Abstract
Mitochondrial genomes with deleterious mutations can replicate in cells along with wild-type genomes in a state of heteroplasmy, and are a cause of severe inherited syndromes, such as mitochondrial myopathy, encephalopathy, lactic acidosis, and stroke (MELAS), neuropathy, ataxia, retinitis pigmentosa-maternally inherited Leigh syndrome (NARP-MILS), and Leber's hereditary optic neuropathy (LHON). The cytosolic E3 ligase, Parkin, commonly mutated in recessive familial parkinsonism, translocates to depolarized mitochondria and induces their autophagic elimination, suggesting that Parkin may signal the selective removal of defective mitochondria within the cell. We report that long-term overexpression of Parkin can eliminate mitochondria with deleterious COXI mutations in heteroplasmic cybrid cells, thereby enriching cells for wild-type mtDNA and restoring cytochrome c oxidase activity. After relieving cybrid cells of Parkin overexpression, a more favorable wild-type to mutant mitochondrial genome ratio is stably maintained. These data support the model that Parkin functions in a mitochondrial quality control pathway. Additionally, they suggest that transiently increasing levels of Parkin expression might ameliorate certain mitochondrial diseases.
Keywords: autophagy, neurodegeneration, Parkinson disease, PINK1, mitochondria
Mitochondrial DNA mutations are responsible for a number of severe syndromes, with symptoms ranging from epilepsy and encephalopathy to lactic acidosis and diabetes (1, 2). In addition, somatically acquired mtDNA mutations have been linked to the pathogenesis of common diseases, such as cancer, diabetes mellitus, and neurodegenerative disorders. For example, patients with sporadic Parkinson disease have a greater number of functionally deleterious mtDNA mutations in their substantia nigral neurons compared with age-matched controls (3, 4), and increased mtDNA deletions, as is observed in patients with multiple mtDNA deletion syndromes, appears to be sufficient to cause parkinsonism (5).
Within the cells of a patient affected with a mitochondrial disease, mutated mtDNA typically coexists with wild-type mtDNA. In this heteroplasmic state, wild-type and mutant mtDNA are packed in separate nucleoids and rarely mix (6). The severity of cellular dysfunction and disease caused by a given mtDNA mutation depends on the ratio of mutant mtDNA to wild-type mtDNA in the cell. Experimentally shifting a population of mtDNA away from the mutant DNA toward wild-type mtDNA improves mitochondrial function within the cell and tissue, and represents a promising therapeutic strategy for diseases in which mtDNA mutations contribute to the pathogenesis (7).
We recently found that the cytosolic E3 ligase, Parkin, which is commonly mutated in autosomal-recessive juvenile parkinsonism (8, 9) and has been linked to mitochondrial maintenance in Drosophila (10–13) and in mouse (14, 15), can translocate to depolarized mitochondria and activate their elimination by autophagy. In cells containing a mixed population of functional and dysfunctional mitochondria, Parkin selectively localizes to uncoupled mitochondria, suggesting that Parkin may function in a mitochondrial quality control process (16). Here we show that overexpression of Parkin can increase the ratio of wild-type to mutant mtDNA in heteroplasmic cybrid cells and that cytochrome c oxidase activity is restored in cybrid cells enriched for wild-type mtDNA. These results indicate that within a mixed population, Parkin can selectively target the defective mitochondria and mediate their elimination.
Results
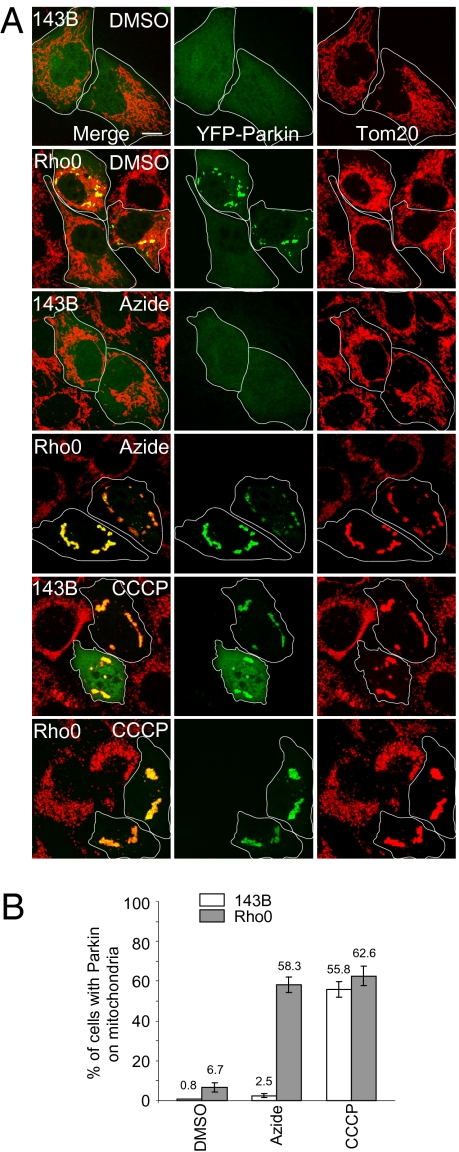
Parkin Translocation to Mitochondria in Rho0 Cells.
Parkin has been shown to translocate to impaired mitochondria and induce their mitophagy (16). We therefore examined Parkin localization in 143B Rho0 cells that lack mtDNA, display mitochondrial deficiencies, and have a lower membrane potential than the parental human osteosarcoma cell line (143B cells) (17–19). YFP-Parkin transiently expressed in Rho0 cells localized to mitochondria in 6.7 ± 2.3% (mean ± SD) of cells, somewhat more than in wild-type 143B cells (0.8 ± 0.1%) but much less than carbonyl cyanide m-chorophenylhydrazone (CCCP)-treated wild-type and Rho0 cells that display Parkin on mitochondria in more than 50% of cells (Fig. 1). The 143B Rho0 cells maintain some level of mitochondrial membrane potential above that found upon uncoupling with CCCP treatment (Fig. S1), thereby retaining certain essential mitochondrial activities, such as the import of mitochondrial proteins encoded by nuclear genes and required for catalytic and biosynthetic pathways (18). The mitochondrial membrane potential of Rho0 cells requires adenine nucleotide translocator and F1-ATPase activity (17, 18). When parental 143B cells were treated with azide to inhibit F1-ATPase activity (17, 18), the membrane potential of parental 143B remained normal as the proton gradient is maintained by oxidative phosphorylation. By contrast, when the F1-ATPase activity of Rho0 cells was inhibited with azide, the mitochondrial membrane potential collapsed, as these cells lack a functional electron transport chain (Fig. S1) (18). Following azide treatment of Rho0 cells, YFP-Parkin localized to mitochondria in 58.3 ± 3.9% of cells, almost as high as in CCCP-treated cells (62.6 ± 4.8%), whereas only 2.5 ± 0.9% of control 143B cells treated with azide displayed mitochondrial YFP-Parkin (Fig. 1). These results indicate that Parkin targets mitochondria lacking mtDNA when regeneration of membrane potential by reversal of the F1-ATPase activity is prevented.
Fig. 1.
YFP-Parkin translocates to mitochondria in 143B Rho0 cells. (A) 143B Rho0 and parental 143B cells were transfected with YFP-Parkin (green) and treated with DMSO, 100 μM sodium azide or 10 μM CCCP for 4 h. Cells were fixed and immunostained for Tom20 (mitochondria, red). (Scale bar, 10 μm.) (B) Cells were scored for YFP-Parkin on mitochondria following treatment with DMSO, azide, or CCCP as described in A. Greater than 90 cells were counted for each condition. The mean and SD were calculated from three replicates and the experiment was repeated twice.
Twinkle Mutant Expression Increases Parkin Translocation to Mitochondria.
To explore the hypothesis that Parkin can selectively target mitochondria with deleterious mtDNA mutations, we examined Parkin translocation to mitochondria in cells transiently expressing a catalytically inactive form of the mitochondrial DNA helicase, Twinkle. Mutations in Twinkle, which disrupt mtDNA replication and lead to multiple mtDNA deletions, can cause dominant progressive external opthalmoplegia (20), parkinsonism (21), and other symptoms (22). In addition, overexpression of catalytically inactive Twinkle mutants (such as Twinkle G575D) in cell culture leads to acute loss of mtDNA and mitochondrial dysfunction (23). Expressing the Twinkle G575D mutant caused a proportion of mitochondria to lose membrane potential and display selective recruitment of Parkin (Fig. S2A) in 13.1 ± 2.3% of HeLa cells relative to 6.7 ± 1.2% in wild-type Twinkle-expressing control cells (Fig. S2B). It is also noteworthy that mitochondria usually appeared reduced in number and clumped upon Parkin translocation in the Twinkle G575D-expressing cells (Fig. S2B), consistent with mitochondrial phenotypes observed in response to Parkin upon mitochondrial uncoupling (16). These results suggest that acute depletion of wild-type mtDNA by mutant Twinkle expression induces Parkin recruitment to the resulting dysfunctional mitochondria.
Parkin Translocation to Mitochondria in Heteroplasmic COXICA65 Cybrid Cells.
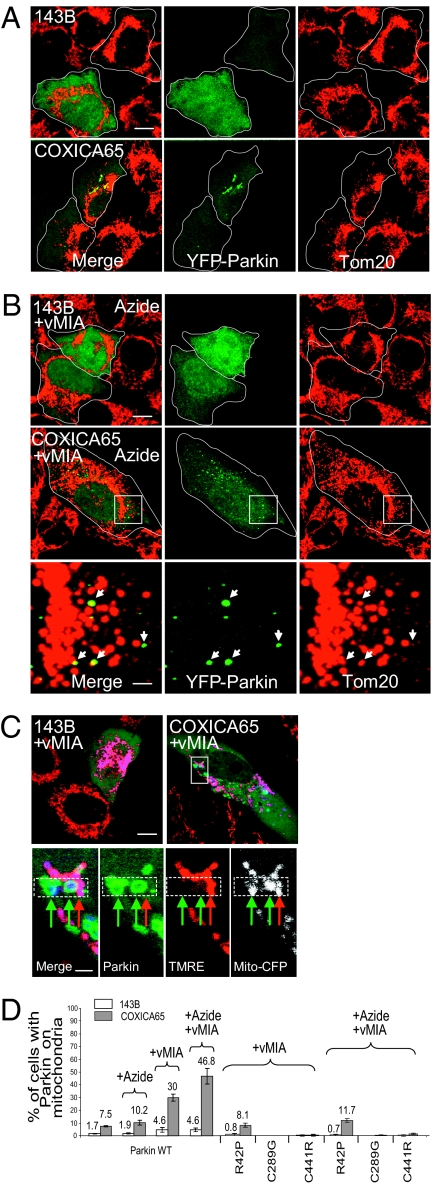
We also examined Parkin translocation to mitochondria in cybrid cells possessing a stable mixture of wild-type and mutant mtDNA genomes. In the wild-type parental 143B line, YFP-Parkin is located on mitochondria in less than 2% of the cells (Fig. 2 A and D). In a cybrid cell line containing a heteroplasmic mixture of ~10% wild-type mtDNA and ~90% mtDNA mutated in the cytochrome b gene (Cytb3.0) (24), only 2.1 ± 0.5% of cells displayed mitochondrial YFP-Parkin, not significantly different from that of the parental 143B cell line (1.3 ± 1.2%) (P = 0.32, Student's t test) (Fig. S3A). However, in a cybrid line containing mtDNA mutated in the cytochrome c oxidase subunit I gene (COXICA65) (25), a significant increase in cells displayed Parkin constitutively localized on mitochondria relative to the parental 143B cells (7.5 ± 0.6% vs. 1.7 ± 0.4%, P = 0.00017) (Fig. 2 A and D). We also observed that LC3 colocalized with Parkin and mitochondria in COXICA65 cybrid cells and not in parental 143B cells (Fig. S4), consistent with previous observations (18). Assessed by flow cytometry, COXICA65 cybrid cells have a lower mean TMRE intensity (45.3 ± 1.5%) relative to 143B cells, whereas Cytb3.0 cybrid cells showed 62.8 ± 6.5% of parental cell tetramethylrhodamine ethyl ester (TMRE) intensity (Fig. S3B), perhaps accounting for the lack of Parkin translocation in Cytb3.0 cells. The absolute difference in membrane potential between the cell lines is likely greater as differences in TMRE intensity underestimate differences in membrane potential (26). The mitochondrial kinase PINK1 accumulates on uncoupled mitochondria and mediates Parkin recruitment to dysfunctional mitochondria (27–30). Cytb3.0 cybrid cells display less PINK1 accumulation relative to COXICA65 cybrid cells following CCCP treatment (Fig. S3C), suggesting that the lower PINK1 accumulation in Cytb3.0 cybrid cells may contribute to the lower Parkin translocation to mitochondria relative to the COXICA65 cybrid cells.
Fig. 2.
YFP-Parkin accumulates on a portion of mitochondria in COXICA65 cybrid cells. The parental 143B cell with 100% wild-type mtDNA and COXICA65 cybrid cell with ~75% mutant mtDNA (G->A transition at 6,930 nt in cytochrome c oxidase subunit I) transfected with YFP-Parkin (green) (A) or YFP-Parkin plus vMIA-myc (B) were incubated without (A) or with 100 μM sodium azide (B) for 4 h. Cells were fixed and immunostained for Tom20 (mitochondria, red). Arrows in B represent YFP-Parkin localized to a subset of mitochondria in zoom image of white box of COXICA65 cells. (C) 143B and COXICA65 coexpressing YFP-Parkin (green), vMIA-myc and mito-CFP (white; blue in the merged image) were stained with 2.5 nM of the potentiometric mitochondrial dye TMRE (red) for 1 h. (D) Cells in each condition were scored for the presence of YFP-tagged wild-type (WT) or mutant Parkin on mitochondria. Greater than 70 cells were counted in each sample. The mean and SD were calculated from three replicates. (Scale bars, 10 μm in the full-size images and 2 μm in the magnified images.)
Blocking F1-ATPase Activity and Mitochondrial Fusion Promotes Parkin Translocation.
Azide inhibition of the F1-ATPase can prevent the generation of mitochondrial membrane potential in impaired mitochondria (Fig. S1) (17, 18) and promotes Parkin translocation to mitochondria in Rho0 cells (Fig. 1). However, azide exposure did not significantly increase Parkin translocation in COXICA65 cybrid cells (no treatment 7.5 ± 0.6% vs. plus azide 10.2 ± 2.0%, P = 0.08) (Fig. 2 A and D and Fig. S5A). We considered that wild-type mitochondria present in cybrid cells (and not present in Rho0 cells) might compensate for defects in mutant mitochondria by the transfer of wild-type RNA or protein during frequent rounds of mitochondrial fusion and fission that intermittently combine multiple nucleoids in elongated tubular organelles. To assess if fusion with wild-type mitochondria may compensate for mutant mitochondrial deficiencies and prevent Parkin translocation to mutant mitochondria in heteroplasmic cells, we inhibited mitochondrial fusion by expression of the human cytomegalovirus protein viral mitochondrial inhibitor of apoptosis (vMIA) (31). The vMIA expression increased the percentage of cells with Parkin on mitochondria in wild-type and COXICA65 cybrid lines, reaching 30 ± 2.7% of cells in the latter case (Fig. 2D and Fig. S5B). Reducing the interconnectivity of COXICA65 cell mitochondria by vMIA expression revealed YFP-Parkin accumulation selectively on mitochondria that displayed lower membrane potential detected by TMRE staining (Fig. 2C), consistent with previous results obtained in Mfn1 and Mfn2 double knock-out cells (16). This finding suggests that inhibiting mitochondrial fusion in heteroplasmic cells may physically isolate mitochondria containing mutant mtDNA from mitochondria containing wild-type mtDNA, and prevent functional complementation among organelles. Treatment of cybrid cells with azide to prevent the generation of a membrane potential by the ATP hydrolysis activity of the F1-ATPase, combined with blocking mitochondrial fusion with vMIA, further increased Parkin translocation to 46.8 ± 6.0% of COXICA65 cells but not in parental 143B cells (Fig. 2 B and D). Therefore, mixing mitochondrial content by fusion appears to compensate for the COXI mutation in heteroplasmic cells. Preventing mitochondrial fusion and blocking the F1-ATPase augments Parkin translocation to mitochondria in COXI cybrid cells. We also examined the mitochondrial recruitment of Parkin patient mutants, R42P, C289G, and C441R in COXICA65 cells in the presence of vMIA and with or without azide treatment. In contrast to wild-type Parkin (Fig. 2 B and D and Fig. S5B), which localizes to mitochondria in 46.8% of cybrid cells in the presence of vMIA and azide, C289G and C441R mutants of Parkin localize to mitochondria in less than 1% of cells and R42P Parkin localizes to mitochondria in 11.7% of cells (Fig. 2D and Fig. S5 C and D). Thus, patient mutations in Parkin are defective to localizing to mitochondria in cybrid cells.
Overexpressing Parkin in COXICA65 Cybrid Cells.
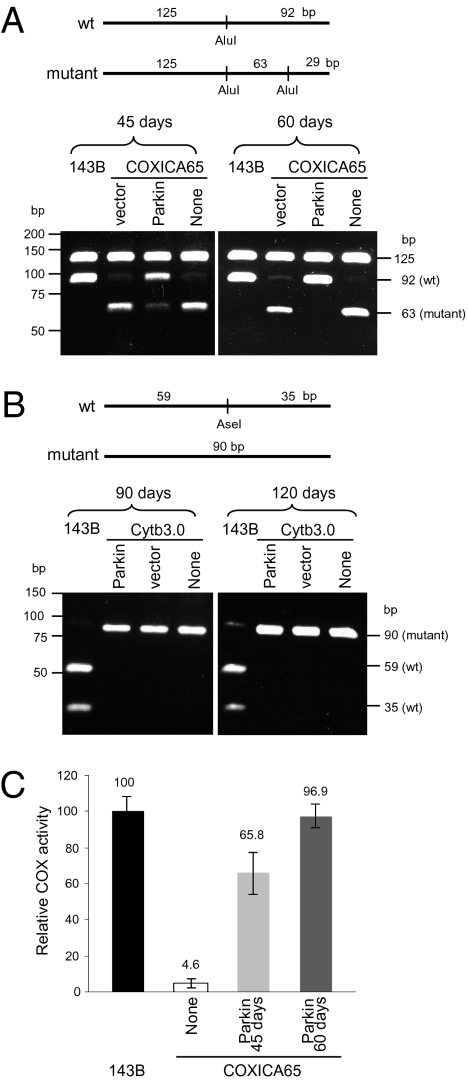
To test the hypothesis that Parkin translocation specifically to impaired mitochondria mediates selective elimination of dysfunctional mitochondria, we used restriction fragment-length polymorphism analysis of PCR products (PCR-RFLP) to analyze the ratio of wild-type to mutant mtDNA in the COXICA65 cybrid cells before and several weeks following YFP-Parkin expression (Fig. 3A). Parental 143B cells display a single band at 92 bp of digested PCR product amplified from wild-type mtDNA, whereas untransfected (none) cybrid cells display a minor band at 92 bp and a major band at 63 bp, reflecting a mixture of wild-type and mutant mtDNA (Fig. 3A). Following transfection and FACS to select for YFP-transfected cells, COXICA65 cybrid cells expressing the YFP vector (YFP-N1) for 45 d also display a minor band at 92 bp and a major band at 63 bp, consistent with the ratio of wild-type and mutant mtDNA of untransfected cybrid cells (Fig. 3A). In contrast, cybrid cells expressing YFP-Parkin for 45 d display an increase in wild-type DNA and a decrease in mutant DNA (Fig. 3A). After 60 d of culturing YFP-Parkin-expressing cells, only a very minor band of mutant mtDNA at 63 bp was detected, indicative of a strong selection against the mutant DNA (Fig. 3A).
Fig. 3.
YFP-Parkin promotes the selective elimination of mutant mtDNA in COXICA65 cybrid cells. (A) COXICA65 cybrid cells were transfected with YFP-Parkin (Parkin), YFP vector (vector), or left untransfected (None). The transfected cells were enriched for YFP-Parkin by FACS, and the ratio of wild-type to mutant mtDNA was assessed 45 (Parkin 45 d) and 60 d (Parkin 60 d) posttransfection by PCR-RLFP. A 217-bp fragment was amplified from wild-type and mutant mtDNA by PCR. Following AluI digestion, the wild-type mtDNA (which possess one AluI site) was cleaved into 125- and 92-bp fragments, although the mutant mtDNA (which possess two AluI sites) was cleaved into 125-, 63-, and 29-bp fragments. The 29-bp fragment was dim by EtBr staining and is not shown here. (B) Cytb3.0 cybrid cells with ~90% mutant mtDNA (4-bp deletion in cytochrome b gene) were transfected with YFP-Parkin (Parkin), YFP vector (vector), or left untransfected (None). The transfected cells were enriched with YFP signal by FACS after 90 and 120 d posttransfection. The 94 and 90-bp fragments were amplified from wild-type and mutant mtDNA, respectively. Following AseI digestion, the wild-type mtDNA (which possess one AseI site) showed 59- and 35-bp fragments and mutant mtDNA (which possess no AseI sites) showed a 90-bp fragment. (C) Cytochrome c oxidase activity (COX) activity was measured for each sample. COX activity is reported as a percent of the 143B parent-cell line, which contains 100% wild-type mtDNA. The mean and SD were calculated from three experiments.
Examining Cytb mutant-cybrid cells containing ~10% wild-type mtDNA that is visible in overexposed gels (Fig. S6) revealed no such selection for wild-type DNA after overexpressing Parkin (Fig. 3B). This failure of Parkin-mediated selection for wild-type DNA in Cytb3.0 cybrid cells is consistent with the lack of Parkin recruitment to mitochondria in Cytb3.0 cybrid cells relative to that seen in COXICA65 cybrid cells (Fig. S3A), the greater maintenance of mitochondrial membrane potential in Cytb3.0 cybrid cells compared with that seen in COXICA65 cybrid cells (Fig. S3B), and the lower accumulation of PINK1 in depolarized mitochondria than in COXICA65 cybrid cells (Fig. S3C).
Cytochrome c oxidase enzyme activity was analyzed in parental, cybrid, and Parkin-expressing COXICA65 cybrid cells (Fig. 3C). Whereas COXICA65 cybrid cells express only 4.64 ± 2.8% of the COX activity of parental cells, cybrid cells expressing YFP-Parkin for 60 d had COX activity restored to that of the parental 143B cells (96.90 ± 8.49%). Consistent with the increased level of wild-type mtDNA (Fig. 3A), COX activity in cybrid cells expressing YFP-Parkin for 45 d showed a level of COX activity (65.8 ± 16.1%) intermediate between parental and 60-d YFP-Parkin cybrid cultures (Fig. 3C). These results indicate that Parkin overexpression has the capacity to selectively eliminate dysfunctional mitochondria and allow the repopulation of cells with functional mtDNA genomes.
Mutant mtDNA Reaccumulation in the COXICA65 Cybrid Cells.
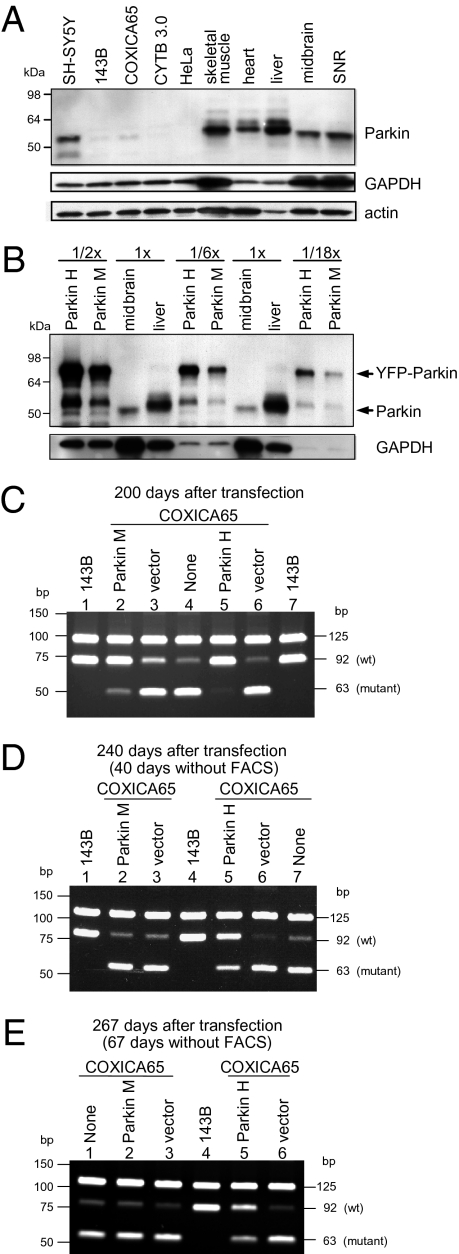
In two additional independent experiments, COXICA65 cybrid lines with moderate and high levels of YFP-Parkin expression (hereafter called Parkin M and Parkin H, respectively) were obtained by FACS sorting of transfected cells. Endogenous Parkin in 143B and the two cybrid cell lines was barely detectable by Western blot and much lower than SH-SY5Y cells and several mouse tissues examined (Fig. 4A). Total Parkin protein levels in Parkin M and Parkin H cell lines are approximately 2- and 6-fold higher than the level of endogenous Parkin in mouse liver (Fig. 4B). [As the monoclonal antibody against Parkin (PRK8) appears to be slightly more reactive to human tissues compared with mouse tissues, these are likely overestimates of the actual fold differences (32).] Although midbrain and substantia nigral mouse tissue expressed significantly lower levels of Parkin than mouse liver and muscle, this finding may reflect the heterogeneity of cell types in brain tissue that express different Parkin levels. Following 200 d of culturing, we found that cybrid cells overexpressing YFP-Parkin became enriched for wild-type mtDNA relative to COXI mutant mtDNA (Fig. 4C). Quantification of mtDNA by 32P labeling showed that the percent wild-type DNA of the total mtDNA ranged from 15.4 to 21.3% in untransfected and YFP vector-transfected cells in the two experiments (Fig. S7). However, Parkin-overexpressing cells showed an increase in the percentage of wild-type mtDNA from ~20% in the cybrid cells before Parkin transfection to 73.3% after 200 d of Parkin overexpression (Fig. S7, lane 1) in the first experiment and to 90.3% after 200 d (Fig. S7, lane 4) of Parkin overexpression in the second experiment.
Fig. 4.
Mutant mtDNA reaccumulates in the absence of Parkin-mediated selection. COXICA65 cybrid cells were transfected with YFP-Parkin (Parkin), YFP vector (vector), or left untransfected (None). In two independent experiments, cells were sorted by YFP signal over the course of 200 d. In the first experiment, a moderate level of YFP-Parkin expression (Parkin M) was achieved; in the second experiment, a high level of YFP-Parkin expression (Parkin H) was achieved. (A) Thirty micrograms of lysates from the indicated human cell lines and mouse tissues (SNR, substantia nigra) were run on SDS gels, transferred to nitrocellulose membranes, and immunoblotted for Parkin, GAPDH, or actin. (B) Thirty micrograms (1×) of lysates from mouse midbrain and liver were run on SDS gels with 15, 5, or 1.67 μg (represented as 1/2×, 1/6×, and 1/18×) of lysates from Parkin H and M cell lines. (C) The wild-type and mutant mtDNA were analyzed by PCR-RFLP for the parent 143B cell line (which is homoplasmic for wild-type mtDNA), Parkin H and M cell lines, and the COXICA65 143B heteroplasmic cell line transfected with vector or left untransfected for 200 d. (D and E) 143B and COXICA65 expressing YFP-Parkin (Parkin M and Parkin H) or YFP vector (vector) analyzed in A were continually cultured for 40 d (240 d after transfection) (D) and 67 d (267 d posttransfection) (E) in the absence of FACS selection.
Using these two cybrid cell lines enriched for wild-type mtDNA (Fig. S7, lanes 1 and 4), we assessed how durably wild-type mtDNA is maintained in the absence of selection for YFP-Parkin expression. After culturing the cybrid cells enriched in wild-type mtDNA by YFP-Parkin expression for a further 40 and 67 d in the absence of FACS selection for YFP-Parkin, considerable reversion toward mutant mtDNA was observed (Fig. 4 D and E) relative to day 0 after the initial 200-d YFP-Parkin selection (Fig. 4C). Cells with greater enrichment in wild-type mtDNA (Fig. 4C, lane 5) displayed more durable maintenance of wild-type mtDNA (Fig. 4 D and E, lane 5). When greater than 90% of mutant mtDNA was eliminated by Parkin (Fig. S7, lane 4), long-term enrichment of wild-type mtDNA was maintained following 67 d of culturing with more than 50% wild-type relative to total mtDNA (Fig. S7, lane 10). However, cells that displayed enrichment to 73.7% wild-type mtDNA (Fig. 4C, lane 2 and Fig. S7, lane 1) reverted by 40 d to the ~20% of wild-type mtDNA content displayed in the untransfected or vector-transfected cybrid cell lines (Fig. 4D, lane 2) and remained stable at this ratio of untransfected control cybrid cells after 67 d (Fig. 4E, lane 2 and Fig. S7, lane 7).
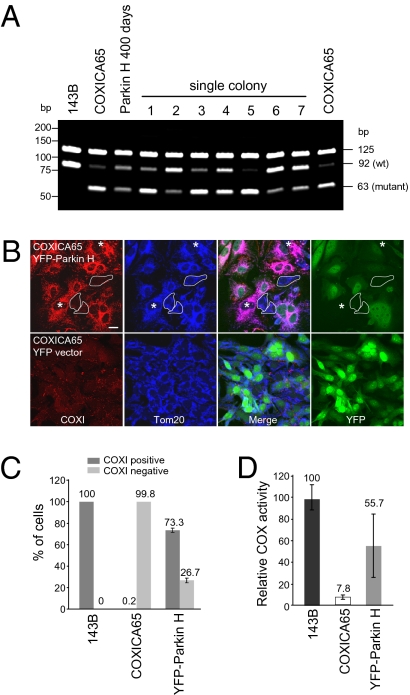
Following further culturing of the cybrid cells that were enriched to ~50% wild-type mtDNA by Parkin overexpression, we subcloned the cells to examine the range of heteroplasmy heterogeneity in these cells. Of the seven clones examined after 200 d of relief from selection for Parkin overexpression, three subclones displayed heteroplasmy similar to that of the original cybrid cells, three subclones displayed mostly but not entirely wild-type mtDNA, and one clone displayed approximately equal levels of wild-type and mutant mtDNA (Fig. 5A). These findings are consistent with previous reports, which demonstrate independent COXI cybrid clones can have relatively stable yet different ratios of wild-type to mutant mtDNA (25), although this phenomenon may depend on the type of mtDNA mutation and the nuclear DNA background (33). We immunostained cybrid cells enriched for wild-type mtDNA by Parkin overexpression, followed by 67 d of culturing without selection for YFP-Parkin, to assess if restored cytochrome c oxidase subunit I (COXI) is maintained (Fig. 5 B and C, and Fig. S8). In the Parkin H cell line, which stably expressed 50% wild-type mtDNA postselection, 73% of the cells enriched in wild-type mtDNA and that displayed undetectable YFP-Parkin expression had clear COXI immunoreactivity (hereafter, COXI-positive), although COXI protein expression was below the limit of immunodetection for 27% of the cells (hereafter, COXI-negative) (Fig. 5B). COXI positivity was seen both in cells still expressing YFP-Parkin (despite removal of selective pressure) and in cells no longer expressing YFP-Parkin (Fig. 5B). By contrast, 99.8% of the original cybrid cells were COXI-negative (Fig. 5C and Fig. S8), despite retaining 4 to 8% COX enzyme activity (Figs. 3C and 5D). We also assayed COX activity in these cells stably expressing ~50% wild-type mtDNA (Fig. 4 D and E, lane 5, and Fig. S7, lane 10). The cells exhibited 55.7 ± 29.2% of the COX activity of the parental 143B cell line (Fig. 5D). Thus, durable enrichment of wild-type mtDNA and restoration of COXI activity can be maintained following negative selection of mutant mtDNA by Parkin overexpression.
Fig. 5.
Partially reverted COXICA65 expressing YFP-Parkin (Parkin H) contains a mixed population. (A) 143B, COXICA65, Parkin H cybrid cells following transfection for 400 d (200 d in the absent of FACS selection) and seven single colonies isolated from the Parkin H cells in Fig. 4E were analyzed for the ratio of wild-type and mutant mtDNA by PCR-RFLP. (B) The COXICA65 cells Parkin-enriched for wild-type mtDNA [Parkin H, 67 d postenrichment (Fig. 4E)] were fixed and stained with Tom20 antibody (mitochondria, blue) and COXI antibody (red). YFP-Parkin is green. Cells with neither YFP-Parkin nor COXI signal were circled. Cells without YFP-Parkin but with COXI signal were marked with *. (Scale bar, 20 μm.) (C) The percentage of COXI-positive and -negative cells were scored in untransfected 143B, COXICA65 cybrid cell lines and COXICA65 cybrid cells enriched to 90% wild-type mtDNA by YFP-Parkin expression, followed by 67 d release from Parkin selection (Parkin H 67 d postenrichment) (A, Upper) considering only YFP-Parkin-negative cells. More than 110 cells lacking detectable YFP-Parkin signal were counted in each sample. (D) Cytochrome c oxidase activity (COX) assay. COX activity for each sample is reported relative to 143B, which contains 100% wild-type DNA.
Discussion
Artificially increasing mitochondrial fission with vMIA appears to augment Parkin identification of impaired mitochondria in cybrid cells, presumably by preventing functional complementation between mitochondria with mutant genomes and mitochondria with wild-type genomes. Additionally, inhibiting the F1-ATPase increases Parkin recruitment to mitochondria in Rho0 cells and in cybrid cells, indicating that the mitochondrial membrane potential maintained by the F1-ATPase is sufficient to prevent Parkin recruitment. Thus, two compensatory mechanisms appear to limit the ability of Parkin to target mitochondria with deleterious mtDNA in heteroplasmic cells. Nevertheless, even in the absence of experimentally promoting mitochondrial fission or chemically depolarizing mitochondria, over a period of months Parkin can identify most of the mitochondria harboring mutant COXI mtDNA and activate their elimination. This process may occur during normal fission and fusion cycles, when mutant mitochondria are segregated by chance from wild-type mitochondria or specifically through an asymmetric division to isolate damaged segments from the otherwise healthy mitochondrion (34). If the elimination of mutant mtDNA is only partial, in the absence of selection for Parkin overexpression, the wild-type mtDNA-enriched cells may revert to some extent toward the original cybrid ratio of wild-type to mutant mtDNA. However, with strong Parkin-mediated enrichment to over 90% wild-type mtDNA, stable maintenance of wild-type mtDNA is apparent for at least 6 months.
It was previously found that Parkin can translocate selectively to a subset of impaired mitochondria in a cell and that overexpression of Parkin can eliminate all mitochondria by mitophagy when they are chemically uncoupled (16). On the basis of these findings, it was hypothesized that Parkin may mediate an organelle quality control pathway. The experiments presented here, designed to test this hypothesis, show that Parkin overexpression has the capacity to selectively eliminate mitochondria containing high levels of COXI mutant mtDNA. These results support the proposal that Parkin may normally select for mitochondria containing wild-type mtDNA by mediating the elimination of dysfunctional mitochondria containing mutant mtDNA. Loss of Parkin function in the substantia nigra may cause early onset parkinsonism by allowing excessive accumulation of deleterious mutant mtDNA. Importantly, these findings also indicate that endogenous Parkin levels may be limiting for the negative selection of dysfunctional mitochondria in at least some cell types, and that up-regulation of Parkin expression may be therapeutically beneficial for hereditary and somatically acquired mitochondrial diseases.
Materials and Methods
Cells and Culture.
The Cytb3.0 cybrid cell line was a gift from Carlos Moraes (University of Miami School of Medicine, Miami) (24) and the COXICA65 and 143B Rho0 cell lines has been described previously (25). The 143B (ATCC), cybrid, and Rho0 cells were cultured in DMEM containing high glucose (4.5 g/L), 2 mM sodium pyruvate, 1 mM L-glutamate, and 50 μg/mL of uridine (Sigma). HeLa cells used for expressing Twinkle (a gift from Hans Spelbrink, University of Tampere, Tampere, Finland) were cultured in DMEM medium containing high glucose (4.5 g/L), 1 mM sodium pyruvate, 2 mM L-glutamate, 10 mM hepes, and 1× nonessential amino acids. All cell-culture supplies were obtained from GIBCO unless otherwise indicated.
PCR-RFLP.
PCR-RFLP was performed as described previously (25). Total DNA was extracted from cells by DNeasy blood and tissue kit (QIAGEN). Next, 450 ng of DNA was used as a template for PCR. The forward primer, 5′-ggcttcctagggtttatcgtgtgagcac-3′, and the reverse primer, 5′-ggccacctacggtgaaaagaaagatgaagc-3′, were used for the COXICA65 cybrid. The forward primer, 5′-atgaccccaatacgcaaaattaac-3′, and the reverse primer, 5′-ttcatcatgcggagatgttggatg -3′, were used for the Cytb3.0 cybrid. PCR amplification was performed as follows: the first step at 94 °C for 2 min; 30 cycles each at 94 °C for 20 s, 58 °C for 30 s and 72 °C for 30 s; and final step at 72 °C for 7 min. PCR product was purified using a gel extraction kit (QIAGEN). Next, 300 ng purified PCR product was digested with AluI (COXICA65) and AseI (Cytb3.0) (NEB) in 40 μL. Eight microliters (60 ng) of digested product was loaded and separated in a 10% TBE polyacrylamide gel (Invitrogen). After electrophoresis, the gel was stained with 2 μg/mL ethidium bromide for 10 min, destained with water for 5 min and imaged. For 32P-labeling, 1 μL (20 μCi) of [α-32P]-dCTP (Perkin-Elmer) was added in each sample before the last PCR cycle and processed as the described above. After electrophoresis, the radioactive signals were detected with a phosphoimage system and scanned by STORM 860 (GE Healthcare). The intensity was quantified by the ImageQuant 5.1 program (GE Healthcare).
Cytochrome c Oxidase Activity Assay.
Cells were collected in HBSS (GIBCO) and lysed in extraction buffer [10 mM hepes, 10 mM NaCl2, 1.5 mM MgCl2, 4 mM NaF, 100 μM NaOVac, 1× protease inhibitor mixture (Roche)] by 20 passages through a 25-gauge needle. Samples were spun at 664 × g for 5 min to pellet nuclei. The supernatant was then spun at 6,797 × g for 10 min to pellet the mitochondrial rich heavy membrane fraction. The pellet was washed with extraction buffer once and resuspended in 1× enzyme dilution buffer containing 1 mM n-dodecyl-β-maltoside (CYTOCX1 kit, Sigma). The COXI activity assay was performed according to the manufacturer's instructions. Fresh 10 μg of mitochondrial protein and 10 μM Ferrocytochrome-c substrate were used in each assay. SpectraMax plus384 (Molecular Devices) was used to measure A550 every 10 s for 1 min. The Vmax of each sample was calculated by SOFTmax PRO (Molecular Devices). The mean and SD were calculated from three experiments.
Supplementary Material
Acknowledgments
We thank H. Spelbrink for Twinkle plamids, I. F. de Coo and C. T. Moraes for Cytb3.0 cybrid cells, D. Maric for FACS, and C. Smith for confocal microscopy. This work was supported by the National Institute of Neurological Disorders and Stroke Intramural Research Program at the National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.0914569107/-/DCSupplemental.
References
- 1.Taylor RW, Turnbull DM. Mitochondrial DNA mutations in human disease. Nat Rev Genet. 2005;6:389–402. doi: 10.1038/nrg1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wallace DC, Fan W. The pathophysiology of mitochondrial disease as modeled in the mouse. Genes Dev. 2009;23:1714–1736. doi: 10.1101/gad.1784909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kraytsberg Y, et al. Mitochondrial DNA deletions are abundant and cause functional impairment in aged human substantia nigra neurons. Nat Genet. 2006;38:518–520. doi: 10.1038/ng1778. [DOI] [PubMed] [Google Scholar]
- 4.Bender A, et al. High levels of mitochondrial DNA deletions in substantia nigra neurons in aging and Parkinson disease. Nat Genet. 2006;38:515–517. doi: 10.1038/ng1769. [DOI] [PubMed] [Google Scholar]
- 5.Schapira AH. Mitochondria in the aetiology and pathogenesis of Parkinson's disease. Lancet Neurol. 2008;7:97–109. doi: 10.1016/S1474-4422(07)70327-7. [DOI] [PubMed] [Google Scholar]
- 6.Gilkerson RW, Schon EA, Hernandez E, Davidson MM. Mitochondrial nucleoids maintain genetic autonomy but allow for functional complementation. J Cell Biol. 2008;181:1117–1128. doi: 10.1083/jcb.200712101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bayona-Bafaluy MP, Blits B, Battersby BJ, Shoubridge EA, Moraes CT. Rapid directional shift of mitochondrial DNA heteroplasmy in animal tissues by a mitochondrially targeted restriction endonuclease. Proc Natl Acad Sci USA. 2005;102:14392–14397. doi: 10.1073/pnas.0502896102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kitada T, et al. Mutations in the Parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392:605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- 9.Shimura H, et al. Familial Parkinson disease gene product, Parkin, is a ubiquitin-protein ligase. Nat Genet. 2000;25:302–305. doi: 10.1038/77060. [DOI] [PubMed] [Google Scholar]
- 10.Yang Y, et al. Mitochondrial pathology and muscle and dopaminergic neuron degeneration caused by inactivation of Drosophila Pink1 is rescued by Parkin. Proc Natl Acad Sci USA. 2006;103:10793–10798. doi: 10.1073/pnas.0602493103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Greene JC, et al. Mitochondrial pathology and apoptotic muscle degeneration in Drosophila Parkin mutants. Proc Natl Acad Sci USA. 2003;100:4078–4083. doi: 10.1073/pnas.0737556100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clark IE, et al. Drosophila pink1 is required for mitochondrial function and interacts genetically with Parkin. Nature. 2006;441:1162–1166. doi: 10.1038/nature04779. [DOI] [PubMed] [Google Scholar]
- 13.Park J, et al. Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature. 2006;441:1157–1161. doi: 10.1038/nature04788. [DOI] [PubMed] [Google Scholar]
- 14.Palacino JJ, et al. Mitochondrial dysfunction and oxidative damage in Parkin-deficient mice. J Biol Chem. 2004;279:18614–18622. doi: 10.1074/jbc.M401135200. [DOI] [PubMed] [Google Scholar]
- 15.Stichel CC, et al. Mono- and double-mutant mouse models of Parkinson's disease display severe mitochondrial damage. Hum Mol Genet. 2007;16:2377–2393. doi: 10.1093/hmg/ddm083. [DOI] [PubMed] [Google Scholar]
- 16.Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buchet K, Godinot C. Functional F1-ATPase essential in maintaining growth and membrane potential of human mitochondrial DNA-depleted rho degrees cells. J Biol Chem. 1998;273:22983–22989. doi: 10.1074/jbc.273.36.22983. [DOI] [PubMed] [Google Scholar]
- 18.Appleby RD, et al. Quantitation and origin of the mitochondrial membrane potential in human cells lacking mitochondrial DNA. Eur J Biochem. 1999;262:108–116. doi: 10.1046/j.1432-1327.1999.00350.x. [DOI] [PubMed] [Google Scholar]
- 19.King MP, Attardi G. Human cells lacking mtDNA: Repopulation with exogenous mitochondria by complementation. Science. 1989;246:500–503. doi: 10.1126/science.2814477. [DOI] [PubMed] [Google Scholar]
- 20.Spelbrink JN, et al. Human mitochondrial DNA deletions associated with mutations in the gene encoding Twinkle, a phage T7 gene 4-like protein localized in mitochondria. Nat Genet. 2001;28:223–231. doi: 10.1038/90058. [DOI] [PubMed] [Google Scholar]
- 21.Baloh RH, Salavaggione E, Milbrandt J, Pestronk A. Familial parkinsonism and ophthalmoplegia from a mutation in the mitochondrial DNA helicase twinkle. Arch Neurol. 2007;64:998–1000. doi: 10.1001/archneur.64.7.998. [DOI] [PubMed] [Google Scholar]
- 22.Van Hove JL, et al. Finding twinkle in the eyes of a 71-year-old lady: A case report and review of the genotypic and phenotypic spectrum of TWINKLE-related dominant disease. Am J Med Genet A. 2009;149A:861–867. doi: 10.1002/ajmg.a.32731. [DOI] [PubMed] [Google Scholar]
- 23.Wanrooij S, Goffart S, Pohjoismäki JL, Yasukawa T, Spelbrink JN. Expression of catalytic mutants of the mtDNA helicase Twinkle and polymerase POLG causes distinct replication stalling phenotypes. Nucleic Acids Res. 2007;35:3238–3251. doi: 10.1093/nar/gkm215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rana M, de Coo I, Diaz F, Smeets H, Moraes CT. An out-of-frame cytochrome b gene deletion from a patient with parkinsonism is associated with impaired complex III assembly and an increase in free radical production. Ann Neurol. 2000;48:774–781. [PubMed] [Google Scholar]
- 25.Bruno C, et al. A stop-codon mutation in the human mtDNA cytochrome c oxidase I gene disrupts the functional structure of complex IV. Am J Hum Genet. 1999;65:611–620. doi: 10.1086/302546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scaduto RC, Jr, Grotyohann LW. Measurement of mitochondrial membrane potential using fluorescent rhodamine derivatives. Biophys J. 1999;76:469–477. doi: 10.1016/S0006-3495(99)77214-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Narendra DP, et al. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010;8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsuda N, et al. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J Cell Biol. 2010;189:211–221. doi: 10.1083/jcb.200910140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vives-Bauza C, et al. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc Natl Acad Sci USA. 2010;107:378–383. doi: 10.1073/pnas.0911187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Geisler S, et al. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol. 2010;12:119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- 31.McCormick AL, Smith VL, Chow D, Mocarski ES. Disruption of mitochondrial networks by the human cytomegalovirus UL37 gene product viral mitochondrion-localized inhibitor of apoptosis. J Virol. 2003;77:631–641. doi: 10.1128/JVI.77.1.631-641.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pawlyk AC, et al. Novel monoclonal antibodies demonstrate biochemical variation of brain Parkin with age. J Biol Chem. 2003;278:48120–48128. doi: 10.1074/jbc.M306889200. [DOI] [PubMed] [Google Scholar]
- 33.Shoubridge EA. Segregation of mitochondrial DNAs carrying a pathogenic point mutation (tRNA(leu3243)) in cybrid cells. Biochem Biophys Res Commun. 1995;213:189–195. doi: 10.1006/bbrc.1995.2115. [DOI] [PubMed] [Google Scholar]
- 34.Twig G, et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008;27:433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.