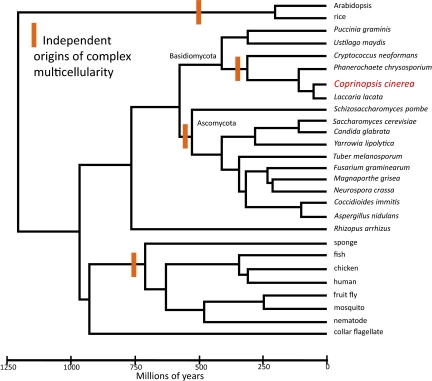
The PNAS paper by Stajich et al. (1) describes a superbly assembled and annotated genome of one of the most morphologically complex fungi, the “inky cap” mushroom Coprinopsis cinerea, and illustrates the accomplishments that can be made when working with a genetic model organism and the challenges that lie ahead if we are to truly understand how genomes relate to the evolution of organism complexity (Fig. 1).
Fig. 1.
Bayesian relaxed clock tree based on amino acid sequence for 50 loci and linked to the fossil record at four divergences (20). Current hypotheses on the origin of complex, multicellular organisms are marked in orange. Comparison of Copriniopsis with Basidiomycota macrofungi Laccaria or Phanerochaete, or with Ascomycota macrofungus Tuber, could reveal commonalities associated with complex multicellularity. If microfungi that are complex and multicellular share these commonalities, useful comparisons could include Ascomycota on the clade that contains Neurospora and Aspergillus [Reproduced with permission from ref. 20 (Copyright 2010, Elsevier)].
Plants and animals may be the first to come to mind when thoughts turn to the evolution of complexity, but within the fungi, there is an amazing amount of morphological diversity, from unicellular yeasts to large “fruiting bodies” capable of producing trillions of spores (2). That diversity, and the relatively small genomes of this group, have helped create the most comprehensive set of whole-genome sequences for any major Eukaryotic lineage in both the number and phylogenetic breadth of taxon sampling. Furthermore, the high incidence of convergent evolution of similar forms creates an ideal system for using phylogenomic comparative methods to study the evolution of development within the fungi. However, given the wealth of fungal genomes, fungal evo-devo is in its infancy, because most of the fungi with well-assembled and annotated genomes are unicellular (3). Enter C. cinerea, which develops into a macroscopic mushroom and is well-characterized genetically.
Stajich et al. (1) assembled the 36-Mb genome sequence of C. cinerea into 13 chromosomes, 9 of which include both telomeres, and identified centromeres by their high concentration of retrotransposons. Annotation was improved by access to expressed sequence tags (EST) and serial analysis of gene expression (SAGE) data. Comparison of the physical chromosome to a genetic map that covered 86% of the genome allowed the authors to observe that recombination varies along the chromosomes and that the highest rates are achieved within their distal 15%. In regions of low recombination, retrotransposons, which outnumber DNA transposons 10:1, are missing. Genes unique to C. cinerea, as well as single-copy genes, are more abundant, whereas genes with paralogs are rare. In chromosomal regions where recombination is high, multigene families are younger and tend to be tandemly oriented compared with areas where recombination is low. Synteny between C. cinerea and another mushroom-producing fungus, Laccaria lacata, with which C. cinerea shared an ancestor some 200 Ma, is recognizable over 40% of the genome. The estimate of the rate of genome rearrangement between these fungi is 3-fold higher than that seen in Saccharomyces, and is high, in general, for Eukaryotes. Stajich et al. (1) also discovered several blocks of synteny that are much larger than expected if rearrangements occur randomly, suggesting that gene order in these regions is being conserved by purifying selection. In these large blocks, genes are unusually closely packed; genes with core, eukaryotic functions are common; the density of transcription factors is elevated; and transposons are absent. In almost all of these attributes, C. cinerea is like other eukaryotes where well-assembled and annotated genomes have been analyzed with access to good genetic data, e.g., worms (4) and other fungi, such as yeast (5), Candida (6), and fission yeast (7). The picture that has emerged is of a highly dynamic genome where chromosomal regions frequently duplicate, and those rare duplications that facilitate adaptation are retained (8), with a bias toward subtelomeric regions. It is a generality that can be extended to the bacteria, where genomes can be sorted into stable regions, with genes coding for core activities, and dynamic regions, where selfish elements and genes responsible for local adaptation are more likely to be found (9, 10).
A product of the dynamic genome, which has been implicated as a means of rapid adaptation, is change in gene family size (11), and Stajich et al. (1) have identified three examples: genes with protein kinase domains, p450 genes, and hydrophobins (proteins that facilitate development in an environment where surface tension is the dominant force) (12). In C. cinerea, there are 380 genes with protein kinase domains, including many not found in yeast and some previously found only in animals. The largest such family contains the FunK1 domain, which is found in complex Basidiomycota and Ascomycota and is absent in other fungi. Almost one half, 59, of the proteins with this domain are packed into one subtelomeric region of one chromosome. A reasonable hypothesis is that FunK1 domains are important to the development of complex, multicellular fungi and that selection has been involved in retaining these genes over numerous, tandem gene duplications. Other similar and equally reasonable hypotheses could be framed for the P450 and hydrophobin genes. However, despite significant advances related to the mechanisms of genome evolution that emerged from this analysis of the C. cinerea genome, the goal of understanding, with confidence, the genomic basis of developmental complexity remains elusive.
What, then, will it take to understand the genome evolution that led to mushrooms and other complex, fungal fruiting bodies? One tack, surely, is to use the molecular biological tools available to Coprinopsis researchers to test hypotheses about FunK1 kinases, p450 genes, and hydrophobins. Another tack is to generate more hypotheses, and can there be a better hypothesis generator than comparative genomics? Which comparisons might be most informative? If mushroom producers
Within the fungi, there is an amazing amount of morphological diversity.
have common genome features, comparison of C. cinerea to other macroscopic Basidiomycota, e.g., Phanerochaete (13) or Laccaria (14), would be useful. However, each of these fungi has a different approach to nutrition (living and reproducing in dung, wood, or the roots of living plants, respectively), and this diversity could confound comparisons aimed only at the development of morphological complexity. In this regard, comparison of C. cinerea with other dung fungi that are complex and multicellular, e.g., the complex dung fungus Ascobolus (Ascomycota) and the simple dung fungus Phycomyces (Mucoromycota), might control for nutritional adaptation. Similarly, comparison of Laccaria lacata with another mycorrhizal fungus, one that makes the complex reproductive structures known as truffles, Tuber (Ascomycota) (15), might separate multicellular development from nutritional mode, as might comparison of macroscopic Basidiomycota and Ascomycota, which cause plant disease (e.g., Armillaria and Sclerotinia).
Between fungi that produce large reproductive structures (e.g., mushrooms and truffles) and those that make no reproductive structures apart from the meiocytes themselves (e.g., yeasts) lie most species of the higher fungi [e.g., Neurospora (16), Aspergillus (17), Coccidioides (18), Fusarium, and the lichenized fungi]. If size does not matter, and the developmental complexity and tissue differentiation seen in these microscopic fungi reflect that seen in mushrooms and truffles, comparison of genetically tractable species with C. cinerea and relatives should also pay dividends. A pair of membranous organelles worth investigating might be the Woronin body of Ascomycota and the septal pore complex of Basidiomycota, which function to seal damaged hyphae and might have been instrumental in allowing fungi to make complex, multicellular structures (19). Making the comparisons suggested herein, most of which can be attempted with existing genome assemblies, might not only unravel the basis of fungal complexity, but point the way for studies of plants and animals, as more of the much larger genomes found in those kingdoms become available.
Footnotes
The authors declare no conflict of interest.
See companion article on page 11889.
References
- 1.Stajich JE, et al. Insights into evolution of multicellular fungi from the assembled chromosomes of the mushroom. Coprinopsis cinerea (Coprinus cinereus). Proc Natl Acad Sci USA. 2010;107:11889–11894. doi: 10.1073/pnas.1003391107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stajich JE, et al. The fungi. Curr Biol. 2009;19:R840–R845. doi: 10.1016/j.cub.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liti G, Louis EJ. Yeast evolution and comparative genomics. Annu Rev Microbiol. 2005;59:135–153. doi: 10.1146/annurev.micro.59.030804.121400. [DOI] [PubMed] [Google Scholar]
- 4.Duret L, Marais G, Biémont C. Transposons but not retrotransposons are located preferentially in regions of high recombination rate in Caenorhabditis elegans. Genetics. 2000;156:1661–1669. doi: 10.1093/genetics/156.4.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barton AB, Pekosz MR, Kurvathi RS, Kaback DB. Meiotic recombination at the ends of chromosomes in Saccharomyces cerevisiae. Genetics. 2008;179:1221–1235. doi: 10.1534/genetics.107.083493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Butler G, et al. Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature. 2009;459:657–662. doi: 10.1038/nature08064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khair L, Subramanian L, Moser BA, Nakamura TM. Roles of heterochromatin and telomere proteins in regulation of fission yeast telomere recombination and telomerase recruitment. J Biol Chem. 2010;285:5327–5337. doi: 10.1074/jbc.M109.078840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koszul R, Caburet S, Dujon B, Fischer G. Eucaryotic genome evolution through the spontaneous duplication of large chromosomal segments. EMBO J. 2004;23:234–243. doi: 10.1038/sj.emboj.7600024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bentley SD, Parkhill J. Comparative genomic structure of prokaryotes. Annu Rev Genet. 2004;38:771–792. doi: 10.1146/annurev.genet.38.072902.094318. [DOI] [PubMed] [Google Scholar]
- 10.Treangen TJ, Abraham AL, Touchon M, Rocha EPC. Genesis, effects and fates of repeats in prokaryotic genomes. FEMS Microbiol Rev. 2009;33:539–571. doi: 10.1111/j.1574-6976.2009.00169.x. [DOI] [PubMed] [Google Scholar]
- 11.Koonin EV. Darwinian evolution in the light of genomics. Nucleic Acids Res. 2009;37:1011–1034. doi: 10.1093/nar/gkp089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wösten HAB. Hydrophobins: Multipurpose proteins. Annu Rev Microbiol. 2001;55:625–646. doi: 10.1146/annurev.micro.55.1.625. [DOI] [PubMed] [Google Scholar]
- 13.Martinez D, et al. Genome sequence of the lignocellulose degrading fungus Phanerochaete chrysosporium strain RP78. Nat Biotechnol. 2004;22:695–700. doi: 10.1038/nbt967. [DOI] [PubMed] [Google Scholar]
- 14.Martin F, et al. The genome of Laccaria bicolor provides insights into mycorrhizal symbiosis. Nature. 2008;452:88–92. doi: 10.1038/nature06556. [DOI] [PubMed] [Google Scholar]
- 15.Martin F, et al. Périgord black truffle genome uncovers evolutionary origins and mechanisms of symbiosis. Nature. 2010;464:1033–1038. doi: 10.1038/nature08867. [DOI] [PubMed] [Google Scholar]
- 16.Galagan JE, et al. The genome sequence of the filamentous fungus Neurospora crassa. Nature. 2003;422:859–868. doi: 10.1038/nature01554. [DOI] [PubMed] [Google Scholar]
- 17.Galagan JE, et al. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature. 2005;438:1105–1115. doi: 10.1038/nature04341. [DOI] [PubMed] [Google Scholar]
- 18.Sharpton TJ, et al. Comparative genomic analyses of the human fungal pathogens Coccidioides and their relatives. Genome Res. 2009;19:1722–1731. doi: 10.1101/gr.087551.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ng SK, Liu FF, Lai JL, Low W, Jedd G. A tether for Woronin body inheritance is associated with evolutionary variation in organelle positioning. PLoS Genet. 2009;5:e1000521. doi: 10.1371/journal.pgen.1000521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berbee ML, Taylor JW. Dating the molecular clock in fungi—how close are we? Fungal Biol Rev. 2010 10.1016/j.fbr.2010.03.001. [Google Scholar]