Mitchell et al. (1) recently reported a single heterozygous variant (R199W) in exon 7 of D amino oxidase (DAO) gene in a family with amyotrophic lateral sclerosis (ALS) that showed a probable linkage to the 12q chromosome (Zmax of 2.7 for d12s1646 by bipoint and 1.9 for d12S330 by multipoint analysis) (1). Characteristics of this variant were in favor of its causative role in this family: it was found in three affected members and one obligate carrier, it was absent in 780 controls matched for ethnic background, and it was relatively well-conserved among species. Moreover, cellular studies performed with lentiviral vectors expressing mutant or wild-type DAO protein showed that R199W DAO led to ubiquitin aggregate formation and decreased cellular viability in vitro. Overall, the identification of a second rare variant in another familial amyotrophic lateral sclerosis (FALS) would be of great interest to firmly incriminate DAO in ALS. In an effort to strengthen these data, we sequenced the 10 coding exons (exons 2–11) of DAO and exon–intron junctions (Ensembl reference sequence ENSG00000110887) in 126 unrelated patients living in France presenting probable or definite ALS with at least one ALS-affected relative. Spinal and bulbar onset was noticed in 80.2% and 29.8% of these patients, respectively. The mean disease onset was 58 ± 9 y, and the mean disease duration was 3 ± 2 y. Protocols were approved by the Medical Research Ethics Committee of Assistance Publique-Hôpitaux de Paris. All participants signed a consent form for the research. Mutations in copper/zinc superoxide dismutase (SOD1), angiogenin (ANG), vesicle associated membrane protein associated protein B (VAPB), TAR DNA binding protein 43 (TARDBP), and fused in sarcoma (FUS) have been excluded in these patients (2).
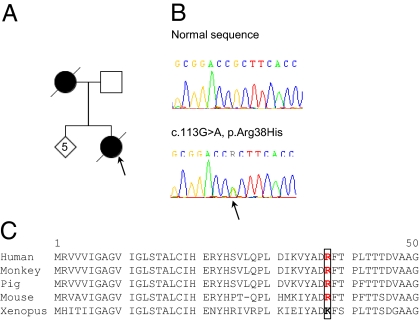
We identified another heterozygous variant in exon 2 of DAO (c.113G > A; p.Arg38His) (Fig. 1B) affecting a residue that is relatively conserved (Fig. 1C) in a woman who presented lower limb disease onset at 42 y and had a rapid disease duration of 7 mo. Her affected mother died at 67 y (Fig. 1A). Exon 2 of DAO was sequenced in control samples obtained from 508 healthy individuals of Caucasian origin. The c.113G > A variant was detected in a man (1/1,016 chromosomes) aged 66 y, suggesting that this variant is a rare polymorphism. We did not identify any other mutation in our FALS patients. Our data do not confirm that mutations in DAO cause ALS. However, they do not exclude that rare ALS families may be associated to mutations in DAO. This rarity has to be underlined. Indeed, our population of FALS initially was of 162 FALS. We have previously identified a mutation for 36 FALS of this cohort (20 in SOD1, 7 in TARDBP, 7 in FUS, 1 in ANG, and 1 in VAPB) (2). Taking into account that Mitchell et al. (1) identified only the R199W variant among 199 tested FALS patients, the mutational frequency of this gene is, until now, ∼0.3% (1/199 + 126) in the FALS population without mutation in already known genes. This finding questions the place of DAO screening in molecular diagnosis of FALS. Moreover, the pathogenicity of the R199W mutation would be definitively confirmed by developing an animal model (knock-in) carrying this variant.
Fig. 1.
Detection of the c.113G > A variant in one FALS. (A) Pedigree of the patient carrying the c.113G > A variant (arrow). DNA was available for the index case only. (B) Part of chromatogram of exon 2 showing the c113G > A variant. (C) Sequence alignment of part of the DAO amino acids (1–50) from diverse species. The position of the Arg38 (boxed) is in red. Sequences used include Homo sapiens (ENSP00000228476), Macaca mulatta (ENSMMUP00000009050), Sus scrofa (ENSSSCP00000010609), Mus musculus (ENSMUSP00000107911), and Xenopus tropicalis (ENSXETP00000048267).
Footnotes
The authors declare no conflict of interest.
References
- 1.Mitchell J, et al. Familial amyotrophic lateral sclerosis is associated with a mutation in D-amino acid oxidase. Proc Natl Acad Sci USA. 2010;107:7556–7561. doi: 10.1073/pnas.0914128107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Millecamps S, et al. SOD1, ANG, VAPB, TARDBP, and FUS mutations in familial amyotrophic lateral sclerosis: Genotype-phenotype correlations. J Med Genet. 2010 doi: 10.1136/jmg.2010.077180. doi:10.1136/jmg.2010.077180. [DOI] [PubMed] [Google Scholar]