Abstract
The mushroom Coprinopsis cinerea is a classic experimental model for multicellular development in fungi because it grows on defined media, completes its life cycle in 2 weeks, produces some 108 synchronized meiocytes, and can be manipulated at all stages in development by mutation and transformation. The 37-megabase genome of C. cinerea was sequenced and assembled into 13 chromosomes. Meiotic recombination rates vary greatly along the chromosomes, and retrotransposons are absent in large regions of the genome with low levels of meiotic recombination. Single-copy genes with identifiable orthologs in other basidiomycetes are predominant in low-recombination regions of the chromosome. In contrast, paralogous multicopy genes are found in the highly recombining regions, including a large family of protein kinases (FunK1) unique to multicellular fungi. Analyses of P450 and hydrophobin gene families confirmed that local gene duplications drive the expansions of paralogous copies and the expansions occur in independent lineages of Agaricomycotina fungi. Gene-expression patterns from microarrays were used to dissect the transcriptional program of dikaryon formation (mating). Several members of the FunK1 kinase family are differentially regulated during sexual morphogenesis, and coordinate regulation of adjacent duplications is rare. The genomes of C. cinerea and Laccaria bicolor, a symbiotic basidiomycete, share extensive regions of synteny. The largest syntenic blocks occur in regions with low meiotic recombination rates, no transposable elements, and tight gene spacing, where orthologous single-copy genes are overrepresented. The chromosome assembly of C. cinerea is an essential resource in understanding the evolution of multicellularity in the fungi.
Keywords: basidiomycete, dikaryon formation, gene families, kinase, meiotic recombination
Although the number of fungal genome-sequencing projects has increased dramatically over the last few years, there is a surprising lack of complete chromosome assemblies from these projects. Sequencing and analysis of chromosome assemblies from ascomycete fungi have revealed blocks of shared ancestral gene order (1, 2), the consequences and sorting of gene pairs after whole-genome duplication (3), and the preferential location of large gene families near chromosome ends (4, 5). Comparable complete-genome analyses in basidiomycete fungi are essential complements to whole-genome shotgun (WGS) assemblies to increase the utility of the sequence information and to learn if unique features of genome organization are associated with the increased complexity of multicellular fungi.
Coprinus cinereus (now known as Coprinopsis cinerea) (6) was chosen by the Fungal Genome Initiative (http://www.broadinstitute.org/science/projects/fungal-genome-initiative) as a key species in a cohesive genome-sequencing strategy designed to increase our understanding of the biology, evolution, and biomedical implications of the entire fungal kingdom. Studies on this fungus using tools such as DNA-mediated transformation (7) and RNAi silencing (8) have provided important insights into the regulation of multicellular development, mushroom fruiting, mating pheromone, and receptor signaling pathways (9–11). Haploid homokaryotic hyphae of basidiomycete fungi fuse to form dikaryons if different alleles are present at both A and B mating-type loci. Dikaryons form mushroom fruiting bodies under appropriate light input, temperature, and CO2 levels. The major controlling elements of the mating cascade have been uncovered with genetic tools, but the full complement of genes that respond during these conditions still is not well understood.
Studies using C. cinerea also have provided important insights into meiotic chromosome behavior because of several experimental advantages. Meiosis occurs with a high degree of synchrony, and meiotic mutants have been obtained and analyzed using a variety of cytological and molecular tools. Pioneering studies exploited the relatively small nuclear volume of C. cinerea meiocytes and used serial sectioning and three-dimensional reconstruction to describe the karyotype, the sites of synaptic initiation, the dynamics of recombination nodule assembly, and chiasmata formation (12). More recently, meiotic mutants have been obtained and analyzed using a variety of cytological and molecular tools including indirect immunofluorescence (13).
We report here the complete genome sequence and chromosome assembly of C. cinerea (SI Text). The genome was assembled into 13 chromosomes of 36 megabases (Mb) (AACS02000000; Dataset S1, Tables S1 and S2) consistent with cytological (12) and genetic (14) evidence and supported by BAC fingerprinting (Dataset S1, Table S1). We used the assembly to investigate rates of meiotic recombination along the chromosomes and the distribution, expression, and evolutionary conservation of single-copy genes and gene families in this model basidiomycete.
Results
High-Resolution Genetic Map.
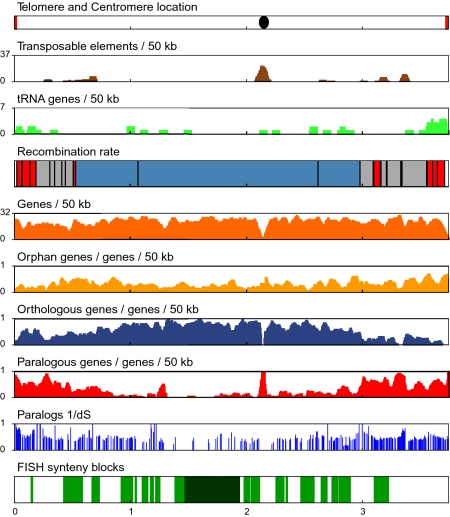
Cytological evidence has indicated that meiotic exchanges are highly enriched in subtelomeric regions of the 13 chromosomes in C. cinerea (12), suggesting that recombination rates might be non-uniform across the genome. To examine crossover distribution, we used 133 simple sequence repeat (SSR) markers evenly distributed across the genome and four additional markers to construct a genetic map (Fig. 1, SI Text, Fig. S1, and Dataset S1, Table S3). Examination of the marker genotypes of the progeny revealed regions of average, high, and low recombination. The total genetic map length of the 31-Mb physical genome that could be mapped is 948 centimorgans (cM), indicating an average frequency of exchange of 33 kb/cM. However, the “hot” regions (8% of the genome) exhibit an elevated rate of recombination (6 kb/cM on average), whereas the “cold” regions (44% of the genome) exhibit very little recombination (198 kb/cM).
Fig. 1.
Summary plots of chromosome II of C. cinerea. The plot shows the location of (Top panel) telomeres in red and centromere as black oval; (Second panel) the density of transposable elements (brown); (Third panel) tRNA genes (light green); (Fourth panel) recombination rates (the position of the SSR markers is indicated by vertical black bars, white is unmapped, red is high recombination, gray is average recombination, blue is low recombination); (Fifth panel) the density of all genes (orange); (Sixth panel), the density of orphan genes (light orange); (Seventh panel), the density of orthologous genes (blue); (Eighth panel) the density of paralogous genes (red); (Ninth panel) similarity of paralog families represented as 1/dS; (10th panel) syntenic regions (all regions of synteny between C. cinerea and L. bicolor are indicated in green, and blocks with >15 anchors are indicated in dark green). Vertical scales are defined for each bar in the bar title. Horizontal scale is Mb.
The hot regions are located predominantly in subtelomeric regions (16/18 are within the 15% of the nearest telomere; Fisher's exact test, P = 0.0002) (Fig. S1). Although the frequency of chiasmata (cross-overs) per bivalent ranges from 1 to 12 in both fungi and other organisms (reviewed in ref. 15), species with both low and high levels of exchange exhibit an elevated rate of recombination in subtelomeric regions (16–18). This elevation may reflect associations between the initiation of chromosome synapsis at subterminal regions and chiasma formation in these species (15, 19).
Retrotransposon Distribution.
Transposon sequences (2.5% of genomic sequence) are not distributed uniformly across the genome (Fig. 1, Fig. S1, and Dataset S1, Tables S4a and S4b). Each chromosome contains a distinctive internal transposon cluster whose position is highly correlated (R2 = 0.89) with the cytological centromere (12) on the nine chromosomes that extend telomere to telomere (Fig. S1). These transposon clusters (20% of all transposon-related sequences) could represent sequence-independent centromeres that are common in other fungi such as Neurospora crassa and Cryptococcus neoformans (20, 21). Many of these transposon clusters lie within regions that are cold for meiotic recombination. With the exception of the transposon clusters, the cold regions lack retrotransposon-related sequences. They contain only 3 of the 44 full-length retrotransposons (χ2= 23.7, P < 0.001), and all of the cold regions contain extensive stretches (over 1 Mb on the larger chromosomes) that lack any retrotransposon-related sequences (Fig. S1). The complexity of factors influencing the genome-wide distribution of transposons has been noted (22, 23), but the pattern we see in C. cinerea with an exclusion of retrotransposons from most but not all regions of low recombination has not been reported previously. Retrotransposons outnumber DNA transposons by a factor of 10 in C. cinerea (SI Text). Retrotransposon-related sequences are found either in the presumed centromere clusters or in regions of average or high recombination, whereas the distribution of DNA transposons is more uniform.
Ortholog and Paralog Distribution.
To predict 13,342 protein-coding genes, 267 tRNA genes, and 10 snRNA genes (Dataset S1, Table S5), we used computational tools in combination with evidence that included proteins from related species and 5,612 ESTs (SI Text). Gene calls were confirmed further by 5′ serial analysis of gene expression (SAGE) from two tissue types (5,130 gene models; Dataset S1, Table S6). Comparative analyses of the available basidiomycete genomes reveals a dramatic increase in gene number from less than 7,000 genes in C. neoformans and Ustilago maydis to more than 13,000 in the sequenced Agaricomycete fungi including C. cinerea (Dataset S1, Table S7). To understand better the mechanism of the observed gene increase and to ask if expansions of gene families are equally likely at different chromosomal locations, we constructed gene families based on sequence similarity using TribeMCL (24) (SI Text and Dataset S1, Table S8). We determined which C. cinerea genes are orphan (no homologs), orthologous (single-copy in C. cinerea and at least one Laccaria bicolor ortholog), or paralogous (multicopy in C. cinerea). We plotted the distribution of these three categories along the chromosomes (Fig. 1 and Fig. S1). It is striking that, whereas the distribution of orphan genes is relatively uniform, orthologous single-copy genes are overrepresented in regions with low rates of meiotic recombination, and paralogous multicopy genes are found primarily in regions with average or high rates of meiotic recombination (Table 1). We conclude that several factors, including high recombination rates, tolerance of transposable elements, telomere proximity, and chromatin structure, potentially could contribute positively to the creation and maintenance of duplicated paralogous genes in the discrete regions that we observe in C. cinerea.
Table 1.
Non-uniform gene distribution in chromosome regions with different rates of meiotic recombination
| Chromosomal region* |
KS comparison, P value |
||||
| Gene class | Hot | Average | Cold | Hot − cold | (Hot + average) − cold |
| Orphans | 5.5 | 5.5 | 5.0 | 0.6512 | 0.175 |
| Orthologs | 5.6 | 7.1 | 11.4 | 1.32 E-12 | 2.20 E-16 |
| Paralogs | 7.7 | 6.2 | 3.9 | 3.08 E-9 | 2.55 E-12 |
KS, Kolmogorov-Smirnov.
*Values reported are the mean number of features/50 kb in each type of chromosome region as defined by recombination rate categories
Paralogous Gene Families.
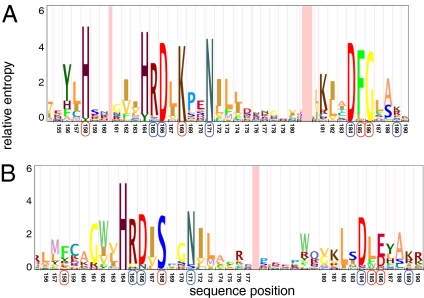
Inspection of Dataset S1, Table S8 reveals that the largest paralogous expansion involves genes encoding proteins with protein kinase domains. Because kinases in many organisms mediate sophisticated control mechanisms essential for complex structure and developmental patterns, we focused on this family in more detail. BLAST and hidden Markov model (HMM) screening of the C. cinerea transcriptome indicates the presence of 380 kinases (SI Text and Dataset S1, Tables S9a and S9b), including 12 classes not present in Saccharomyces cerevisiae, three of which have not been previously observed outside the Metazoa (Dataset S1, Table S9c). The largest family, with 133 members, FunK1, has unusual modifications in conserved kinase motifs, expanding the documented diversity of this important catalytic domain (Fig. 2). Catalytic residues D166, N171, and D184 are conserved [1ATP.pdb numbering (25)], suggesting that FunK1 family members use a catalytic mechanism similar to that of conventional protein kinases and are enzymatically active. There are some notable changes in the FunK1 motifs. A highly conserved lysine corresponding to K168 is replaced by an invariant serine in FunK1. This lysine, which donates a hydrogen bond to the transferred gamma phosphate group, presumably is replaced by a basic residue from elsewhere in the FunK1 sequence if efficient catalysis is to be retained. Several very highly conserved residues without direct catalytic roles also are missing from FunK1, including H158, F185, and G186. However, conservation of R165 and K189, which form intramolecular interactions with phosphorylated amino acids in some protein kinases, suggests that members of the FunK1 family are regulated by phosphorylation (25). The FunK1 family has homologs in the Agaricomycotina and Pezizomycotina, but not in other fungi, suggesting a potential link between this kinase family and the multicellularity of these fungi. We observed that the left subterminal region of chromosome IX contains 59 FunK1 kinases in all orientations (head to head, head to tail, and tail to tail) with only two transposon clusters and very few interspersed nonkinase genes. In contrast, the distribution of protein kinases from families with widely distributed orthologs is more scattered, with the vast majority occurring in regions with low rates of meiotic recombination (Fig. S2).
Fig. 2.
HMM logos of protein kinase active site regions. (A) Conventional protein kinases. (B) FunK1 protein kinases. Conserved residues positions in both conventional and FunK1 kinases are indicated with blue circles. Residue positions that are nearly universally conserved in conventional ePK domains but are altered in FunK1 kinases are indicated with red circles.
There also are copy-number changes within conserved fungal kinase families. Phylogenetic analysis of the MAPK cascade revealed Basidiomycota-specific duplications in the MAPK genes (S. cerevisiae FUS3/KSS1) which are involved in the pheromone response, whereas HOG1 a p38 kinase, is single copy throughout the sampled fungi (Fig. S3).
We examined two additional gene families of significant size (cytochrome P450 and hydrophobins) to ask if duplicated paralogs from these families also are found in restricted chromosome regions. The P450 gene family (125 genes contain the Pfam domain) includes genes with metabolic roles in monooxygenase metabolism, and the family is implicated in the degradation capabilities of Phanerochaete chrysosporium on substrates ranging from lignin to diesel fuel (26, 27). Phylogenetic analysis shows that the expansion of the family was independent in P. chrysosporium and C. cinerea (SI Text and Fig. S4). Although scattered members of the P450 and other expanded families are found within cold regions, tandem repeats of these genes are found exclusively in genomic regions with higher recombination rates. In contrast to the FunK1 family, many P450 gene paralogs are found in head-to-tail orientation with a maximum of four copies in any one chromosomal location. A similar pattern is observed for the hydrophobin gene family (34 genes). Hydrophobins are small, secreted proteins that self-assemble at hydrophilic–hydrophobic interfaces to form amphipathic films (28, 29). These films help hyphae emerge from moist substrates to form aerial structures and also line internal cavities in the fruiting body (28). We have found that C. cinerea has the largest described hydrophobin gene family for any fungus. Phylogenetic analysis (Fig. S5) shows independent expansions of the gene family in P. chrysosporium, L. bicolor, and C. cinerea. The hydrophobin paralogs often are found in head-to-tail orientation with a maximum of seven genes in one cluster. These clusters are found in regions with high rates of recombination, whereas hydrophobins that are unique within their contigs occur in regions with low rates of recombination.
The patterns of gene duplication have important implications for adaptation and evolution in some species where pathogenic factors like adhesins and cell-surface variation genes tend to be found in the highly recombining regions near chromosome ends (4, 30). In C. cinerea, gene ontology (GO) terms are correlated with local recombination rates along the chromosomes (SI Text and Dataset S1, Table S10). In addition, we have found that the age of gene duplicates, as measured by calculating synonymous substitution rates (dS), correlates well with the recombination rate classes. We identified 3,796 duplicated gene pairs and found that pairs residing in cold regions are significantly older (P < 1E-16, Kolmogorov-Smirnov test; median dS 2.2) than the pairs that reside in regions of average or elevated recombination (median dS 1.95), as illustrated in Fig. 1 and Fig. S1. The genomic orientation of paralogous gene pairs also is highly nonrandom. Overall, 49% of all adjacent genes are in tandem orientation, as expected from a random distribution, whereas 88% of adjacent paralogous genes are in tandem orientation (P < 1E-16; Fisher's exact test). The increased age in the cold regions of the genome indicates a lower generation rate of gene duplicates and confirms that an unequal rate of sequence evolution is a general property of all gene families in C. cinerea.
Transcriptional Program Associated with Mating Behavior.
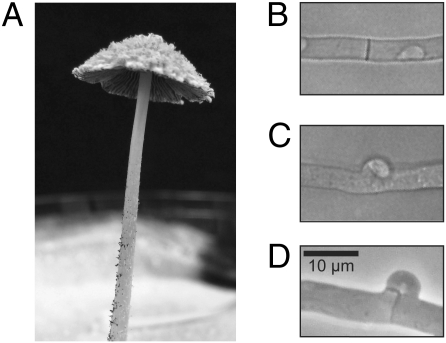
Coordinated gene expression of adjacent FunK1 kinases, P450 genes, and hydrophobins might provide a selective advantage that contributes to the maintenance of the clusters. Alternatively, individual members of these paralogous gene families may play important roles at discrete stages in C. cinerea development. The ease of culture and synchronous development (under the control of light and nutritional cues) of C. cinerea greatly facilitates studies of transcriptional regulation. Accordingly, we designed a 13,230-feature microarray that includes at least one 70-mer oligonucleotide for each known and predicted gene, EST, and repeated element. We used the validated arrays to investigate the transcriptional program that occurs during mating (SI Text). In the typical basidiomycete life cycle, nuclear fusion does not occur immediately after mating cell fusion. Instead, a nucleus from each mating partner is maintained in a common cytoplasm (the “dikaryotic cell”), and all tissues (except for the multinucleate stipe cells) of the highly differentiated mushroom fruiting bodies of C. cinerea (Fig. 3A) are composed of dikaryotic cells. The formation of the dikaryotic mycelium in mushrooms is initiated by fusion of undifferentiated hyphal cells and is maintained by a complex cell division in which both nuclei divide in synchrony in the tip cell, and daughter nuclei then are partitioned equally into the new tip and subterminal cells. Partitioning involves the formation of a structure known as the “clamp connection” (Fig. 3D) through which one of the daughter nuclei must pass. The steps of dikaryon formation are controlled by two sets of unlinked, multiallelic mating-type genes called “A” and “B” (Fig. S6). Many details of the process, including the different steps that are under the control of the A genes (which encode homeodomain proteins that heterodimerize in a compatible mating) and the B genes (which encode pheromones and receptors that activate each other in a compatible mating) are understood from classical genetics and more recent molecular studies (10).
Fig. 3.
Photograph and micrographs of C. cinerea. (A) Mature C. cinerea fruiting body that is shedding spores. The upper surface of the cap has loosely adhering “veil cells.” The lower surface of the cap is composed of “gills” which support the basidia (meiotic cells). The cap is elevated several centimeters above the Petri dish by the “stipe” (stalk). (B) Simple septum between two cells in a monokaryotic hypha. (C) “False clamp” between two cells in an “A-on” hypha. (D) True clamp connection between two cells in a dikaryotic hypha (“A-on B-on”). Magnification in B–D is the same.
We examined mycelia in which targets of the A locus (the “A-on” strain; Fig. 3C) and the B locus (the “B-on” strain, which has a morphology similar to the strain shown in Fig. 3B) were expressed separately and compared these with transcripts expressed in the dikaryon (Fig. 3D) and in the unmated monokaryotic strain (Fig. 3B). We observed 877 transcripts with significant differences in expression levels in one or more of these conditions (SI Text and Dataset S1, Table S11). Of particular interest was a FunK1 kinase (CC1G_04033) with an ortholog in L. bicolor, which was significantly up-regulated in the A-on mycelium, along with a FunK1 paralog (CC1G_13267), suggesting that these may have a unique role in cell signaling in the A-regulated part of the pathway that requires synchronization of nuclear division. Other differentially regulated genes of interest include the previously characterized genes clp1 and pcc1, 12 transcription factors, five additional kinases including S. cerevisiae HOG1 and RCK2 orthologs, STE3 receptors, major facilitator superfamily transporters, and many genes involved in cell-cycle regulation, the cytoskeleton, and cell wall biogenesis (SI Text and Dataset S1, Table S11). A MAPK signaling complex, which includes the HOG1 homolog Fus3, plays a central role in the yeast pheromone response (31). However, the Fus3 MAPK cascade does not occur outside of the Saccharomycetales (32). We did observe that HOG1 a p38 MAPK, and RCK2 a calcium/calmodulin-dependent protein kinase, are both strongly down-regulated in A-on and B-on cells. Orthologs of these protein kinases are involved in the hyperosmotic response of yeast (33, 34). HOG1 is down-regulated in S. cerevisiae a1/α2 cells (35), but in contrast to what we observe in C. cinerea, its target, RCK2 (34), is not (35). It has been suggested that when FUS3 transcription is shut down in a1/α2 cells, HOG1 is down-regulated so it is not spuriously activated in the site vacated by its homolog (35). In C. cinerea, down-regulation of HOG1 and its target RCK2 must serve a different purpose, because the Fus3 is absent (32). We conclude that our high-throughput approach to identifying potential regulators and targets in the A-on and B-on pathways is essential for understanding this complex morphogenetic process, especially because very few of the downstream targets of the mating factors have been identified in any basidiomycete (36).
Overall, we found very few examples of coordinated regulation of adjacent paralogous gene duplicates in these experiments. Despite the presence of 59 adjacent FunK1 family members in the genome, scattered members of this cluster were coordinately regulated during dikaryon formation. We did observe significantly coordinated expression of a single paralogous tandem array of hydrophobins and of a paralogous tandem array of P450 genes during dikaryon formation. However, examination of expression patterns of all paralogous pairs indicated that adjacent pairs were no more likely to be coordinately regulated than nonadjacent pairs. We conclude that adjacent gene duplicates have diverged in expression timing and perhaps function.
Synteny.
Although there is larger number of genes in the currently sampled Agaricomycotina genomes (~10,000–15,000) than in Cryptococcus or Ustilaginomycotina (~6,000), there is no evidence for a whole-genome duplication, because no substantial region of duplication was identified through dot-plot or gene-based comparisons of self-vs.-self of the C. cinerea genome. A comparative approach also was used to ask if the key genomic features present in C. cinerea, particularly the potential for similar large genomic clusters with limited meiotic recombination, are represented in other Agaricomycetes. The genome of L. bicolor (37) provides an appropriate test case, because these Agaricomycetes last shared a common ancestor 200 Mya (SI Text). L. bicolor is an important ectomycorrhizal symbiont of hardwood and conifer species, although it also can adopt a transient saprotrophic lifestyle similar to that of C. cinerea. The genome of L. bicolor is 1.8 times larger than the C. cinerea genome, contains 1.6 times the number of predicted gene models, and is estimated to contain 13.65 Mb of transposons and transposon relics (in contrast to the 0.86 Mb in the assembled C. cinerea genome).
To identify blocks of synteny between these species, we employed the program (for “Fast Identification of Segmental Homology”), because the Manhattan distance metric it employs allows very asymmetric intervals between the syntenic “anchors” (38) (SI Text). We found that 39% of the assembled C. cinerea genome is syntenic with L. bicolor (Fig. S1 and Dataset S1, Table S12a). To estimate the total number of chromosomal rearrangement events that have occurred since their split from a common ancestor, we fit our data to the Nadeau-Taylor model (39, 40), which assumes that genes and chromosomal rearrangement breakpoints are uniformly distributed at random along the chromosomes. We calculate a rate of 3.5–4.5 chromosomal rearrangements per million years have accumulated along each lineage since separation. This rate is at the high end of the range described previously for eukaryotes (41) and is approximately 3-fold higher than in S. cerevisiae (42).
Despite the prevalence of rearrangements in these lineages, we observed 10 blocks with more than 15 anchors (SI Text and Dataset S1, Table S12b). Because these are highly unlikely (P < 0.0016) if rearrangements are tolerated equally, it was of interest to determine the nature of these chromosomal regions that are unusually refractory to rearrangement in mushrooms. These regions (3.4 Mb) are found primarily in genomic regions with low meiotic recombination rates on the five largest chromosomes (Fig. 1 and Fig. S1). GO analysis of these regions revealed that they are enriched (P < 0.0005 to P < 0.01) in genes annotated to basic structures and processes such as nitrogen metabolism, the cytoskeleton, and metabolic regulation, as well as in particular G protein-coupled receptors (Dataset S1, Table S13). These regions contain 2.4 times the number of expected transcription factors (χ2 = 67.7, P < 0.001) and lack transposable elements. Interestingly, the 1,378 genes in these blocks are spaced on average only 872 bp apart, in contrast to the average gene spacing in the genome (1,261 bp) and in sharp contrast to the average gene spacing in regions that display elevated rates of recombination (1,655 bp).
Discussion
Our gap closure, telomere and centromere identification, and sequence anchoring to the genetic map have increased the value of the draft C. cinerea genome sequence. The revised gene calls supported by ESTs and 5′SAGE profiling reported here (SI Text) enabled accurate annotation and comparative analyses. We conclude that regions of the genome that have persisted intact over evolutionary time exhibit low rates of meiotic recombination and also lack sequences such as tandemly repeated members of large gene families and transposable elements that could promote ectopic recombination. The presence of transcription factors in the syntenic blocks may be an important developmental control that is interdependent and cannot easily be disrupted. Studies focusing on the phenotypes of knockdown or knockouts of these genes now can be undertaken to determine if they play important roles in mushroom development. Further profiling of the C. cinerea gene expression during specific tissue and developmental time points, enabled by the completion and annotation of the genome, will help address these questions.
We also observed that paralogous gene families are overrepresented near chromosome ends, in regions of average or high meiotic recombination. The finding of independent expansions in several Agaricomycotina genomes, likely driven by tandem duplications in both the P450 and hydrophobin families, suggests a common mechanism for rapid family expansions. The diversification of these families may indicate acquisition of new or partitioned function (43) and may indicate an increased importance of these families in the cellular processes of Agaricomycotina fungi. The further identification of protein kinase FunK1 family (a family that is only found in multicellular fungi) expansions in C. cinerea suggests experimental approaches to the identification of signaling pathways that control cellular development and differentiation processes specific to multicellular fungi.
Our expression studies to date suggest an important role for specific FunK1 family members in C. cinerea development. They also provide no evidence for coordinate regulation of adjacent FunK1 family members or for the vast majority of duplicated paralogous genes in the C. cinerea genome. Because many aspects of genome organization are open to experimental manipulation in C. cinerea, the potential influence of chromosome position on expression of key members of this important family can be evaluated systematically.
Materials and Methods
Genome Sequencing, Assembly, and Annotation.
The haploid Okayama 7 #130 strain was sequenced by WGS sequencing to a level of 10× coverage of the predicted 36-Mb genome, assembled with Arachne (44). Genome finishing into chromosomes was undertaken by manual inspection of the assembly, PCR from ends of WGS contigs, identification of telomeric repeats, and incorporation of BAC mapping data (SI Text). The protein-coding genes (13,342 sequences) were identified and confirmed using a combination of gene prediction and evidence-based tools including ESTs and 5′SAGE (SI Text). The initial sequence, assembly, and annotation can be accessed at http://www.broadinstitute.org/annotation/genome/coprinus_cinereus.
Dikaryon Formation (Mating) and Microarray analysis.
Four strains were used to examine genes under the control of the A pathway (“A-on”; AmutB43), the B pathway (“B-on”; A43Bmut), and both pathways (“A-on, B-on”; AmutBmut) in comparison with the reference (“A-off, B-off”; A43B43). The 70-mer oligonucleotide arrays include 13,230 probes designed for all predicted genes and ESTs. Methods for hybridization, data capture, and analysis are described in SI Text, and data have been deposited in GEO (GSE20628). Annotations, ESTs, SAGE tags, SSRs, and linkage data, and oligonucleotides on the microarray platform can be accessed at http://genome.semo.edu/ccin.
Supplementary Material
Acknowledgments
We thank the Center for Genomics and Bioinformatics at Indiana University for microarray production and Susan Whitfield for assistance with the figures. This article benefited greatly from input from C. D. Jones, J. Heitman, G. May, G. R. Fink, J. W. Taylor, and members of the J. W. Taylor laboratory. Support to finish, annotate, and map the C. cinerea genome was provided to P.J.P., F.S.D., A.C.G., W.W.L., S.K.W., J.B.H., and A.H by National Science Foundation (NSF) Grant EF 0412016. J.D. and T.J.V. were supported by NSF Grant DBI0227314 (to T.J.V.). M.L.F. was supported by NSF Grant MCB 0135462, and C.J.R. was supported by an NSF graduate fellowship. J.E.S. was supported by a NSF graduate fellowship and by a postdoctoral fellowship from the Miller Institute for Basic Research in Science. M.B., A.L., and V.T.-H. were supported by National Institutes of Health (NIH) Grant HG00783 (to M.B.). G.M. was supported by NIH Grant HG004164. M.E.Z., C.B., and H.P. were supported by NIH Grant GM43930 (to M.E.Z.). T.K. was supported by a Grant-in-Aid for Scientific Research from the Ministry of Education, Science, Sports, and Culture of Japan. U.K., P.J.H., S.K., and R.V. were supported by the Deutsche Bundesstiftung Umwelt (to U.K.). H.S.K., C.H.A., and C.K.C. were supported by the Research Grants Council of Hong Kong SAR, China, reference number 466608 (to H.S.K.). F.M. was supported by an Institut National de la Recherche Agronomique Grant “AIP BioResources.” Support for BAC end-sequencing was provided to H.M. by the Biotechnology Center, Akita Prefectural University. D.A., B.C., and A.T. were funded by the Swedish Research Council. Array construction was supported by the Indiana METACyt Initiative of Indiana University, funded in part through a major grant from the Lilly Endowment, Inc. (to P.J.P. and M.E.Z.). The University of North Carolina at Chapel Hill's Libraries provided support for open access publication and page charges were paid by startup funds to J.E.S. from the University of California, Riverside.
Footnotes
The authors declare no conflict of interest.
See Commentary on page 11655.
Data deposition: Assembly accession: AACS02000000; Expression data in GEO: GSE20628; EST library accessions: DR774668-DR775517, DN591505-DN593171, DN593172-DN593841, DR752715-DR753264, DN593842-DN593917, DR753265-DR753301, DR753303-DR753936, FG068230-FG068291, DR421062-DR421601, DR752151-DR752714, DR753937-DR753939, DR907568-DR908072.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1003391107/-/DCSupplemental.
References
- 1.Wong S, Butler G, Wolfe KH. Gene order evolution and paleopolyploidy in hemiascomycete yeasts. Proc Natl Acad Sci USA. 2002;99:9272–9277. doi: 10.1073/pnas.142101099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Galagan JE, et al. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature. 2005;438:1105–1115. doi: 10.1038/nature04341. [DOI] [PubMed] [Google Scholar]
- 3.Scannell DR, et al. Independent sorting-out of thousands of duplicated gene pairs in two yeast species descended from a whole-genome duplication. Proc Natl Acad Sci USA. 2007;104:8397–8402. doi: 10.1073/pnas.0608218104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Butler G, et al. Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature. 2009;459:657–662. doi: 10.1038/nature08064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fabre E, et al. Comparative genomics in hemiascomycete yeasts: Evolution of sex, silencing, and subtelomeres. Mol Biol Evol. 2005;22:856–873. doi: 10.1093/molbev/msi070. [DOI] [PubMed] [Google Scholar]
- 6.Redhead SA, Vilgalys R, Moncalvo J, Johnson J, Hopple JS. Coprinus Persoon and the disposition of Coprinus species sensu lato. Taxon. 2001;50:203–241. [Google Scholar]
- 7.Binninger DM, Skrzynia C, Pukkila PJ, Casselton LA. DNA-mediated transformation of the basidiomycete Coprinus cinereus. EMBO J. 1987;6:835–840. doi: 10.1002/j.1460-2075.1987.tb04828.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wälti MA, et al. Targeted gene silencing in the model mushroom Coprinopsis cinerea (Coprinus cinereus) by expression of homologous hairpin RNAs. Eukaryot Cell. 2006;5:732–744. doi: 10.1128/EC.5.4.732-744.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kamada T. Molecular genetics of sexual development in the mushroom Coprinus cinereus. Bioessays. 2002;24:449–459. doi: 10.1002/bies.10083. [DOI] [PubMed] [Google Scholar]
- 10.Casselton LA, Kües U. In: Sex in Fungi: Molecular Determination and Evolutionary Implications. Heitman J, Kronstad JW, Taylor JW, Casselton LA, editors. Washington: American Association for Microbiology Press; 2007. pp. 283–300. [Google Scholar]
- 11.Kües U. Life history and developmental processes in the basidiomycete Coprinus cinereus. Microbiol Mol Biol Rev. 2000;64:316–353. doi: 10.1128/mmbr.64.2.316-353.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holm P, Rasmussen S, Zickler D, Lu B, Sage J. Chromosome pairing, recombination nodules and chiasma formation in the basidiomycete Coprinus cinereus. Carlsberg Res Commun. 1981;46:305–346. [Google Scholar]
- 13.Acharya SN, et al. Coprinus cinereus rad50 mutants reveal an essential structural role for Rad50 in axial element and synaptonemal complex formation, homolog pairing and meiotic recombination. Genetics. 2008;180:1889–1907. doi: 10.1534/genetics.108.092775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muraguchi H, Ito Y, Kamada T, Yanagi SO. A linkage map of the basidiomycete Coprinus cinereus based on random amplified polymorphic DNAs and restriction fragment length polymorphisms. Fungal Genet Biol. 2003;40:93–102. doi: 10.1016/s1087-1845(03)00087-2. [DOI] [PubMed] [Google Scholar]
- 15.Burns C, Pukkila PJ, Zolan ME. In: Cellular and Molecular Biology of Filamentous Fungi. Borkovich KA, Ebbole DJ, editors. Washington: American Society of Microbiology Press; 2010. pp. 81–95. [Google Scholar]
- 16.Barnes TM, Kohara Y, Coulson A, Hekimi S. Meiotic recombination, noncoding DNA and genomic organization in Caenorhabditis elegans. Genetics. 1995;141:159–179. doi: 10.1093/genetics/141.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jensen-Seaman MI, et al. Comparative recombination rates in the rat, mouse, and human genomes. Genome Res. 2004;14:528–538. doi: 10.1101/gr.1970304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barton AB, Pekosz MR, Kurvathi RS, Kaback DB. Meiotic recombination at the ends of chromosomes in Saccharomyces cerevisiae. Genetics. 2008;179:1221–1235. doi: 10.1534/genetics.107.083493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zickler D. From early homologue recognition to synaptonemal complex formation. Chromosoma. 2006;115:158–174. doi: 10.1007/s00412-006-0048-6. [DOI] [PubMed] [Google Scholar]
- 20.Galagan JE, et al. The genome sequence of the filamentous fungus Neurospora crassa. Nature. 2003;422:859–868. doi: 10.1038/nature01554. [DOI] [PubMed] [Google Scholar]
- 21.Loftus BJ, et al. The genome of the basidiomycetous yeast and human pathogen Cryptococcus neoformans. Science. 2005;307:1321–1324. doi: 10.1126/science.1103773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Charlesworth B, Sniegowski P, Stephan W. The evolutionary dynamics of repetitive DNA in eukaryotes. Nature. 1994;371:215–220. doi: 10.1038/371215a0. [DOI] [PubMed] [Google Scholar]
- 23.Duret L, Marais G, Biémont C. Transposons but not retrotransposons are located preferentially in regions of high recombination rate in Caenorhabditis elegans. Genetics. 2000;156:1661–1669. doi: 10.1093/genetics/156.4.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Enright AJ, Van Dongen S, Ouzounis CA. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002;30:1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zheng J, et al. 2.2 A refined crystal structure of the catalytic subunit of cAMP-dependent protein kinase complexed with MnATP and a peptide inhibitor. Acta Crystallogr D Biol Crystallogr. 1993;49:362–365. doi: 10.1107/S0907444993000423. [DOI] [PubMed] [Google Scholar]
- 26.Martinez D, et al. Genome sequence of the lignocellulose degrading fungus Phanerochaete chrysosporium strain RP78. Nat Biotechnol. 2004;22:695–700. doi: 10.1038/nbt967. [DOI] [PubMed] [Google Scholar]
- 27.Doddapaneni H, Chakraborty R, Yadav JS. Genome-wide structural and evolutionary analysis of the P450 monooxygenase genes (P450ome) in the white rot fungus Phanerochaete chrysosporium: Evidence for gene duplications and extensive gene clustering. BMC Genomics. 2005;6:92. doi: 10.1186/1471-2164-6-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wessels JG. Hydrophobins: Proteins that change the nature of the fungal surface. Adv Microb Physiol. 1997;38:1–45. doi: 10.1016/s0065-2911(08)60154-x. [DOI] [PubMed] [Google Scholar]
- 29.Wösten HA. Hydrophobins: Multipurpose proteins. Annu Rev Microbiol. 2001;55:625–646. doi: 10.1146/annurev.micro.55.1.625. [DOI] [PubMed] [Google Scholar]
- 30.Verstrepen KJ, Reynolds TB, Fink GR. Origins of variation in the fungal cell surface. Nat Rev Microbiol. 2004;2:533–540. doi: 10.1038/nrmicro927. [DOI] [PubMed] [Google Scholar]
- 31.Elion EA. Pheromone response, mating and cell biology. Curr Opin Microbiol. 2000;3:573–581. doi: 10.1016/s1369-5274(00)00143-0. [DOI] [PubMed] [Google Scholar]
- 32.Mody A, Weiner J, Ramanathan S. Modularity of MAP kinases allows deformation of their signalling pathways. Nat Cell Biol. 2009;11:484–491. doi: 10.1038/ncb1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O'Rourke SM, Herskowitz I, O'Shea EK. Yeast go the whole HOG for the hyperosmotic response. Trends Genet. 2002;18:405–412. doi: 10.1016/s0168-9525(02)02723-3. [DOI] [PubMed] [Google Scholar]
- 34.Teige M, Scheikl E, Reiser V, Ruis H, Ammerer G. Rck2, a member of the calmodulin-protein kinase family, links protein synthesis to high osmolarity MAP kinase signaling in budding yeast. Proc Natl Acad Sci USA. 2001;98:5625–5630. doi: 10.1073/pnas.091610798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Galgoczy DJ, et al. Genomic dissection of the cell-type-specification circuit in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2004;101:18069–18074. doi: 10.1073/pnas.0407611102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kahmann R, Schirawski J. In: Sex in Fungi: Molecular Determintation and Evolutionary Implications. Heitman J, Kronstad JW, Taylor JW, Casselton LA, editors. Washington: American Association for Microbiology Press; 2007. [Google Scholar]
- 37.Martin F, et al. The genome of Laccaria bicolor provides insights into mycorrhizal symbiosis. Nature. 2008;452:88–92. doi: 10.1038/nature06556. [DOI] [PubMed] [Google Scholar]
- 38.Calabrese PP, Chakravarty S, Vision TJ. Fast identification and statistical evaluation of segmental homologies in comparative maps. Bioinformatics. 2003;19(Suppl 1):i74–i80. doi: 10.1093/bioinformatics/btg1008. [DOI] [PubMed] [Google Scholar]
- 39.Nadeau JH, Taylor BA. Lengths of chromosomal segments conserved since divergence of man and mouse. Proc Natl Acad Sci USA. 1984;81:814–818. doi: 10.1073/pnas.81.3.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sankoff D, Ferretti V, Nadeau JH. Conserved segment identification. J Comput Biol. 1997;4:559–565. doi: 10.1089/cmb.1997.4.559. [DOI] [PubMed] [Google Scholar]
- 41.Eichler EE, Sankoff D. Structural dynamics of eukaryotic chromosome evolution. Science. 2003;301:793–797. doi: 10.1126/science.1086132. [DOI] [PubMed] [Google Scholar]
- 42.Gordon JL, Byrne KP, Wolfe KH. Additions, losses, and rearrangements on the evolutionary route from a reconstructed ancestor to the modern Saccharomyces cerevisiae genome. PLoS Genet. 2009;5:e1000485. doi: 10.1371/journal.pgen.1000485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ohno S. Evolution by Gene Duplication. Heidelberg: Springer-Verlag; 1970. [Google Scholar]
- 44.Batzoglou S, et al. ARACHNE: A whole-genome shotgun assembler. Genome Res. 2002;12:177–189. doi: 10.1101/gr.208902. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.