Abstract
The first step in assembly of membrane and storage glycerolipids is acylation of glycerol-3-phosphate (G3P). All previously characterized membrane-bound, eukaryotic G3P acyltransferases (GPATs) acylate the sn-1 position to produce lysophosphatidic acid (1-acyl-LPA). Cutin is a glycerolipid with omega-oxidized fatty acids and glycerol as integral components. It occurs as an extracellular polyester on the aerial surface of all plants, provides a barrier to pathogens and resistance to stress, and maintains organ identity. We have determined that Arabidopsis acyltransferases GPAT4 and GPAT6 required for cutin biosynthesis esterify acyl groups predominantly to the sn-2 position of G3P. In addition, these acyltransferases possess a phosphatase domain that results in sn-2 monoacylglycerol (2-MAG) rather than LPA as the major product. Such bifunctional activity has not been previously described in any organism. The possible roles of 2-MAGs as intermediates in cutin synthesis are discussed. GPAT5, which is essential for the accumulation of suberin aliphatics, also exhibits a strong preference for sn-2 acylation. However, phosphatase activity is absent and 2-acyl-LPA is the major product. Clearly, plant GPATs can catalyze more reactions than the sn-1 acylation by which they are currently categorized. Close homologs of GPAT4-6 are present in all land plants, but not in animals, fungi or microorganisms (including algae). Thus, these distinctive acyltransferases may have been important for evolution of extracellular glycerolipid polymers and adaptation of plants to a terrestrial environment. These results provide insight into the biosynthetic assembly of cutin and suberin, the two most abundant glycerolipid polymers in nature.
Keywords: cutin, suberin, lysophosphatidic acid phosphatase, bifunctional
Glycerol-3-phosphate acyltransferase (GPAT) (EC 2.3.1.15) catalyzes the initial step of glycerolipid synthesis, the incorporation of an acyl group from acyl-CoA onto the sn-1 position of sn-glycerol-3-phosphate (G3P) to yield 1-acyl-lysophosphatidic acid (1-acyl-LPA). This reaction has been extensively characterized in bacteria, fungi, animals, and plants (1–5). A family of eight plant GPAT acyltransferase genes in Arabidopsis was first identified based on sequence similarity to known nonplant GPAT enzymes. When expressed in yeast, several members of the family were shown to catalyze acyl transfer to G3P, although the position on glycerol that was acylated was not determined (6).
Cutin and suberin are lipophilic barriers found associated with the plant cell wall. They have broadly similar functions in that they control small molecule fluxes and act as protective barriers. The major components of both cutin and the aliphatic domain of suberin are ω-oxidized fatty acids, namely ω-hydroxy fatty acids (ω-OHFAs) and α,ω-dicarboxylic acids (DCAs), with varying amounts of glycerol. Thus, cutin and suberin are also glycerolipids (7, 8). Although it is one of the most abundant lipid polymers in nature, with an area five times earth's land surface (9), the specific enzyme reactions in cutin assembly are largely unknown. We recently reported that four GPAT enzymes participate in cutin or suberin biosynthesis in Arabidopsis and control both the quantity and composition of cutin or suberin (10–12). Our evidence was based on results of mutant analyses and the phenotypes obtained from overexpression in transgenic plants. Arabidopsis leaf and stem cutin contains largely DCAs. GPAT4 and GPAT8 are predominantly expressed in leaves and stems, and gpat4/gpat8 double knockout mutants have major reductions in C18 unsaturated DCA cutin monomers (11). GPAT5 is required for the accumulation of suberin aliphatics, especially C22:0 and C24:0 ω-OHFA and DCA monomers (10). In Arabidopsis flowers, cutin is dominated by C16 monomers. GPAT6 is strongly expressed in flowers, and gpat6 mutants are substantially reduced in all C16 cutin monomers (DCA, 16-hydroxy- and 10,16-dihydroxypalmitates), while overexpression of GPAT6 increases these monomers (12). In addition, C16:0 DCA becomes the major cutin monomer of flowers when the midchain hydroxylase CYP77A6 is inactive (12).
To further characterize the reactions catalyzed by these acyltransferases, GPAT4, GPAT5 and GPAT6 were expressed in a yeast GPAT-deficient mutant (gat1Δ) and/or a wheat germ cell-free translation system. The reactions were studied using α,ω-dicarboxylic acid acyl-CoAs (DCA-CoAs) as acyl donors and [14C]G3P as acyl acceptor. Analysis of the products of these reactions indicated that all three acyltransferases esterify acyl groups preferentially to the sn-2 position of G3P. In addition, GPAT4 and GPAT6, but not GPAT5, are bifunctional enzymes that catalyze a phosphatase reaction resulting in sn-2 monoacylglycerols (MAGs) as products. The phosphatase activity of GPAT6 was reduced almost completely by site-directed mutagenesis of putative active-site residues, and the major product became 2-acyl-LPA. Thus, the plant GPATs exhibit a diversity of reactions not reported previously. The distinctive enzymology is discussed in terms of the roles of GPATs in the biosynthesis and evolution of cutin and aliphatic suberin.
Results
Previous in vitro characterizations of plant GPAT enzymes have been in the context of studies on triacylglycerol and membrane glycerolipid biogenesis, and thus have used acyl-CoAs without ω-oxidized groups as acyl donors. However, ω-OHFAs and DCAs are the common acyl monomers of cutin and suberin (13, 14). To test whether these ω-oxidized fatty acids are effective substrates for the GPAT enzymes, ω-OHFA and DCA-CoA substrates were chemically synthesized (15), purified to >90%, and the products characterized by TLC and by electrospray ionization tandem mass spectrometry (ESI-MS/MS) (SI Appendix, Table S1). DCA-CoA products were the mono-CoA thioesters.
Initial studies of in vitro substrate specificity of these acyltransferases expressed in yeast demonstrated that GPAT4 was active with C18:1 ω-oxidized acyl-CoAs, GPAT6 was active with C16:0 ω-oxidized acyl-CoAs, and GPAT5 was active with C22:0 normal and ω-oxidized acyl-CoAs. These substrate preferences match the monomers that are most altered in mutants disrupted in the respective acyltransferases. Among the ω-oxidized acyl-CoA substrates we tested, enzyme activities (measured by 14C-incorporation from [14C]G3P) were highest with DCA-CoAs. Therefore, we focused on DCA substrates for further characterization of GPAT activity. Enzymes were assayed under conditions where product formation was linear with respect to protein concentration and time.
LPA is the expected product of the GPAT reaction. With DCA-CoA as substrate, LPA will possess an additional carboxylic acid group. Such amphipathic products have significant aqueous solubility. To prevent product loss through traditional solvent extraction procedures, analysis of the products was therefore performed by direct application of quenched reactions to TLC plates. This procedure assured that both water-soluble and nonpolar enzyme products would be quantitatively recovered and analyzed.
Monoacylglycerol Is a Product of GPAT4 and GPAT6 but Not of GPAT5.
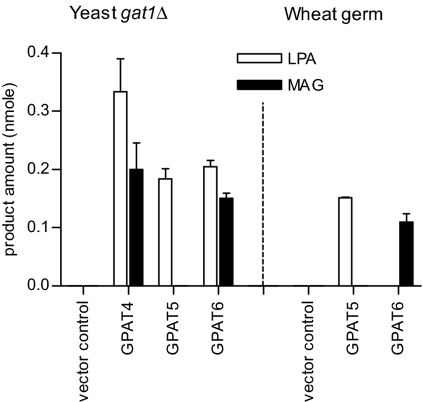
When microsomes from yeast expressing GPAT4 or GPAT6 were assayed with C18:1 or 16:0 DCA-CoAs, a second labeled product was observed in addition to LPA (Fig. 1). This product migrated slightly after C18:1 MAG, but substantially ahead of C18:1-LPA (SI Appendix, Fig. S1A). We therefore considered the possibility that the 14C-labeled product was DCA-MAG, because the extra carboxylic acid group would impart additional polarity when compared with C18:1 MAG. The identification of this nonpolar 14C-product as MAG is described in the next section. By contrast, when GPAT5 was assayed with C22:0 DCA-CoA, LPA was the major product and MAG was not observed (Fig. 1 and SI Appendix, Fig. S1A).
Fig. 1.
Product distribution of GPAT assays in either GPAT-transformed yeast (gat1Δ) microsomes or wheat germ cell-free translation system. GPAT assays were conducted using 14C-G3P and DCA-CoA substrates as described under Methods. C18:1/22:0/16:0 DCA-CoAs were acyl donors for GPAT4/5/6 assays, respectively, with empty vector as control. Products (nmol) were LPA and MAG. Values are mean ± SD (n = 3) for yeast assays, and duplicates were done in wheat germ system (error bars represent range).
Because removal of the phosphate to produce MAG could be a consequence of the recombinant GPAT reaction or alternatively of an endogenous yeast phosphatase, GPAT5 and GPAT6 were also expressed in a wheat germ cell-free translation system and the GPAT assays repeated. In this system, almost no LPA was produced in the GPAT6 reaction and instead the predominant product was MAG (Fig. 1 and SI Appendix, Fig. S1B). It is possible that more complete conversion to MAG in wheat germ compared with yeast expression occurs because a more “native” conformation (e.g., protein folding, posttranslational modification) is achieved for the plant membrane enzymes. The reaction products observed for GPAT5 were similar to the yeast expression, with no formation of MAG (Fig. 1 and SI Appendix, Fig. S1B). Taken together, the production of MAG as a major product with GPAT4/GPAT6 and the production of LPA as major product with GPAT5 were observed in both yeast and wheat germ cell-free translation system (Fig. 1). These observations support the conclusion that the MAG product is a result of the plant GPAT rather than an endogenous phosphatase present in yeast microsomes. Two other lines of evidence support this conclusion. First, MAG formation was linear with time with no detectable lag (SI Appendix, Fig. S2). This is consistent with GPAT-bound LPA intermediate being acted on by GPAT itself rather than LPA being released and hydrolyzed by an endogenous phosphatase. Second, neither LPA nor MAG formation was observed when 14C-glycerol was used as an acyl acceptor, providing evidence of a biosynthetic route from G3P to LPA to MAG, rather than from G3P to glycerol to MAG.
GPAT4 and GPAT6 Catalyzed Acylations Occur Predominantly at the sn-2 Position of G3P.
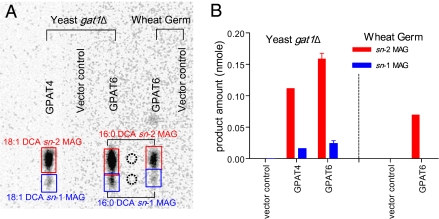
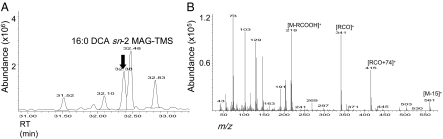
All previously studied membrane-bound eukaryotic GPAT enzymes have been reported to acylate G3P predominantly at the sn-1 position to produce 1-acyl-LPA. However, the major products of Arabidopsis GPAT4 and GPAT6 were MAGs. To identify the regiospecificity of these MAGs, the quenched assay mixture from either yeast or wheat germ expression was directly spotted onto borate-TLC plates. Such plates allow resolution of MAG α and β isomers with minimal acyl migration (16). To confirm the identity of the products, sn-1 and sn-2 MAGs were chemically synthesized from C16:0 and C18:1 DCAs (SI Appendix, SI Methods) and their structures were confirmed by GC-MS (SI Appendix, Fig. S3). The major labeled neutral lipid product from either the GPAT4 or GPAT6 reaction comigrated with the DCA sn-2 MAG standards (Fig. 2A). The ratio of sn-2:sn-1-labeled products was 5:1 or greater (Fig. 2B). To confirm the identification of MAG products, large-scale GPAT assays were performed. The bands corresponding to the sn-2 MAG products of GPAT4 or GPAT6 assays were recovered from the borate-TLC plate, silylated, and analyzed by GC-MS. By comparison of their EI-MS spectra and retention times with the synthetic DCA-MAG standards, the major products of the GPAT6 and GPAT4 assays were respectively confirmed as C16:0 DCA sn-2 MAG (Fig. 3) and C18:1 DCA sn-2 MAG (SI Appendix, Fig. S4). No MAG products were observed when microsomes from yeast transformed with empty vector were used as source of enzyme or when acyl-CoA or G3P was omitted from the reaction (SI Appendix, Fig. S5).
Fig. 2.
Regiospecificity of DCA-MAGs formed in GPAT4 and GPAT6 assays. GPAT assays were conducted using 14C-G3P and DCA-CoA substrates as described under Methods. (A) Separation of MAGs by borate-TLC (solvent system, chloroform:acetone 1:1). GPAT4 assay was performed in yeast system with C18:1 DCA-CoA as acyl donor. GPAT6 was assayed in both yeast and wheat germ systems with C16:0 DCA-CoA as acyl donor. MAGs were identified by comigration with C18:1 DCA sn-1 and sn-2 MAG standards (dotted circle). (B) Quantification of GPAT products. Values are mean ± SD (n = 3) for GPAT6 yeast assays. DCA sn-2 MAGs predominate over sn-1 MAGs in both GPAT4 and GPAT6 assays.
Fig. 3.
Identification of C16:0 DCA sn-2 MAG from GPAT6 assay by GC-MS. GPAT6 assay product C16:0 DCA sn-2 MAG was isolated and identified as described under Methods and SI Appendix, SI Methods. (A) GC chromatogram of TLC fraction containing C16:0 DCA sn-2 MAG. (B) Corresponding EI-MS spectrum of C16:0 DCA sn-2 MAG-Tris-trimethylsilyl derivative (peak at 32.38 min). See SI Appendix, Fig. S3 for spectrum of standard.
sn-2-Acyl-LPA Is the Major Product of GPAT5.
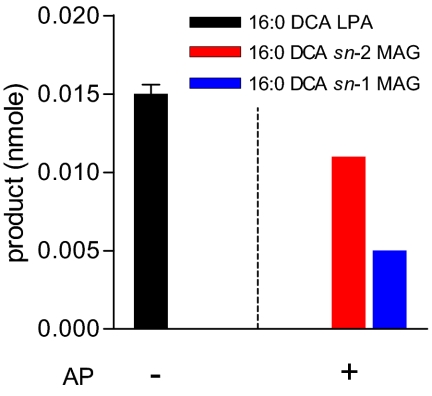
In addition to the preference for longer chain lengths, we observed that GPAT5 can also use C16:0 DCA-CoA as substrate to form C16:0 DCA-LPA, with no further conversion to MAG. This result is consistent with a several fold increase in C16:0 DCA in stem cutin monomers after ectopic coexpression of GPAT5 with CYP86A1 in Arabidopsis (11). Therefore, we chose C16:0 DCA-LPA formed by GPAT5 to examine its regiospecificity. Enzyme reactions were immediately dephosphorylated with alkaline phosphatase (AP) (17) to minimize any DCA-LPA product loss or acyl migration. The resulting C16:0 DCA-MAGs were then analyzed by borate-TLC (SI Appendix, Fig. S6). Fig. 4 shows the results before and after AP treatment. Before AP treatment, C16:0 DCA-LPA is the major product and MAG production is not observed. However, after AP treatment, quantitative conversion of C16:0 DCA-LPA to MAG was observed. Once again, the sn-2 MAG isomer was predominant (ratio 2:1). The above estimates of sn-2/sn-1 ratios are minimal because some acyl migration may have occurred during dephosphorylation and sample analysis. Therefore, the above result indicates that acylation of G3P by GPAT5 to form LPA also occurs predominantly at the sn-2 position. A similar sn-2 preference was observed with C22:0-CoA (SI Appendix, Fig. S7).
Fig. 4.
Positional analysis of GPAT5 assay product C16:0 DCA-LPA. GPAT assays were conducted using 14C-G3P and C16:0 DCA-CoA as substrates. Immediately after assay, the reaction mixture was either directly spotted onto TLC plate to separate and quantify labeled LPA, or incubated with 0.25 unit of alkaline phosphatase (AP) before TLC analysis. The resulting MAGs were separated on borate-TLC and identified by comparison with C18:1 DCA sn-1 and sn-2 MAG standards. Products quantification was performed as described under Methods. Values are mean ± SD (n = 3) for GPAT5 yeast assays without AP treatment.
GPAT4 and GPAT6 Retain Highly Conserved Amino Acid Residues Required for Phosphatase Activity, Whereas GPAT5 Does Not.
Analysis of the amino acid sequences of the eight members of the GPAT family revealed that all eight members have a plsC acyltransferase domain in the C-terminal region and a second domain in the N-terminal region that is homologous to conserved motifs of the HAD-like hydrolase superfamily. This superfamily includes phosphatases and indeed the N-terminal 240 amino acids of GPAT4-8 align best with phosphoserine phosphatase from Methanococcus jannaschii (MJ-PSP) based on sequence and predicted secondary structure.
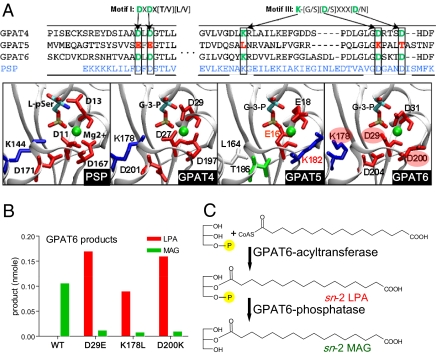
As shown in Fig. 5A, alignments of the sequences of Arabidopsis GPAT4, -5, and -6 with MJ-PSP indicate that several residues known to be essential for phosphatase activity and Mg2+ binding (18–20) are conserved between MJ-PSP and the plant GPAT enzymes. In particular, two residues demonstrated to be essential for catalytic activity of phosphoserine phosphatase are conserved in GPAT4 and GPAT6, but not in GPAT5. This includes the first ASP in motif I that forms a phosphoenzyme intermediate and when mutated to GLU in human phosphoserine phosphatase leads to complete loss of activity. In addition, the conserved first ASP in motif III is a key coordinator to the Mg2+ ion and is essential for phosphatase activity. Structural modeling of the N-terminal regions of GPAT4, -5, and -6 based on the MJ-PSP template (Fig. 5A) indicates highly similar structures near the MJ-PSP active site and a similar overall fold (SI Appendix, Fig. S8). In GPAT4 and GPAT6 there is a similar clustering of the residues critical for catalysis and Mg2+ binding while the LYS substitution of the first ASP in motif III in GPAT5 creates a significantly different electrostatic environment at the site where Mg2+ binds in MJ-PSP. Furthermore, (L-)G3P (or LPA) has the same stereochemical configuration as L-phosphoserine, the natural substrate of PSP (Fig. 5A), so it is reasonable that there is a close active-site relationship between GPAT and MJ-PSP. Site-directed mutagenesis of any of these three conserved AA residues on GPAT6 (i.e., mutation of D29 to E29, or K178 to L178, or D200 to K200) led to the reduction of the phosphatase activity by at least 85% (Fig. 5B and SI Appendix, Fig. S9A) and resulted in sn-2 LPA as the major product (Fig. 5C and SI Appendix, Fig. S9 B and C). These experiments confirm that the predicted phosphatase domain of GPAT6 is responsible for MAG products from the enzyme reaction.
Fig. 5.
Site-directed mutagenesis analysis of GPAT6. (A) Sequence alignment of GPAT4, -5, and -6 with MJ-PSP and active-site close-up for MJ-PSP and predicted structures of N-terminal domain of GPAT4, -5, and -6. The critical amino acid residues required for PSP activity in motif I and III are labeled with arrows. Those amino acid residues are conserved in GPAT4 and GPAT6, but not in GPAT5. MJ-PSP structure from 1F5S is shown with L-phosphoserine ligand from MJ-PSP-ligand complex from Protein Data Bank structure 1L7P. The GPAT models shown were generated in the absence of any ligand but are shown with G3P aligned in active site. Acidic residues are shown in red, basic residues in blue, polar residues in green, and nonpolar residues in white. Mg2+ is shown in green. (B) Product distribution of GPAT6 assay in wheat germ translation system. Assays were conducted using 14C-G3P and C16:0 DCA-CoA as the substrates for three GPAT6 single mutants (D29E, K178L, or D200K) and GPAT6-WT control. Products (nmol) were predominantly sn-2 MAG for GPAT6-WT, whereas sn-2 LPA is the major product of GPAT6-phosphatase mutants (see SI Appendix, Fig. S9 for regiospecific data).Values are duplicates, error bars represent range. (C) Reactions catalyzed by the bifunctional enzyme GPAT6 to produce sn-2 MAG.
The C-terminal acyltransferase domain of the GPAT family possesses the classic H(X)4D motif of PlsC class acyltransferases and a CPEGT sequence also similar to a second FPEGT motif of PlsC (21). A model for the C-terminal region can be constructed by using the known crystal structure of the squash plastid G3P acyltransferase as a template (SI Appendix, Fig. S8). The phosphatase and acyltransferase domains are connected with a 60-residue linker of unknown structure (SI Appendix, Fig. S10).
Discussion
The Black Box of Cutin and Suberin Biosynthesis.
Although cutin and suberin are the most abundant lipid polymers that occur in nature, comparatively little is known about their detailed molecular structure and the biosynthetic steps that lead to assembly from their monomers (13). Unknowns include the extent of assembly within or outside the cell, the number of monomers that constitute the polymer, and the degree of cross-linking between monomers and other cell wall structures. This lack of knowledge occurs despite the recent identification of several proteins that clearly function in cutin or suberin biosynthesis. Chief among this growing list are P450s of the CYP86 (11, 12, 22–25) and CYP77 (12) families, GPATs (10–12), and members of LACS (26–28) and BAHD gene families (25), whereas a number of proteins including ABC transporters (29), HOTHEAD (27, 30, 31) and BODYGUARD (32–36) have more uncertain biosynthetic functions. Adding to the complexity of these polymers, even within Arabidopsis, it is possible to define several distinct cutins and suberins based on monomer composition. GPAT6 plays an essential role in cutins rich in C16 monomers (12). These cutins are typically rich in 10,16-dihydroxypalmitic acid and are found in the epidermis of Arabidopsis petals and sepals (12). GPATs 4 and 8 play essential but overlapping roles for cutins rich in C18 unsaturated DCA, which are present in leaf and stem, and which contribute to the stomatal ledges (11). Finally, GPAT5 is required for the deposition of C22:0 and C24:0 ω-OHFA and DCA suberin monomers such as found in the root and in seed coat outer integument (10, 37).
In the present study, GPAT4, -5, and -6 were expressed in vitro, and their products were characterized. None of these GPATs demonstrated sn-1 acyl transfer as the major activity, which is surprising as this is the activity detected in all membrane-bound eukaryotic GPATs that have been previously characterized. Instead, our results unexpectedly indicated that each of the above acyltransferases preferentially transfer acyl groups to the sn-2 position of G3P. Also surprisingly, GPAT4 and GPAT6 were found to be bifunctional G3P acyltransferase/phosphatases, producing sn-2 MAG (not LPA) as a major product (Fig. 5C). Thus, we conclude that cutin glycerolipid biosynthesis proceeds via a different set of initial reactions than membrane or storage glycerolipid synthesis. The subcellular location of such reactions remains uncertain. Our modeling data indicate that GPAT phosphatase and acyltransferase domains are connected by a region with membrane helix characteristics (SI Appendix, Fig. S10). It is possible that the 60-aa linker region between the two catalytic domains is involved in membrane interactions, but further analysis is required to better understand this aspect. Furthermore, it remains to be investigated whether in epidermal cells GPAT proteins could have a more precise localization, perhaps to an ER subdomain associated with the plasma membrane. In this regard, GPAT8, the closest homolog to GPAT4, has been demonstrated to be localized in the ER with both N and C termini in the cytosol (38).
Possible Roles of sn-2 MAG and GPATs in Polyester Biosynthesis.
The fact that GPAT4 and GPAT6 can directly synthesize sn-2 MAG in vitro suggests that sn-2 MAG may be a building block for Arabidopsis polyester assembly. Although the scenarios below are speculative, the elucidation of the sn-2 acyl transfer and the bifunctional nature of GPAT4 and GPAT6 allow additional hypotheses for cutin assembly. For example, assuming sn-2 MAG is an intermediate (or synthon) in cutin biosynthesis, it could be envisaged that the free carboxyl group of DCA-MAG undergoes further activation to CoA by a long-chain acyl-CoA synthetase. In this sense, DCA-MAG would be recognized as an analog of ω-OHFA, with COOH at one end of the aliphatic chain, and not one but two primary OH groups at the other end of the chain. It may then be possible for DCA-MAG-CoA to undergo a second acyl transfer to a free hydroxyl group resulting in oligomers synthesized within the cell. Alternatively, as MAG can clearly be exported by epidermal cells (39), and because the ABC transporter WBC11 has been suggested to play a direct role in cutin formation (29), MAG might also be considered as one form in which precursors for cutin assembly are exported. Regardless, future experiments to test the ability of sn-2 MAG to participate in additional reactions are likely to be informative.
Analysis of the seed suberin monomer profile in gpat5 mutants has shown that C22:0 and C24:0 very long-chain fatty acid (VLCFA), ω-OHFA and DCA monomer accumulation depends on GPAT5 (10, 37). In addition, ectopic expression of GPAT5 produces MAG with VLCFA as a component of epicuticular surface waxes (39). These results indicate that GPAT5 was indeed acting in vivo as a VLCFA acyltransferase to a glycerol-based acceptor. Of particular importance in these studies is that the extracellular MAG is predominantly the thermodynamically less stable sn-2 isomer. Thus, our current result showing that GPAT5 preferentially transfers acyl groups to the sn-2 position of G3P in vitro (Fig. 4) is consistent with the in vivo production of sn-2 MAG previously observed after GPAT5 ectopic expression in Arabidopsis. The in vitro GPAT5 enzyme assays produced sn-2 LPA instead of sn-2 MAG as the major product, regardless of acyl-CoA substrate (FA/ω-OHFA/DCA) (SI Appendix, Fig. S7). Thus, the formation of extracellular MAG observed in vivo (39) may result from activity of endogenous phospholipases/lipases of epidermal cells that convert LPA to MAG. Alternatively, we cannot rule out that GPAT5 may participate in a phosphatase reaction if it interacts with other factors.
sn-2 G3P Acyltransferase/Phosphatase May Have Evolved Specifically for Plant Extracellular Lipid Biosynthesis.
Analysis of the amino acid sequences of the eight members of the GPAT family revealed that all eight members have a plsC acyltransferase domain in the C-terminal region. However, only GPAT4, -6, and -8 possess all of the conserved amino acids at critical positions of the N-terminal region that are essential for phosphatase activity in phosphoserine phosphatases. Several other candidate bifunctional acyltransferase/phosphatases can be detected by searches of sequence databases; however, to our knowledge, none of these putative acyltransferase/phosphatases have been characterized or the bifunctional activities confirmed. Nevertheless, identification and characterization of some bifunctional enzymes containing phosphatase activity have been reported (40–43), such as a soluble epoxide hydrolase with lipid phosphate phosphatase activity (41). Although the formation of sn-2 LPA had been observed in vitro in a glycerophosphate acyltransferase system of Brevibacterium (44) and a solubilized microsomal GPAT from spinach leaves (45), no specific GPAT enzyme with sn-2 specificity has previously been cloned and characterized.
Based on fossil records and other evidence, the evolution of cutin and suberin is associated with adaptations that occurred as marine plants adapted to life on land >400 million years ago. In support of this hypothesis, the GPAT family that is required for lipid polyester biosynthesis is a plant-specific lineage as evidenced by the fact that no homologs with >28% identity (BlastP E < 10−5) can be detected in animals or microorganisms (including algae). However, seven homologs (E < 10−100) occur in the genome of the primitive moss, Physcomitrella patens, and two homologs in the genome of the spikemoss, Selaginella moellendorffii, a nonvascular early ancestor of land plants. Most of these homologs have all of the conserved residues associated with phosphatase activity described above (SI Appendix, Fig. S11). The absence of homologs in algae, but their presence in the most primitive extant land plants supports a hypothesis that the bifunctional GPATs may have evolved specifically for biosynthesis of extracellular lipid structures that contribute to survival on land. A similar conclusion can be made based on occurrence of the CYP86 family of fatty acyl ω-oxidases that occur in P. patens and higher plants, but not in algae or other organisms (46).
Although knowledge of the biosynthesis of the monomers of cutin and suberin has advanced greatly in recent years, understanding how these become assembled into an insoluble extracellular polymer matrix outside the plasma membrane of plant cells remains a major challenge of plant biochemistry. This study has uncovered an unexpected bifunctional enzyme process that perhaps is an early step leading to the assembly of the polymer. Further exploration of how sn-2 MAG may participate in reactions inside or outside the cell should yield important insights into the acyl transfer reactions leading to the formation of plant glycerolipid polymers.
Methods
Materials.
Details of plasmid constructs for expression in yeast and wheat germ, synthesis of acyl-CoAs and MAGs, and their identification are described in SI Appendix, SI Methods. Unless stated otherwise, reagents were from Sigma-Aldrich. Borate TLC plates (Partisil K6 Silica gel 60 Å, 250 μm; Whatman) were prepared by dipping in 10% boric acid in methanol and activation at 110 °C, 30 min before use. [14C(U)]Glycerol-L-3-phosphate was from American Radiolabeled Chemicals. The haploid gene disruption strain YKR067w::kanMX4 (BY4742; Mat α; his3Δ1; leu2Δ0; lys2Δ0; ura3Δ0; YKR067w::kanMX4) was purchased from EUROSCARF.
Transformation and Expression of GPATs and Their Vector Controls in Yeast gat1Δ Strain.
Transformation of empty vector (pYES-DEST52 or pYES2/CT) and the recombinant plasmids into gat1Δ strain was performed according to Invitrogen manual. The induction of GPAT expression was as described by Zheng et al. (4) except that induction time was 21 h, after which cells were harvested by centrifugation (1,500 × g for 5 min). The resulting cell pellets were washed with 10 vol of ice-cold 20 mM Tris·HCl (pH 7.9) and resuspended in buffer [20 mM Tris·HCl (pH 7.9), 10 mM MgCl2, 1 mM EDTA, 5% (vol/vol) glycerol, 1 mM DTT, 0.3 M ammonium sulfate]. After adding 0.5-mm Silica beads (BioSpec Products), homogenates were prepared with a Retsch MM301 Ball Mill (frequency: 30/s, twice for 2.5 min). After centrifugation (3,000 × g for 5 min at 4 °C), the supernatant was further centrifuged at 30,000 × g for 15 min and pellets were resuspended in 50 mM Tris·HCl (pH 7.9) buffer containing 20% (vol/vol) glycerol and 1mM DTT, and stored at −80 °C for GPAT assay.
Wheat Germ Cell-Free Protein Translation.
Translation of GPAT5 and GPAT6 from construct GPAT5/pIVEX1.4WG and GPAT6/pIVEX1.4WG was achieved by using RTS 100 Wheat Germ CECF Kit (Roche), according to the manufacturer's instructions. Liposomes (10 mg/mL) were prepared from acetone-washed soy lethicin (L-α-phosphatidylcholine) and added at 60 μg per reaction. The reaction mixtures containing translated proteins were stored at −80 °C before direct use for GPAT assay.
GPAT Activity Assay.
G3P acyltransferase activity was assayed at room temperature for 10 min in a 30-μL reaction mixture containing 37.5 mM Tris·HCl (pH 7.5), 0.5 mM G3P (with 0.1 μCi [14C]G3P), 45 μM acyl-CoA, 2 mM MgCl2, 4 mM NaF, 1 mM DTT, and 0.1% BSA (fatty acid free) (47, 48). Yeast microsomes (20 μg) or wheat germ translation reaction mixture (2 μL) was used as the enzyme source. Reactions were stopped by 5 μL of acetonitrile:acetic acid (4:1), and the entire reaction mixture was loaded immediately onto a K6 TLC plate, developed with CHCl3:CH3OH:HAc:H2O (85:15:10:3.5). The gat1Δ strain containing empty vector pYES-DEST52 or pYES2-CT was used as GPAT5 and GPAT4/6 assay control, respectively. Radiolabeled products, LPA or MAG, were identified by comigration with C18:1-LPA and C18:1 MAGs. Quantification was done by autoradiography on a Packard Instant Imager.
Regiospecificity of DCA-MAGs.
To minimize isomerization due to acyl migration, the quenched GPAT assay mixture containing DCA-MAG products was directly analyzed by borate-TLC, developed with chloroform:acetone (1:1) (16). The borate TLC system will separate β (sn-2)-MAGs from α (sn-1 and sn-3)-MAGs. Because the substrate is sn-glycerol-3-phosphate it is assumed that there is no sn-3 MAG production. DCA-MAGs were identified by comigration with synthesized standards of C18:1 DCA sn-1 and sn-2 MAGs. To confirm the structures of DCA sn-2 MAGs, large-scale GPAT4 and GPAT6 assays were performed. Details of product recovery and identification are described for MAGs in SI Appendix, SI Methods.
Regiospecificity of DCA-LPA.
Immediately after GPAT assay, 0.1 M borate buffer (pH 7.5, 1 mL) and 0.1 M Tris·HCl (pH 7.5, 1 mL) were added. Dephosphorylation (17) was started by adding 1 μL of Escherichia coli alkaline phosphatase (0.25 unit) and continued for 30 min at room temperature. Products were extracted by diethyl ether, and MAGs were separated by borate-TLC. The identification of DCA-MAGs was as described above.
Prediction of N- and C-Terminal Structures of GPATs.
GPAT sequences were submitted to a variety of sequence alignment and fold recognition services through the bioinfo.pl meta server (49). The best template across all GPATs, phosphoserine phosphatase from Methanococcus jannaschii (Protein Data Bank entry 1F5S) from the FUGUE server (50), was then used to model the N-terminal structure for GPAT5. N-terminal structures for the other GPATs were then constructed by using the GPAT5 model as a template. Template-based modeling, including the prediction of missing loops, was performed with Modeler (version 8) (51) in conjunction with the MMTSB Tool Set (52). The C-terminal domain was modeled in a similar fashion using squash glycerol-3-phosphate (1)-acyltransferase (Protein Data Bank entry 1K30) as the template.
Site-Directed Mutagenesis of GPAT6.
GPAT6 single mutants (D29E, K178L, and D200K) were synthesized by Geneart and inserted into wheat germ His6-tag vector pIVEX 1.4 WG with NcoI/PstI.
Supplementary Material
Acknowledgments
We thank Jilian Fan for construction of GPAT5 and GPAT6 expression vectors and wheat germ cell-free protein translation and the Michigan State University Research Technology Support Facility and Mass Spectrometry Facility. This work was supported by US Department of Agriculture Grant 2005-35318-15419, National Science Foundation Grant MCB-0615563, and Bayer CropScience.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.0914149107/-/DCSupplemental.
References
- 1.Wendel AA, Lewin TM, Coleman RA. Glycerol-3-phosphate acyltransferases: Rate limiting enzymes of triacylglycerol biosynthesis. Biochim Biophys Acta. 2009;1791:501–506. doi: 10.1016/j.bbalip.2008.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang YM, Rock CO. Acyltransferases in bacterial glycerophospholipid synthesis. J Lipid Res. 2008;49:1867–1874. doi: 10.1194/jlr.R800005-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gimeno RE, Cao J. Mammalian glycerol-3-phosphate acyltransferases: New genes for an old activity. J Lipid Res. 2008;49:2079–2088. doi: 10.1194/jlr.R800013-JLR200. [DOI] [PubMed] [Google Scholar]
- 4.Zheng Z, Zou J. The initial step of the glycerolipid pathway: Identification of glycerol 3-phosphate/dihydroxyacetone phosphate dual substrate acyltransferases in Saccharomyces cerevisiae. J Biol Chem. 2001;276:41710–41716. doi: 10.1074/jbc.M104749200. [DOI] [PubMed] [Google Scholar]
- 5.Murata N, Tasaka Y. Glycerol-3-phosphate acyltransferase in plants. Biochim Biophys Acta. 1997;1348:10–16. doi: 10.1016/s0005-2760(97)00115-x. [DOI] [PubMed] [Google Scholar]
- 6.Zheng Z, et al. Arabidopsis AtGPAT1, a member of the membrane-bound glycerol-3-phosphate acyltransferase gene family, is essential for tapetum differentiation and male fertility. Plant Cell. 2003;15:1872–1887. doi: 10.1105/tpc.012427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Graça J, Santos S. Suberin: A biopolyester of plants’ skin. Macromol Biosci. 2007;7:128–135. doi: 10.1002/mabi.200600218. [DOI] [PubMed] [Google Scholar]
- 8.Franke R, Schreiber L. Suberin—A biopolyester forming apoplastic plant interfaces. Curr Opin Plant Biol. 2007;10:252–259. doi: 10.1016/j.pbi.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 9.Simonich SL, Hites RA. Vegetation-atmosphere partitioning of polycyclic aromatic-hydrocarbons. Environ Sci Technol. 1994;28:939–943. doi: 10.1021/es00054a028. [DOI] [PubMed] [Google Scholar]
- 10.Beisson F, Li Y, Bonaventure G, Pollard M, Ohlrogge JB. The acyltransferase GPAT5 is required for the synthesis of suberin in seed coat and root of Arabidopsis. Plant Cell. 2007;19:351–368. doi: 10.1105/tpc.106.048033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Y, et al. Identification of acyltransferases required for cutin biosynthesis and production of cutin with suberin-like monomers. Proc Natl Acad Sci USA. 2007;104:18339–18344. doi: 10.1073/pnas.0706984104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li-Beisson Y, et al. Nanoridges that characterize the surface morphology of flowers require the synthesis of cutin polyester. Proc Natl Acad Sci USA. 2009;106:22008–22013. doi: 10.1073/pnas.0909090106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pollard M, Beisson F, Li Y, Ohlrogge JB. Building lipid barriers: Biosynthesis of cutin and suberin. Trends Plant Sci. 2008;13:236–246. doi: 10.1016/j.tplants.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 14.Bernards MA. Demystifying suberin. Can J Bot. 2002;80:227–240. [Google Scholar]
- 15.Kawaguchi A, Yoshimura T, Okuda S. A new method for the preparation of acyl-CoA thioesters. J Biochem. 1981;89:337–339. doi: 10.1093/oxfordjournals.jbchem.a133207. [DOI] [PubMed] [Google Scholar]
- 16.Thomas AE, Scharoun JE, Ralston H. Quantitative estimation of isomeric monoglycerides by thin-layer chromatography. J Am Oil Chem Soc. 1965;42:789–792. [Google Scholar]
- 17.Bertrams M, Heinz E. Positional specificity and fatty acid selectivity of purified sn-glycerol 3-phosphate acyltransferases from chloroplasts. Plant Physiol. 1981;68:653–657. doi: 10.1104/pp.68.3.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Collet JF, Stroobant V, Pirard M, Delpierre G, Van Schaftingen E. A new class of phosphotransferases phosphorylated on an aspartate residue in an amino-terminal DXDX(T/V) motif. J Biol Chem. 1998;273:14107–14112. doi: 10.1074/jbc.273.23.14107. [DOI] [PubMed] [Google Scholar]
- 19.Collet JF, Stroobant V, Van Schaftingen E. Mechanistic studies of phosphoserine phosphatase, an enzyme related to P-type ATPases. J Biol Chem. 1999;274:33985–33990. doi: 10.1074/jbc.274.48.33985. [DOI] [PubMed] [Google Scholar]
- 20.Wang WR, Kim R, Jancarik J, Yokota H, Kim SH. Crystal structure of phosphoserine phosphatase from Methanococcus jannaschii, a hyperthermophile, at 1.8 A resolution. Structure. 2001;9:65–71. doi: 10.1016/s0969-2126(00)00558-x. [DOI] [PubMed] [Google Scholar]
- 21.Lewin TM, Wang P, Coleman RA. Analysis of amino acid motifs diagnostic for the sn-glycerol-3-phosphate acyltransferase reaction. Biochemistry. 1999;38:5764–5771. doi: 10.1021/bi982805d. [DOI] [PubMed] [Google Scholar]
- 22.Xiao FM, et al. Arabidopsis CYP86A2 represses Pseudomonas syringae type III genes and is required for cuticle development. EMBO J. 2004;23:2903–2913. doi: 10.1038/sj.emboj.7600290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Höfer R, et al. The Arabidopsis cytochrome P450 CYP86A1 encodes a fatty acid ω-hydroxylase involved in suberin monomer biosynthesis. J Exp Bot. 2008;59:2347–2360. doi: 10.1093/jxb/ern101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Compagnon V, et al. CYP86B1 is required for very long chain ω-hydroxyacid and α, ω -dicarboxylic acid synthesis in root and seed suberin polyester. Plant Physiol. 2009;150:1831–1843. doi: 10.1104/pp.109.141408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Molina I, Li-Beisson Y, Beisson F, Ohlrogge JB, Pollard M. Identification of an Arabidopsis feruloyl-coenzyme A transferase required for suberin synthesis. Plant Physiol. 2009;151:1317–1328. doi: 10.1104/pp.109.144907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schnurr J, Shockey J, Browse J. The acyl-CoA synthetase encoded by LACS2 is essential for normal cuticle development in Arabidopsis. Plant Cell. 2004;16:629–642. doi: 10.1105/tpc.017608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bessire M, et al. A permeable cuticle in Arabidopsis leads to a strong resistance to Botrytis cinerea. EMBO J. 2007;26:2158–2168. doi: 10.1038/sj.emboj.7601658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weng H, Molina I, Shockey J, Browse J. Organ fusion and defective cuticle function in a lacs1 lacs2 double mutant of Arabidopsis. Planta. 2010;231:1089–1100. doi: 10.1007/s00425-010-1110-4. [DOI] [PubMed] [Google Scholar]
- 29.Bird D, et al. Characterization of Arabidopsis ABCG11/WBC11, an ATP binding cassette (ABC) transporter that is required for cuticular lipid secretion. Plant J. 2007;52:485–498. doi: 10.1111/j.1365-313X.2007.03252.x. [DOI] [PubMed] [Google Scholar]
- 30.Krolikowski KA, Victor JL, Wagler TN, Lolle SJ, Pruitt RE. Isolation and characterization of the Arabidopsis organ fusion gene HOTHEAD. Plant J. 2003;35:501–511. doi: 10.1046/j.1365-313x.2003.01824.x. [DOI] [PubMed] [Google Scholar]
- 31.Kurdyukov S, et al. Genetic and biochemical evidence for involvement of HOTHEAD in the biosynthesis of long-chain alpha-, omega-dicarboxylic fatty acids and formation of extracellular matrix. Planta. 2006;224:315–329. doi: 10.1007/s00425-005-0215-7. [DOI] [PubMed] [Google Scholar]
- 32.Tanaka T, Tanaka H, Machida C, Watanabe M, Machida Y. A new method for rapid visualization of defects in leaf cuticle reveals five intrinsic patterns of surface defects in Arabidopsis. Plant J. 2004;37:139–146. doi: 10.1046/j.1365-313x.2003.01946.x. [DOI] [PubMed] [Google Scholar]
- 33.Kurdyukov S, et al. The epidermis-specific extracellular BODYGUARD controls cuticle development and morphogenesis in Arabidopsis. Plant Cell. 2006;18:321–339. doi: 10.1105/tpc.105.036079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chassot C, Nawrath C, Métraux JP. Cuticular defects lead to full immunity to a major plant pathogen. Plant J. 2007;49:972–980. doi: 10.1111/j.1365-313X.2006.03017.x. [DOI] [PubMed] [Google Scholar]
- 35.Tang DZ, Simonich MT, Innes RW. Mutations in LACS2, a long-chain acyl-coenzyme A synthetase, enhance susceptibility to avirulent Pseudomonas syringae but confer resistance to Botrytis cinerea in Arabidopsis. Plant Physiol. 2007;144:1093–1103. doi: 10.1104/pp.106.094318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Voisin D, et al. Dissection of the complex phenotype in cuticular mutants of Arabidopsis reveals a role of SERRATE as a mediator. PLoS Genet. 2009;5:e1000703. doi: 10.1371/journal.pgen.1000703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Molina I, Ohlrogge JB, Pollard M. Deposition and localization of lipid polyester in developing seeds of Brassica napus and Arabidopsis thaliana. Plant J. 2008;53:437–449. doi: 10.1111/j.1365-313X.2007.03348.x. [DOI] [PubMed] [Google Scholar]
- 38.Gidda SK, Shockey JM, Rothstein SJ, Dyer JM, Mullen RT. Arabidopsis thaliana GPAT8 and GPAT9 are localized to the ER and possess distinct ER retrieval signals: Functional divergence of the dilysine ER retrieval motif in plant cells. Plant Physiol Biochem. 2009;47:867–879. doi: 10.1016/j.plaphy.2009.05.008. [DOI] [PubMed] [Google Scholar]
- 39.Li Y, Beisson F, Ohlrogge JB, Pollard M. Monoacylglycerols are components of root waxes and can be produced in the aerial cuticle by ectopic expression of a suberin-associated acyltransferase. Plant Physiol. 2007;144:1267–1277. doi: 10.1104/pp.107.099432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hasemann CA, Istvan ES, Uyeda K, Deisenhofer J. The crystal structure of the bifunctional enzyme 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase reveals distinct domain homologies. Structure. 1996;4:1017–1029. doi: 10.1016/s0969-2126(96)00109-8. [DOI] [PubMed] [Google Scholar]
- 41.Newman JW, Morisseau C, Harris TR, Hammock BD. The soluble epoxide hydrolase encoded by EPXH2 is a bifunctional enzyme with novel lipid phosphate phosphatase activity. Proc Natl Acad Sci USA. 2003;100:1558–1563. doi: 10.1073/pnas.0437724100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou Y, Bhattacharjee H, Mukhopadhyay R. Bifunctional role of the leishmanial antimonate reductase LmACR2 as a protein tyrosine phosphatase. Mol Biochem Parasitol. 2006;148:161–168. doi: 10.1016/j.molbiopara.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 43.Mukhopadhyay R, Bisacchi D, Zhou Y, Armirotti A, Bordo D. Structural characterization of the As/Sb reductase LmACR2 from Leishmania major. J Mol Biol. 2009;386:1229–1239. doi: 10.1016/j.jmb.2008.07.056. [DOI] [PubMed] [Google Scholar]
- 44.Oh-Hashi Y, et al. Enzymatic bases for the fatty acid positioning in phospholipids of Brevibacterium ammoniagenes. Arch Biochem Biophys. 1986;244:413–420. doi: 10.1016/0003-9861(86)90608-9. [DOI] [PubMed] [Google Scholar]
- 45.Frentzen M. Comparison of certain properties of membrane-bound and solubilized acyltransferase activities of plant microsomes. Plant Sci. 1990;69:39–48. [Google Scholar]
- 46.Rensing SA, et al. The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science. 2008;319:64–69. doi: 10.1126/science.1150646. [DOI] [PubMed] [Google Scholar]
- 47.Ichihara K. sn-Glycerol-3-phosphate acyltransferase in a particulate fraction from maturing safflower seeds. Arch Biochem Biophys. 1984;232:685–698. doi: 10.1016/0003-9861(84)90589-7. [DOI] [PubMed] [Google Scholar]
- 48.Athenstaedt K, Weys S, Paltauf F, Daum G. Redundant systems of phosphatidic acid biosynthesis via acylation of glycerol-3-phosphate or dihydroxyacetone phosphate in the yeast Saccharomyces cerevisiae. J Bacteriol. 1999;181:1458–1463. doi: 10.1128/jb.181.5.1458-1463.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ginalski K, Elofsson A, Fischer D, Rychlewski L. 3D-Jury: A simple approach to improve protein structure predictions. Bioinformatics. 2003;19:1015–1018. doi: 10.1093/bioinformatics/btg124. [DOI] [PubMed] [Google Scholar]
- 50.Shi JY, Blundell TL, Mizuguchi K. FUGUE: Sequence-structure homology recognition using environment-specific substitution tables and structure-dependent gap penalties. J Mol Biol. 2001;310:243–257. doi: 10.1006/jmbi.2001.4762. [DOI] [PubMed] [Google Scholar]
- 51.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 52.Feig M, Karanicolas J, Brooks CL., 3rd MMTSB Tool Set: Enhanced sampling and multiscale modeling methods for applications in structural biology. J Mol Graph Model. 2004;22:377–395. doi: 10.1016/j.jmgm.2003.12.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.