regulates BERP expression in a p53-dependent fashion. p53+/+ and p53−/− mice were sham-injected or injected with 45 mg/kg of PTZ, and histological sections of the cerebellum and hippocampus were subjected to in situ hybridization to detect BERP mRNA. BERP mRNA expression was up-regulated in p53+/+ mouse brains (B and D), whereas levels in p53−/− mouse brains (A and C) were comparable to the endogenous levels present in sham-injected p53−/− and p53+/+ mice (E and F). Results shown are representative of five mice per genotype. (Scale bar: 200 μm.)
BERP-Deficient Mice Show Increased Resistance to PTZ-Induced Seizures.
To investigate the physiological function of BERP, we generated BERP-deficient mice using conventional gene targeting and homologous recombination in murine ES cells (Fig. S2A). Successful germline transmission of the targeted BERP allele was confirmed by PCR and Southern blotting (Fig. S2B). The absence of BERP protein in BERP−/− mice was confirmed by Western blotting of whole-brain lysates using an anti-BERP antibody (Fig. S2C). F1 heterozygotes were backcrossed six generations to C57BL/6 mice, and intercrosses of these animals yielded progeny at the expected gender and Mendelian ratios. BERP−/− mice were viable, healthy, and fertile, with no gross or histological abnormalities.
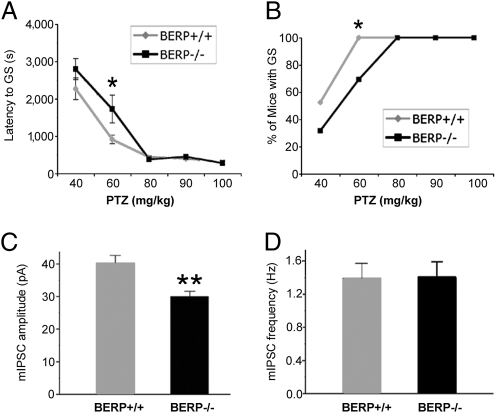
We first examined the susceptibility of BERP-deficient mice to PTZ-induced seizures. PTZ s.c. injection elicits a specific sequence of behavioral seizure responses that can be measured semiquantitatively by documenting their presence and progression during a defined observation period. A more quantitative measure of PTZ susceptibility is obtained by measuring the latency of specific seizure response end points. At a dose of 60 mg/kg of PTZ, there was a significant decrease in the proportion of BERP−/− mice (9 of 13) that exhibited generalized seizures compared with BERP+/+ controls (P < 0.05, Fisher's exact test and χ2 test) (Fig.4A). The difference in mean latency to generalized seizure was also statistically significant at this dose (P < 0.05, Student's t test) (Fig. 4B). Although differences in susceptibility to partial and maximal seizures did not reach statistical significance (Fig. S3), the general trend of these data supported our conclusion that BERP−/− mice were more resistant to PTZ-induced seizures than BERP+/+ controls. These findings suggest that BERP expression confers sensitivity to PTZ.
Fig. 4.
Altered PTZ seizure susceptibility and electrophysiology in BERP-deficient mice. (A and B) Reduced seizure susceptibility. PTZ was administered s.c. to mice at doses of 40 mg/kg (n = 21 for BERP+/+, n = 19 for BERP−/−), 60 mg/kg (n = 15 for BERP+/+, n = 13 for BERP−/−), 80 mg/kg (n = 11 for BERP+/+, n = 9 for BERP−/−), 90 mg/kg (n = 10 for BERP+/+, n = 10 for BERP−/−), and 100 mg/kg (n = 7 for BERP +/+, n = 7 for BERP−/−). (A) Latency. Each data point represents the mean latency ± SEM to a PTZ-induced generalized seizure (GS) in BERP+/+ and BERP−/− mice. *P < 0.05, Student's t test. (B) Dose–response curves. Each data point represents the percentage of BERP+/+ and BERP−/− mice exhibiting GSs at the indicated dose of PTZ. *P < 0.05, Fisher's exact test and χ2 test. (C and D) Reduced neuronal mean mIPSC amplitude. Cortical neurons from untreated BERP+/+ (n = 11) and BERP−/− (n = 13) mice were analyzed by the whole-cell patch-clamp method to determine mean mIPSC frequency (C) and amplitude (D). **P < 0.01, Student's t test.
PTZ is an antagonist of GABAAR signaling. Because our BERP−/− mice showed a significantly reduced rate of PTZ-induced seizures, we reasoned that BERP might affect GABAAR function in synaptic transmission. We therefore used whole-cell patch-clamp recordings to compare mean miniature inhibitory postsynaptic current (mIPSC) frequency and amplitude in cortical neurons from BERP−/− and BERP+/+ mice. We found that the mean mIPSC amplitude was reduced in the mutant cells (Fig.4C) but that the mean mIPSC frequency was equal in BERP−/− and BERP+/+ cells (Fig.4 D). Because changes in the frequency and amplitude of mean mIPSCs correlate with presynaptic (neurotransmitter release) and postsynaptic (neuroreceptor function) factors, respectively (29), these results suggested that BERP deficiency suppresses synaptic GABAAR function and reveal that endogenous BERP plays a role in GABAAR regulation.
BERP−/− Mice Show Decreased Surface Expression of the GABAARγ2-Subunit.
Because changes to GABAAR function could be attributable to altered expression of these channels on the cell surface (27), we examined whether BERP influenced surface expression of γ2-containing GABAARs in murine brain cells. We established cortical neuron cultures from BERP+/+, BERP+/−, and BERP−/− embryonic day 17.5 (E17.5) mouse embryos and carried out surface protein biotinylation assays followed by immunoblotting to detect GABAARγ2 protein, the major component of GABAARs in the brain (26). We found that GABAARγ2 expression was decreased on neurons of BERP−/− mice compared with neurons of BERP+/+ and BERP+/− mice (Fig. 5). In contrast, expression of the AMPA receptor GluR2 subunit (GluR2) was not altered in the absence of BERP, suggesting some specificity of BERP action. To determine whether GABAARγ2 mRNA levels were also decreased under conditions of BERP deficiency, we performed real-time RT-PCR on RNA extracted from brains of BERP−/− and BERP+/+ E17.5 embryos. Comparable levels of GABAARγ2 mRNA were observed (Fig. S4), suggesting that the decrease in GABAARγ2 surface protein expression seen in BERP−/− neurons is a posttranscriptional phenomenon.
Fig. 5.
BERP deficiency decreases surface expression of GABAARs. Cortical neuron cultures were established from BERP+/+, BERP+/−, and BERP−/− E17.5 embryos. (A) Biotinylation of surface proteins was carried out, and extracts of pooled cultures were subjected to Western blot analysis to detect surface GABAARγ2 and GluR2 subunits. GABAARγ2 protein was reduced in the absence of BERP, but GluR2 levels were comparable in all genotypes. Results shown are a single trial representative of four independent experiments. (B) Densitometry analysis of surface GABAARγ2 expression in cortical neuron cultures from BERP+/+, BERP+/−, and BERP−/− embryos expressed in arbitrary units. Results shown are the mean expression ± SEM from four independent experiments. **P < 0.01, Student's t test.
Discussion
In a comprehensive review of 129 known p53-regulated genes, Riley et al. (30) established a set of criteria that a gene would have to meet to be considered p53-regulated. Our data on BERP fulfill these criteria, in that BERP site E bound to p53 in human cells and supported transcription only when p53wt was present. Moreover, a C→A point mutation at position 4 of BERP site E rendered it unable to support transcription regardless of p53 status. In addition, BERP mRNA was up-regulated in 5FU-treated HCT116 p53+/+ cells but not HCT116p53−/− cells. Furthermore, BERP mRNA was increased in the brains of PTZ-treated p53+/+ mice but not in the brains of p53−/− mice. Thus, we believe that BERP undergoes p53-dependent transcriptional activation and is indeed a unique p53-regulated gene.
Seizure susceptibility tests indicated that BERP−/− mice were more resistant to PTZ-induced seizures than BERP+/+ controls, suggesting a possible link between BERP and the CNS. BERP associates with myosin V via its C-terminal domain (17), and myosin V is involved in the intracellular trafficking of organelles and receptors (20, 21). Both BERP and myosin V are members of the cytoskeleton-associated recycling or transport (CART) complex, which is involved in the constitutive recycling of transferrin receptors in HeLa cells (22). It is possible that the CART complex, and therefore BERP, could be involved in the intracellular trafficking of other cell surface receptors that undergo constitutive endocytosis. Our patch-clamp recordings showed decreased mIPSC amplitude in BERP-deficient brains, and our Western blots revealed decreased surface expression of GABAARγ2 in cultured BERP−/− neurons. However, GABAARγ2 mRNA expression was not reduced in the absence of BERP. These data and the literature have led us to hypothesize that BERP may be involved in the intracellular trafficking of GABAARs. If the CART complex also exists in neurons, it is possible that BERP could be involved in the postendocytic recycling of GABAARs. Trinidad et al. (31) identified BERP as a possible phosphoprotein in a large-scale analysis of proteins isolated from murine postsynaptic densities, implying that when in a phosphorylated state, BERP might be localized synaptically. Additional work is required to place BERP in one of the five stages of the GABAAR lifecycle (32).
Because PTZ is a modulator of GABAAR function, PTZ's ability to up-regulate BERP expression in a p53-dependent manner suggests that (i) PTZ is a unique activator of p53, (ii) p53 is downstream of GABAAR function, and (iii) BERP acts downstream of p53. If our hypothesis that BERP is involved in GABAAR trafficking is correct, the following scenario becomes possible. PTZ disrupts the neuroelectrical homeostasis of the neuron and inhibits GABAAR function, which, in turn, triggers p53 activation. p53 up-regulates the expression of its target gene BERP, which then acts in a feedback loop to regulate the cell surface expression of GABAARs. In preliminary experiments designed to validate this scenario, we have found that p53 protein levels are increased in PTZ-treated human glioblastoma cells (Fig. S5), supporting our hypothesis that PTZ can activate p53.
Our work identifies an association between p53, BERP, and GABAARs and implies that p53 may be involved in the regulation of PTZ-induced effects. The implication is that p53, through BERP, may play a unique role in the control of GABAAR signaling and seizure susceptibility in the CNS. Further studies are required to elucidate the precise functions of p53 and BERP in GABAAR signaling.
Materials and Methods
Mice.
The p53-deficient mice used in this study were originally created in the laboratory of Tyler Jacks (MIT, Boston, MA) (33). BERP-deficient mice were generated and genotyped using PCR as described in SI Materials and Methods. All animal work was performed in compliance with the guidelines of the University Health Network Animal Care Committee.
ChIP Assay.
HCT116 p53+/+ and HCT116 p53−/− cells, the kind gifts of B. Vogelstein (Sidney Kimmel Comprehensive Cancer Center, Baltimore, MD), were treated with 5FU (Sigma) for 24 h. Extracts were treated with 37% vol/vol formaldehyde and sonicated [4 × 20-s pulses, 30% duty cycle, output control = 3, S-450A Sonifier (Branson)] to generate fragments of 200–1,000 bp in size. ChIP was performed using the EZ ChIP Kit (Millipore) and anti-p53 antibody (DO-1; Santa Cruz). PCR amplification was carried out using primers specific for the putative BERP p53REs. Primer sequences are shown in SI Materials and Methods.
Luciferase Assay.
pGL3-p21, the kind gift of S. Benchimol (York University, Toronto, ON, Canada) (34), contains the p53RE of the Cdkn1a promoter plus the E1B core promoter sequence subcloned into the pGL3-Basic vector (Promega) just upstream of the luciferase gene. pGL3-TATA is the pGL3-p21 vector with the Cdkn1a p53RE removed. To generate the pGL3-BerpB, pGL3-BerpE, pGL3-BerpF, pGL3-BerpG, and pGL3-BerpM vectors, the Cdkn1a p53RE of pGL3-p21 was replaced by PCR-amplified fragments of HCT116 cell genomic DNA representing the putative BERP p53RE sites B, E, F, G, and M, respectively. Using Fugene 6 (Roche), the constructs were cotransfected into HCT116 p53−/− cells along with a β-gal vector and vectors expressing either p53wt or p53mut (35, 36). At 24 h posttransfection, cells were analyzed using the Luciferase Assay System (Promega). Luciferase activity was normalized to β-gal activity and expressed relative to pGL3-TATA.
Real-Time RT-PCR.
Total RNA was extracted using RNeasy (Qiagen) and reverse-transcribed using the SuperScript First Strand RT-PCR cDNA synthesis system (Invitrogen) according the manufacturers’ protocols. Quantitative PCR was performed on cDNA using TaqMan Gene Expression Assays (Applied Biosystems; SI Materials and Methods) and the 7900HT Fast Real-Time PCR System (Applied Biosystems).
In Situ Hybridization.
p53+/+ and p53−/− male mice (8–12 wk old) were treated with PTZ (45 mg/kg; Sigma) or PBS via a single i.p. injection. At 6 or 24 h postinjection, brains were harvested, fixed for 24 h in 10% vol/vol neutral buffered formalin, dehydrated, embedded in paraffin, and subjected to in situ hybridization to detect BERP mRNA using a previously described protocol (37).
PTZ Seizure Susceptibility Testing.
Mice (six batches) were shipped to the Research Service at the Department of Veterans Affairs Medical Center (Coatesville, PA). Each batch contained BERP+/+ (∼10) and BERP−/− (∼10) mice, and batches were shipped ∼6 wk apart. On arrival, mice were housed under standard conditions for at least 1 wk before testing. Standard conditions were a 14 h (light)/10 h (dark) circadian light cycle with food and water available ad libitum. Seizure tests were conducted on male mice (8–12 wk old) between 9:00 AM and 12:00 PM. Mice received a single freshly prepared s.c. injection of PTZ (Sigma), in a volume equal to 1% of body weight, dissolved in physiological saline at pH 7.0. A range of PTZ doses was studied (40, 60, 80, 90, and 100 mg/kg) and staggered such that several doses were used in testing each batch of mice. Researchers performing seizure tests were blinded to mouse genotypes. Following PTZ injection, mice were placed into a Plexiglas chamber (20 mm wide, 30 mm deep, 40 mm high) with a mesh floor and scored over 45 min for seizure responses. Three end points, as well as the latencies to each end point, were recorded: partial seizure (characterized by a sudden whole-body spasm or a sudden and rapid clonus involving the head, neck, or one forelimb), generalized seizure (characterized by loss of upright posture and bilateral clonus of forelimbs and hind limbs), and maximal seizure (characterized by bilateral tonic extension of the hind limbs).
Brain Slice Preparation and Electrophysiology.
Male mice (3–4 wk old) were anesthetized with 2% vol/vol halothane and decapitated. The methods used for brain slice preparation and mIPSC recording have been described previously (38, 39). Briefly, coronal brain slices (300 μm) were prepared using a vibratome (Vibratome Co.) and ice-cold oxygenated (95% O2, 5% CO2) artificial cerebral spinal fluid, which contained 125 mM NaCl, 2.5 mM KCl, 25 mM NaHCO3, 1.25 mM NaH2PO4, 1 mM MgCl2, 2 mM CaCl2, and 25 mM glucose at pH 7.3–7.4 and osmolarity at 300–320 mOsm. For whole-cell patch-clamp recordings, mIPSCs of cortical pyramidal neurons were recorded at room temperature using an Axopatch 200B (Molecular Devices). Recording electrode resistance was 3–5 MΩ using an electrode solution containing 145 mM CsCl, 1 mM CaCl2, 2 mM MgCl2, 5 mM EGTA, 10 mM Hepes, and 4 mM K2ATP at pH 7.3 and osmolarity at 280–290 mOsm. Signals were filtered at 2 kHz and sampled at 10 kHz. Neurons were voltage-clamped at −70 mV in the presence of TTX (1.0 μM), 6-cyano-7-nitroquinoxaline-2,3-dione (20 μM), and d-2-amino-5-phosphonovalerate (50 μM). mIPSCs were abolished by the addition of 10 mM bicuculline, a GABAAR antagonist.
Murine Cortical Neuron Cultures.
Cultures of primary neurons were derived from the cerebral cortex of E17.5 mouse embryos, as previously described (40). The excised cortex was placed in ice-cold HBSS and gently dissociated. Cells were pelleted by centrifugation at 300 × g × 5′ at 4 °C, resuspended in plating medium [Neurobasal medium (Invitrogen), 2% vol/vol B-27 supplement (Gibco), 10% vol/vol FBS (HyClone), 0.5 mM L-glutamine, 25 μM glutamic acid], and plated on poly-D-lysine–coated Petri dishes (day 0). On day 3, one half of the plating medium was replaced with maintenance medium [Neurobasal medium, 2% vol/vol B-27 supplement, 0.5 mM L-glutamine]. Maintenance medium was exchanged every 3 d thereafter.
Biotinylation Assay of Surface Proteins.
Cortical neurons were cultured for 10 d as above, and surface GABAARγ2 and GluR2 proteins were measured using a Cell Surface Protein Biotinylation and Purification Kit (Thermo Fisher Scientific, Inc.) (41). Briefly, neurons were incubated with Sulfo-NHS-SS-Biotin (Thermo Fisher Scientific, Inc.) at 4 °C for 30 min, followed by addition of lysis buffer. Lysates were incubated with Immobilized NeutrAvidin Gel (Thermo Fisher Scientific, Inc.) for 60 min to isolate biotinylated proteins. Western blotting with anti-GABAARγ2 antibody (Abcam) or anti-GluR2 antibody (Abcam) was performed, followed by densitometry analysis as previously described (42).
Supplementary Material
Acknowledgments
We thank Tom Ferraro and George Smith for expert technical assistance and Mary E. Saunders for scientific editing. C.C.C. is supported by the Canadian Institute of Health Research.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1006529107/-/DCSupplemental.
References
- 1.Vousden KH, Lu X. Live or let die: The cell's response to p53. Nat Rev Cancer. 2002;2:594–604. doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- 2.Levine AJ, Hu W, Feng Z. The P53 pathway: What questions remain to be explored? Cell Death Differ. 2006;13:1027–1036. doi: 10.1038/sj.cdd.4401910. [DOI] [PubMed] [Google Scholar]
- 3.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 4.Daujat S, Neel H, Piette J. MDM2: Life without p53. Trends Genet. 2001;17:459–464. doi: 10.1016/s0168-9525(01)02369-1. [DOI] [PubMed] [Google Scholar]
- 5.Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 6.Momand J, Zambetti GP, Olson DC, George D, Levine AJ. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell. 1992;69:1237–1245. doi: 10.1016/0092-8674(92)90644-r. [DOI] [PubMed] [Google Scholar]
- 7.Stambolic V, et al. Regulation of PTEN transcription by p53. Mol Cell. 2001;8:317–325. doi: 10.1016/s1097-2765(01)00323-9. [DOI] [PubMed] [Google Scholar]
- 8.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 9.Sonoda J, Wharton RP. Drosophila Brain Tumor is a translational repressor. Genes Dev. 2001;15:762–773. doi: 10.1101/gad.870801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frank DJ, Edgar BA, Roth MB. The Drosophila melanogaster gene brain tumor negatively regulates cell growth and ribosomal RNA synthesis. Development. 2002;129:399–407. doi: 10.1242/dev.129.2.399. [DOI] [PubMed] [Google Scholar]
- 11.Betschinger J, Mechtler K, Knoblich JA. Asymmetric segregation of the tumor suppressor brat regulates self-renewal in Drosophila neural stem cells. Cell. 2006;124:1241–1253. doi: 10.1016/j.cell.2006.01.038. [DOI] [PubMed] [Google Scholar]
- 12.Arama E, Dickman D, Kimchie Z, Shearn A, Lev Z. Mutations in the beta-propeller domain of the Drosophila brain tumor (brat) protein induce neoplasm in the larval brain. Oncogene. 2000;19:3706–3716. doi: 10.1038/sj.onc.1203706. [DOI] [PubMed] [Google Scholar]
- 13.Reymond A, et al. The tripartite motif family identifies cell compartments. EMBO J. 2001;20:2140–2151. doi: 10.1093/emboj/20.9.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hulsen T, Huynen MA, de Vlieg J, Groenen PM. Benchmarking ortholog identification methods using functional genomics data. Genome Biol. 2006;7:R31. doi: 10.1186/gb-2006-7-4-r31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moreno-Hagelsieb G, Latimer K. Choosing BLAST options for better detection of orthologs as reciprocal best hits. Bioinformatics. 2008;24:319–324. doi: 10.1093/bioinformatics/btm585. [DOI] [PubMed] [Google Scholar]
- 16.El-Husseini AE, Kwasnicka D, Yamada T, Hirohashi S, Vincent SR. BERP, a novel ring finger protein, binds to alpha-actinin-4. Biochem Biophys Res Commun. 2000;267:906–911. doi: 10.1006/bbrc.1999.2045. [DOI] [PubMed] [Google Scholar]
- 17.El-Husseini AE, Vincent SR. Cloning and characterization of a novel RING finger protein that interacts with class V myosins. J Biol Chem. 1999;274:19771–19777. doi: 10.1074/jbc.274.28.19771. [DOI] [PubMed] [Google Scholar]
- 18.Honda K, et al. Actinin-4, a novel actin-bundling protein associated with cell motility and cancer invasion. J Cell Biol. 1998;140:1383–1393. doi: 10.1083/jcb.140.6.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nikolopoulos SN, et al. The human non-muscle alpha-actinin protein encoded by the ACTN4 gene suppresses tumorigenicity of human neuroblastoma cells. Oncogene. 2000;19:380–386. doi: 10.1038/sj.onc.1203310. [DOI] [PubMed] [Google Scholar]
- 20.Wang Z, et al. Myosin Vb mobilizes recycling endosomes and AMPA receptors for postsynaptic plasticity. Cell. 2008;135:535–548. doi: 10.1016/j.cell.2008.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Provance DW, Mercer JA. Myosin-V: Head to tail. Cell Mol Life Sci. 1999;56:233–242. doi: 10.1007/s000180050425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yan Q, et al. CART: An Hrs/actinin-4/BERP/myosin V protein complex required for efficient receptor recycling. Mol Biol Cell. 2005;16:2470–2482. doi: 10.1091/mbc.E04-11-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ohkawa N, et al. Molecular cloning and characterization of neural activity-related RING finger protein (NARF): A new member of the RBCC family is a candidate for the partner of myosin V. J Neurochem. 2001;78:75–87. doi: 10.1046/j.1471-4159.2001.00373.x. [DOI] [PubMed] [Google Scholar]
- 24.Boulay JL, et al. Loss of heterozygosity of TRIM3 in malignant gliomas. BMC Cancer. 2009;9:71. doi: 10.1186/1471-2407-9-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martins-de-Souza D, et al. Prefrontal cortex shotgun proteome analysis reveals altered calcium homeostasis and immune system imbalance in schizophrenia. Eur Arch Psychiatry Clin Neurosci. 2009;259:161–163. doi: 10.1007/s00406-008-0847-2. [DOI] [PubMed] [Google Scholar]
- 26.Macdonald RL, Olsen RW. GABAA receptor channels. Annu Rev Neurosci. 1994;17:569–602. doi: 10.1146/annurev.ne.17.030194.003033. [DOI] [PubMed] [Google Scholar]
- 27.Lüscher B, Keller CA. Regulation of GABAA receptor trafficking, channel activity, and functional plasticity of inhibitory synapses. Pharmacol Ther. 2004;102:195–221. doi: 10.1016/j.pharmthera.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 28.el-Deiry WS, Kern SE, Pietenpol JA, Kinzler KW, Vogelstein B. Definition of a consensus binding site for p53. Nat Genet. 1992;1:45–49. doi: 10.1038/ng0492-45. [DOI] [PubMed] [Google Scholar]
- 29.Nusser Z, Cull-Candy S, Farrant M. Differences in synaptic GABA(A) receptor number underlie variation in GABA mini amplitude. Neuron. 1997;19:697–709. doi: 10.1016/s0896-6273(00)80382-7. [DOI] [PubMed] [Google Scholar]
- 30.Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol. 2008;9:402–412. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- 31.Trinidad JC, Specht CG, Thalhammer A, Schoepfer R, Burlingame AL. Comprehensive identification of phosphorylation sites in postsynaptic density preparations. Mol Cell Proteomics. 2006;5:914–922. doi: 10.1074/mcp.T500041-MCP200. [DOI] [PubMed] [Google Scholar]
- 32.Jacob TC, Moss SJ, Jurd R. GABA(A) receptor trafficking and its role in the dynamic modulation of neuronal inhibition. Nat Rev Neurosci. 2008;9:331–343. doi: 10.1038/nrn2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jacks T, et al. Tumor spectrum analysis in p53-mutant mice. Curr Biol. 1994;4:1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- 34.Lin Y, Ma W, Benchimol S. Pidd, a new death-domain-containing protein, is induced by p53 and promotes apoptosis. Nat Genet. 2000;26:122–127. doi: 10.1038/79102. [DOI] [PubMed] [Google Scholar]
- 35.Kern SE, et al. Oncogenic forms of p53 inhibit p53-regulated gene expression. Science. 1992;256:827–830. doi: 10.1126/science.1589764. [DOI] [PubMed] [Google Scholar]
- 36.Baker SJ, Markowitz S, Fearon ER, Willson JK, Vogelstein B. Suppression of human colorectal carcinoma cell growth by wild-type p53. Science. 1990;249:912–915. doi: 10.1126/science.2144057. [DOI] [PubMed] [Google Scholar]
- 37.Skinnider BF, et al. Interleukin 13 and interleukin 13 receptor are frequently expressed by Hodgkin and Reed-Sternberg cells of Hodgkin lymphoma. Blood. 2001;97:250–255. doi: 10.1182/blood.v97.1.250. [DOI] [PubMed] [Google Scholar]
- 38.Ning K, et al. Circadian regulation of GABAA receptor function by CKI epsilon-CKI delta in the rat suprachiasmatic nuclei. Nat Neurosci. 2004;7:489–490. doi: 10.1038/nn1236. [DOI] [PubMed] [Google Scholar]
- 39.Wan Q, et al. Recruitment of functional GABA(A) receptors to postsynaptic domains by insulin. Nature. 1997;388:686–690. doi: 10.1038/41792. [DOI] [PubMed] [Google Scholar]
- 40.Shan Y, et al. Regulation of PINK1 by NR2B-containing NMDA receptors in ischemic neuronal injury. J Neurochem. 2009;111:1149–1160. doi: 10.1111/j.1471-4159.2009.06398.x. [DOI] [PubMed] [Google Scholar]
- 41.Ding S, Ingleby L, Ahern CA, Horn R. Investigating the putative glycine hinge in Shaker potassium channel. J Gen Physiol. 2005;126:213–226. doi: 10.1085/jgp.200509287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ning K, et al. Dual neuroprotective signaling mediated by downregulating two distinct phosphatase activities of PTEN. J Neurosci. 2004;24:4052–4060. doi: 10.1523/JNEUROSCI.5449-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.