Abstract
In the study of long-term memory, how memory persists is a fundamental and unresolved question. What are the molecular components of the long-lasting memory trace? Previous studies in Aplysia and Drosophila have found that a neuronal variant of a RNA-binding protein with a self-perpetuating prion-like property, cytoplasmic polyadenylation element binding protein, is required for the persistence of long-term synaptic facilitation in the snail and long-term memory in the fly. In this study, we have identified the mRNA targets of the Drosophila neuronal cytoplasmic polyadenylation element binding protein, Orb2. These Orb2 targets include genes involved in neuronal growth, synapse formation, and intriguingly, protein turnover. These targets suggest that the persistent form of the memory trace might be comprised of molecules that maintain a sustained, permissive environment for synaptic growth in an activated synapse.
Keywords: protein synthesis, synaptic plasticity, memory
Current models for the cellular basis of long-term memory involve both the activity-dependent formation and elimination of synapses as well as alterations to the strength of preexisting synapses. The activity-dependent changes in synapse numbers and synaptic efficacy require the synthesis of a new set of proteins locally at the synapse (1–6). While investigating how synaptic stimulation activates local protein synthesis, we have identified a neuron-specific form of cytoplasmic polyadenylation element binding protein (CPEB) (7).
CPEB belongs to a family of RNA-binding proteins, and some of the family members bind to a U-rich sequence with a general structure UUUUUAU (known as CPE element) in the 3′ end of many cellular mRNAs (8). This RNA binding can either activate or repress the translation of their target mRNAs (8). We have found that in Aplysia, synaptic activity of a neuronal variant of CPEB is not required for the initial serotonin-dependent changes in synaptic efficacy or for the growth of new synapses, but it is essential for maintaining these changes beyond 24 h (7, 9). Furthermore, both Aplysia CPEB and a Drosophila CPEB, Orb2, have the ability to adopt different conformational states, one of which is dominant and self-perpetuating, reminiscent of a prion-like protein (10, 11). Based on these observations, we proposed that the persistence of memory requires the recruitment of a prion-like self-sustaining active form of neuronal CPEB (Orb2) only at activated synapses, and this active form of CPEB, in turn, maintains the memory trace through continued regulation of a specific set of synaptic proteins (12). Consistent with this hypothesis, Keleman et al. (13) have shown that reduction in Drosophila Orb2 levels selectively effects long-term stabilization of memory but not learning or short-term memory. These observations suggested that a long-lasting memory trace is at least partly comprised of Orb2 targets, which are still not known.
In this study, using a candidate gene approach and a genome-wide analysis, we have identified 28 mRNA targets of Drosophila Orb2. A number of these targets are involved in regulating the growth or formation of synapses, which is consistent with the idea that the persistent form of memory requires new synapse formation. In addition, we found that a number of Orb2 targets are proteases or components of the ubiquitin-mediated protein-degradation pathway. These findings raise the possibility that Orb2-dependent regulation of protein-turnover creates a permissive environment for synaptic growth only at the activated synapse.
Results
Drosophila Orb2 Binds to Genes Implicated in Long-Term Memory.
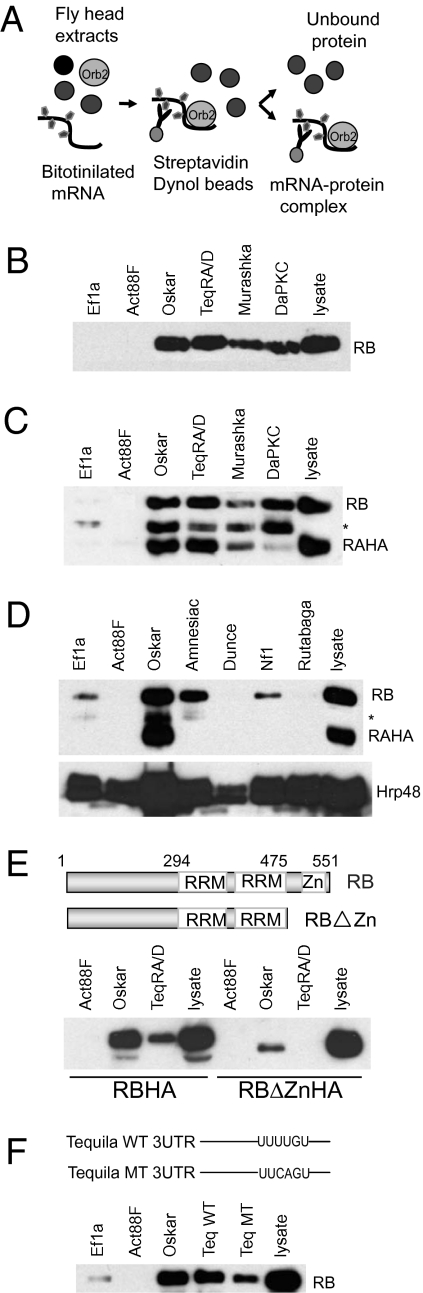
Because Orb2 is involved in regulating memory processes (13), we first took a candidate gene approach and analyzed a number of genes that have previously been implicated in short- or long-term memory in Drosophila (14, 15). We reasoned that bona fide targets of Orb2 should satisfy at least two criteria. First, Orb2 should bind to the 3′UTR of the target mRNA. Second, Orb2 should activate or repress the translation of the target gene. To score for Orb2 binding, we employed a pull-down assay where biotin-labeled mRNA was used to pull down endogenous Orb2 from Drosophila head extract (Fig. 1A). Drosophila Orb2 has two isoforms: Orb2RA and Orb2RB. The two forms differ in their N-terminal sequence, but they have the same RNA-binding domain at the C-terminal end. Whereas the mRNAs for both isoforms are present in the adult Drosophila head, only Orb2RB protein can be readily detected in the naïve brain (Fig. S1). Therefore, we used wild-type flies for Orb2RB (Fig. 1B) and flies expressing a HA-tagged Orb2RA (Elav-Gal4::UAS-Orb2RAHA) (Fig. 1 C and D) for Orb2RA. In the absence of known Orb2 targets, we used the 3′UTR of Oskar gene, a target of the other CPEB family member in Drosophila (Orb1) as a possible positive control (16). The Oskar gene is required for stable long-term memory in Drosophila (14).
Fig. 1.
Drosophila Orb2 binds to the 3′UTR of Oskar, Tequila, DaPKC, and Murashka. (A) A schematic diagram shows the experimental design for the mRNA protein pull-down assay. (B) Endogenous Orb2RB and (C) exogenously expressed Orb2RAHA (Elav-GAL4:: UAS-Orb2RAHA) specifically bind to the 3′UTR of Oskar, Murashka, Tequila, and DaPKC. Specificity of the assay was determined by using nonspecific RNA EF1α and Actin88F. The asterisk indicates an Orb2 immunoreactive polypeptide that is most likely derived from the degradation of Orb2RB, because it was not observed when pull down was performed using orb2 null fly heads (Fig. S1). (D) Orb2RAHA binds only to long-term memory-related genes. The endogenous Orb2RB binds to the intermediate-term memory gene Amnesiac but not the short-term memory genes Dunce, Rutabaga, and Nf1. The amount of Orb2RB bound to Nf1 is similar to control EF1α mRNA and thus, is considered as background binding. Orb2RAHA binds only to the control Oskar mRNA but none of the other genes. The same blot was probed with RNA-binding protein Hrp48 to ensure the presence of the probe RNA. (E) The conserved Zn-finger domain in the C-terminal end of Orb2RB is required for mRNA binding. (Upper) The RRMs and the Zn-finger domain are indicated. The numbers indicate amino acid numbers. (Lower) The pull down from head extracts of Orb2RB lacking the Zn-finger domain (Act-GAL4::UAS-Orb2RBΔZnHA) or wild-type Orb2RB (Act-GAL4:: UAS-Orb2RBHA) were probed with anti-HA antibodies. The lysates show the relative amounts of Orb2 proteins in head extracts used in the pull down. (F) The CPE element is not essential for Orb2 binding. Mutation of the putative CPE element in Tequila 3′UTR UUUUGU (TeqWT) to UUCAGU (TeqMT) reduces but does not abolish the endogenous Orb2RB binding.
Using these binding assays, we obtained three potential Orb2 targets that are believed to be required for stable long-term memory in Drosophila: Tequila, a homolog of human neurotrypsin, DaPKC, a homolog of mouse atypical protein kinase C, and Murashka, a putative E3 ubiquitin ligase (14, 17, 18). When we incubated the biotin-labeled 3′UTRs of Tequila, DaPKC, or Murashka with head extract, all three genes selectively pulled down Orb2RB and Orb2RAHA (Fig. 1 B and C). In contrast, the 3′UTR of short-term memory genes Dunce, Rutabaga, and Nf1 or the intermediate-term memory gene amnesiac did not pull down Orb2RAHA, and only Amnesiac pulled down Orb2RB from total brain lysate (Fig. 1D). The lack of binding is not caused by probe instability, because we found the abundant RNA-binding protein Hrp48 with all mRNA.
Does Orb2 bind directly to the mRNAs, or is it recruited indirectly to the RNA through other proteins? The Orb2 gene, like other CPEB family members, has two RNA recognition motifs (RRM) and a conserved Zn-finger domain (19). Studies with other CPEB family members have shown that removal of the Zn-finger domain alone reduces RNA binding by more than 90% compared with that of wild-type proteins (19). To test direct binding of Orb2, we used flies expressing Orb2RB lacking the Zn-finger domain (RBΔZnHA) and found that, in the absence of the Zn-finger domain (RBΔZnHA), the binding to oskar 3′UTR was greatly reduced and binding to tequila 3′UTR was completely abolished (Fig. 1E); this suggests that, similar to other CPEB family members, Orb2RB directly binds to target mRNA. However, because of the insolubility of the recombinant protein, we could not perform an in vitro binding assay and therefore, cannot rule out the possibility that the Orb2 binding to the mRNA is indirect. The 3′UTRs of the Tequila, Murashka, and DaPKC all contain a putative U-rich CPE element UUUUG/AU. Finally, to test if the U-rich CPE element is necessary for Orb2 binding, we examined the Tequila 3′UTR because of its short length (107 nucleotides), the presence of only one putative CPE element, and its robust binding to both forms of Orb2. However, mutating the UUUUGU to UUCAGU only slightly reduced the binding of Orb2RB to Tequila 3′UTR (Fig. 1F), suggesting that other sequences are involved in Orb2 binding. From these results, we conclude that both Orb2RA and Orb2RB bind to the 3′UTR of genes that are involved in memory formation, particularly long-term memory. Moreover, the binding of Orb2RB to Amnesiac suggests that, in addition to a common set of targets, Orb2RA and Orb2RB might have distinct mRNA targets.
Target mRNA Translation Is Suppressed by Drosophila Orb2 in a Heterologous System.
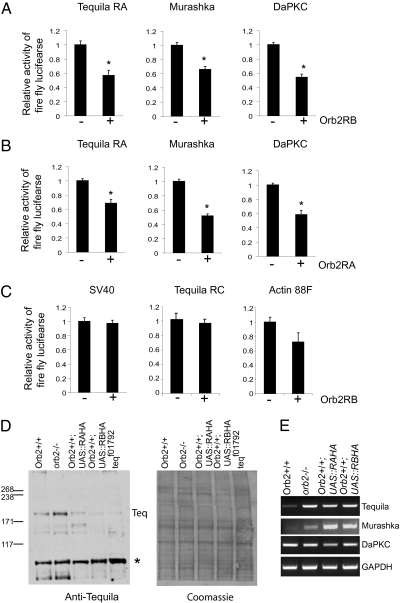
Does binding of Orb2 alter the translation of the target mRNA? To answer this question, we used a dual-luciferase reporter assay in Drosophila S2 cells, which our Western analysis showed lacks endogenous Orb2 protein. The 3′UTR of each candidate target gene was linked to firefly luciferase, and a control SV40 3′UTR was linked to renilla luciferase; the ratio of firefly to renilla luciferase activity in the presence (+) or absence (−) of Orb2RA or Orb2RB was used to measure translational regulation. In the dual-luciferase assay, expression of either Orb2RA or Orb2RB reduced the activity of firefly luciferase bearing the 3′ UTR of all three candidate target mRNAs by ∼50% (Student t test; P value < 0.05) (Fig. 2 A and B). In contrast, the activity of firefly luciferase bearing three control UTRs that did not bind to Orb2 was unchanged, suggesting the Orb2 expression does not cause a general repression in gene expression (Fig. 2C). The reduction of the luciferase activity was caused by translational repression, because the amount of the reporter mRNA was not reduced in the presence of Orb2 (Fig. S2). Thus, both isoforms of Orb2 can act as a repressor of translation when overexpressed in S2 cells. It is possible that repression is an outcome of overexpression in a heterologous cell system rather than a reflection of the endogenous Orb2 activity in the nervous system. Nonetheless, the selective translational repression suggests that Tequila, Murashka, and DapKC mRNAs are indeed targeted by Orb2.
Fig. 2.
The long-term memory-related genes Tequila, DaPKC, and Murashka are translationally repressed by Orb2 in S2 cells. When overexpressed, both Orb2RB (A) and Orb2RA (B) selectively suppress the translation of target mRNA in S2 cells. The relative activity of firefly luciferase over renilla luciferase activity in the presence (+) or absence (−) of Orb2RB (A) or Orb2RA (B) is plotted. (C) The translation of firefly luciferase-bearing control SV40 3′UTR or the 3UTR of Tequila RC isoform or Actin88F were not affected. The Tequila gene has five predicted isoforms of which Tequila RC has its own 3′UTR. Both forms of Orb2 bind to the common 3′UTR but not to the RC 3′UTR. The ratios were obtained from at least three independent experiments for each construct, and each experiment consists of three independent transfections. The significance was tested by Student t test, and a P value < 0.05 was considered significant and is indicated with an asterisk. The error bars indicate SEM. (D) The long-term memory gene Tequila is regulated by Orb2 in the adult fly brain. Western blot using anti-Tequila antibody shows that, in orb2 null mutant (orb2−/−), Tequila protein level is up-regulated, whereas transgenic overexpression of Orb2RA (Orb2+/+;ELAVGAL4:UAS-RAHA) and Orb2RB (Orb2+/+; ELAVGAL4:UAS-RBHA) results in about 50% reduction of Tequila expression levels. Teqf10792 is a hypomorphic allele of the Tequila gene and serves as antibody control. The asterisk indicates a nonspecific immunoreactive band, and coomassie staining of the same membrane serves as a loading control. (E) The steady-state level of Tequila and Murashka mRNA levels are altered on misregulation of Orb2. Tequila and Murashka mRNA levels are increased in orb2 null mutants and in Orb2 overexpressing flies. RT-PCR was used to analyze the Tequila, Murashka, and DaPKC mRNA on total RNA isolated from indicated genetic backgrounds. GAPDH served as PCR control. DaPKC mRNA level was not changed, implying that mRNA levels of all Orb2 targets are not susceptible to changes in Orb2 protein level.
Identification of Putative Orb2 Targets from a Genome-Wide Screen.
Because our candidate gene approach was limited in scope, we next set out to perform an unbiased genome-wide screen to identify Orb2 targets. The conventional biochemical approaches were not successful, owing to the insolubility of recombinant Orb2 and the low amount of endogenous Orb2. Therefore, we took advantage of an observation that we made during the course of this work. When we analyzed the level of Tequila protein in adult Drosophila brains, we observed an up-regulation in the level of Tequila in orb2 null mutants (SI Materials and Methods has the generation and characterization of the orb2 null mutants) and a reduction in Orb2RA and Orb2RB overexpressing flies (relative level compared with wild-type brain: Orb2RA = 0.49 ± 0.08 and Orb2RB = 0.63 ± 0.01; P value < 0.05), consistent with the idea that Orb2 acts as a repressor of Tequila translation (Fig. 2D Left). However, unexpectedly, we also observed an up-regulation of Tequila mRNA in orb2 null as well as Orb2RA and Orb2RB overexpressing flies (Fig. 2E). The change in Tequila mRNA levels, either in the absence or overexpression of Orb2 suggested that systemic chronic alteration of Orb2 levels, in addition to effecting translation, also affects the steady-state level of mRNA of some (but not all) targets either as a result of compensatory transcriptional modulation or changes in stability. Consistent with our Tequila results, we found that Murashka mRNA levels were also altered in orb2 null and overexpressing flies (Fig. 2E). Thus, we reasoned that by profiling the relative mRNA levels of genes in wild-type and orb2 null brains, we might be able to identify additional targets of Orb2. To this end, we purified polyA mRNA from the adult fly heads and compared the transcriptional profiles between wild-type and orb2 null mutants using the Affymetrix Drosophila GeneChip array (version 2).
We found 371 genes from the entire dataset (18,500 loci) with steady-state levels significantly altered (P value < 0.05) in orb2 null brains compared with wild-type brains (details in Materials and Methods). The Fisher's exact test for overrepresentation of gene ontology (GO) terms (20) revealed that functional categories that are overrepresented in orb2 null flies include genes involved in proteolysis, mating behavior, reproduction, and surprisingly, defense and immune responses (Table S1). The defense and immune response genes were also identified by others in an attempt to identify the targets of Drosophila RNA-binding protein Pumilio (21). The significance of the enrichment of immune-response genes in our dataset is not immediately apparent. Moreover, it is also not apparent how many of these changes are caused by secondary consequences of the Orb2 deletion.
To begin to identify potential targets of Orb2 in the adult nervous system, we filtered the dataset by applying several additional criteria and focused on two sets of genes. The restricted expression of Orb2 in the adult nervous system is sufficient to rescue the long-term memory deficit of orb2 mutant flies (13) and Aplysia CPEB is required for the stabilization of the newly grown synapse (9). For the first set of genes, we selected those that are known to be associated with cellular growth or have neuronal function. For the second set of genes, we focused on genes involved in proteolysis for the following reasons. First, the genes involved in proteolysis were significantly altered (P value = 4.69e-4) in the orb2 null mutant compared with wild type (Table S1). Second, in our candidate gene approach, 2 of 3 genes that we identified are involved in proteolysis: Murashka, a Really Interesting New Gene (RING)-finger protein with potential E3-ubiqutin ligase activity and Tequila, a serine-type endopeptidase. Finally, studies on the neuromuscular junction synapse (NMJ) suggested that an ubiquitin-dependent mechanism regulates synaptic growth and function in Drosophila (22). We next verified the gene-chip data by RT-PCR analysis and selected only those genes that confirmed the differences first detected by microarray. Finally, we selected a set of 29 putative Orb2 targets, of which 19 were genes with known function and 10 were uncharacterized genes with functions predicted to be involved in cellular growth or proteolysis (Table S2).
Confirmation of Orb2 Targets.
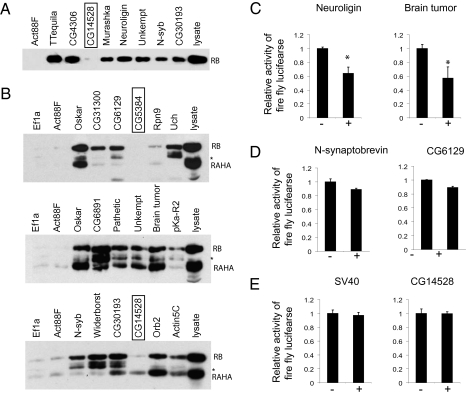
Not all up- or down-regulated genes in the orb2 null animals need to be Orb2 targets, and thus, to obtain the real targets out of these putative targets, we applied stringent criteria. Because the obligatory step in Orb2-dependent translation is binding of the protein to the mRNA, we tested Orb2RA and Orb2RB binding to the 3′UTRs of all putative target genes using the pull-down assay (Fig. 3 A and B and Fig. S3). Of 29 genes tested, we found 24 genes that bound to Orb2, 13 of which are common for both Orb2RA and Orb2RB, 10 are Orb2RB-specific, and 1 (CG14528) is Orb2RA-specific (Fig. 3 A and B and Fig. S3). We observed differences in the extent of Orb2 binding to different mRNAs, which might be a reflection of number of binding sites present in each mRNA.
Fig. 3.
Orb2 binds to the bona fide target mRNA. (A) A representative blot of binding of endogenous Orb2RB protein to the targets isolated from GeneChip analysis. The gene CG14528 did not bind to Orb2RB. (B) The common and distinct targets of Orb2RA and Orb2RB were obtained from GeneChip analysis. The asterisk indicates an Orb2 immunoreactive polypeptide, which is most likely derived from the degradation of Orb2RB. The genes marked with a box did not bind to either form of Orb2. Fig. S3 shows binding to additional targets. (C–E) Orb2RB represses the translation of the majority of targets in S2 cells. (C) Representative plots of Orb2RB-dependent translational suppression of Orb2 targets. Fig. S3 shows additional Orb2 targets. (D) The translation of the majority of the target genes was reduced except Synaptobrevin and CG6129, which showed modest but statistically insignificant suppression. P value < 0.05 was considered significant in a Student t test. (E) The translation of firefly luciferase with SV40 3′UTR or the 3′UTR from CG14528, which did not bind to Orb2RB, was not affected by Orb2RB.
The high percentage of Orb2 binding that we observed in the selected genes is not simply caused by widespread Orb2 binding. In our candidate gene approach, we tried several genes with known synaptic function, and none of them bound to Orb2; subsequently, we found that these genes were also not altered in the microarray. In addition, we tested genes that were identified in long-term memory mutant screens (14) but were unchanged in our microarray study. We selected six genes with a defined 3′UTR greater than 300 nucleotides in length and rich in U-residues. We found that none of these UTRs bound to Orb2RB and only one bound to Orb2RA, suggesting that the use of the microarray and additional filtering indeed enriched for Orb2 targets (Fig. S1). Finally, we observed that, unlike Orb2, Drosophila Orb1, which has RNA-binding RRM domain and Zn-finger domain similar to Orb2, shows more widespread nonspecific RNA binding under similar conditions (Fig. S1). These observations suggest that the Orb2 binds specifically to a select set of RNA. Interestingly, we also found that that both isoforms of Orb2, RA and RB, can bind to their own common 3′UTR (Fig. 3B).
Next, we sought to determine if these genes are expressed in brain structures that are known to be required for long-term memory in Drosophila, such as the mushroom body. To this end, we performed RNA in situ hybridization and observed that brain tumor, unkempt, actin5C, cg4306, cg6129, and cg12769 were expressed in the mushroom body Kenyon cells, antennal nerve, and optic-lobe region (Fig. S4) (7, 13). We failed to detect the rest of the target mRNAs by in situ hybridization, perhaps because of low expression levels.
Finally, we sought to determine if the mRNAs that bound to Orb2 are also translationally regulated by Orb2. Because Orb2RB bound to almost all of the targets, we performed the S2 cell-based translational assay in the presence or absence Orb2RB. We observed that translation of the firefly luciferase mRNAs bearing the 3′UTR of the majority (Fig. 3C and Fig. S3), but not all (Fig. 3D), of the candidate targets were translationally suppressed in the presence of Orb2RB (Student t test; P value < 0.05). Firefly luciferase with a control 3′UTR or harboring the 3′UTR of CG14528, which did not bind to Orb2RB, did not show any change in translation (Fig. 3E). The failure or modest suppression of some of the targets might be caused by weak binding affinity of Orb2RB to some targets in S2 cells or the fact that translational regulation of some mRNAs requires additional components that are missing in S2 cells.
These studies lead us to several conclusions with respect to Orb2 target selection. First, these results confirmed our earlier finding that, in addition to a common set of targets, Orb2RA and Orb2RB can bind a distinct set of mRNAs, although they carry the same RNA-binding domain. Second, unlike the canonical CPEB, the U-rich CPE-like element is not required for Orb2 binding. We observed that a number of targets, but not all, have a U-rich sequence, UUUUG/AU, similar to a CPE element in the 3′UTR (Table S2). However, some genes with the sequence UUUUG/AU did not bind to Orb2. We did not find any additional sequence motifs overrepresented in this set of 28 targets by Multiple Em for Motif Elicitation motif-search analysis. It is possible that, in addition to the primary sequence, secondary structures of RNA might be an important determinant of Orb2 binding, which is the case for mouse CPEB3 and CPEB4 (23). Third, the expression of Orb2, particularly Orb2RB, at every stage of development implies that regulation of some Orb2 targets is required for the proper development of Drosophila and that Orb2 serves additional functions outside of the adult nervous system. Finally, the binding to its own 3′UTR suggests that Orb2 can regulate its own synthesis.
Discussion
In this study, we used a candidate gene approach combined with a genome-wide screen to identify potential Drosophila Orb2 targets (Table 1). Surprisingly, the use of orb2 null flies, despite inherent limitations, turned out to be quite useful in identifying Orb2 targets. Clearly, we have not identified all of the Orb2 targets, and it is yet to be determined if the protein levels of these targets are indeed altered in response to a learning-related stimulus in an Orb2-dependent manner. However, the identification of these targets provides clues to Orb2 function and a plausible molecular makeup of the long-lasting memory trace.
Table 1.
Targets of Drosophila Orb2RA and Orb2RB
| Synaptic growth/stability | Synaptic activity | Proteolysis | Unknown |
| Neuroligin | N-Syb | Uch | CG30193 |
| Still life | Pka-R2 | Rpt1 | CG14528 |
| Brain tumor | Widerborst | Rpt4 | CG6891 |
| Capulet | Pathetic | Rpn9 | CG12769 |
| Glaikit | Unkempt | CG31300 | |
| Act5c | Tequila | CG6129 | |
| Branchless | Murashka | CG4306 | |
| DaPKC | CG10824 | ||
| Orb2 |
The targets are grouped based on their known functions either in the nervous system or in other cell types. The cellular functions of these targets are cell polarity and asymmetric growth, DaPKC, widerbrost (a protein phosphatase) (30), glaikit (a phospholipase D) (31), cytoskeleton remodeling, actin5C, capulet (adenylyl cyclase associated protein) (34), still life (Guanine nucleotide exchange factor) (35), growth factors, branchless (FGF) (39), translation regulators, pathetic (amino acid transporter) (38), brain tumor (37), cell-adhesion molecules, neuroligin (40), synaptic vesicle protein n-synaptobrevin (41), regulators of cAMP signaling, PKA-R2, secretory protease, tequila and components of ubiquitin pathway, unkempt (putative ubiquitin ligase) (42), murashka (putative E3 ubiquitin ligase) (14), ubiquitin hydrolase (43), and components of 20S proteosomes rpt1, rpt4, and rpt9 (44).
These targets can be broadly classified into the following groups: (i) genes involved neuronal growth and synapse formation, (ii) genes involved in synaptic function, (iii) genes involved in proteolysis, and (iv) genes with unknown (predicted) functions (Table 1). Although we lack direct evidence, we postulate that, in the adult fly brain, Orb2-dependent persistent regulation of these target genes in the activated synapse might stabilize altered synaptic function and synapse number and thus, memory in the following ways. First, the suppression of ubiquitin-mediated protein degradation machinery, as well as other proteases, might allow for the accumulation of otherwise unstable molecules. On the other hand, activation of the proteosomal pathway and proteases might remove inhibitory constraints at the synapse to create and maintain a permissive environment for synaptic growth. The involvement of Uch and ubiquitin-mediated protein-turnover pathways in synaptic plasticity has previously been observed in Aplysia and in mice (24–28). Because ubiquitination can act as a posttranslational modification in addition to a signal for degradation, the ubiquitination pathway can, in principle, modulate stability, function, or membrane distribution of proteins at the activated synapse. Second, the cell polarity and asymmetric growth genes, such as DaPKC (29), widerborst (30), and glaikit (31), might allow the activated synapse to capture or modify the globally distributed gene product selectively at the activated synapse. In Drosophila, DaPKC is required for the formation of glutamatergic synapses in the NMJ (29). DaPKC is believed to be a homolog of mouse atypical protein kinase C (PKMζ), and in mice, the continuous activity of PKMζ is necessary for the persistence of memory for months (32). Interestingly, in mice, PKMζ mRNA is localized in the dendrites (33). Taken together, these observations in flies and mice raise the possibility that Orb2-dependent regulation of DapKC/PKMζ at the activated synapse contributes to memory-related synaptic growth. Finally, the regulation of the cytoskeleton-remodeling complexes, such as Capulet (34), Still-life (35), and Actin 5C, initiators of de novo synapse formation, such as Neuroligin (36), and growth regulators, such as Brain tumor (37), Pathetic (38), and Branchless (39), might regulate the maintenance of newly grown synapse for a long time. Future studies should be directed to addressing these postulations.
Materials and Methods
Elav-GAL4 (stock #458), Act-GAL4 (stock #0.4414), and tequila f01792 (stock #18473) were obtained from Bloomington Stock Center. The UAS-Orb2RAHA and UAS-Orb2RBHA lines were generated by the authors. To generate flies expressing Orb2RB lacking Zn-finger domain (OrbRBΔZn), 66 amino acids were deleted from the C-terminal end of Orb2RB by PCR (primers in SI Materials and Methods) and cloned into an upstream activating sequence vector with a C-terminal HA tag (additional information in SI Materials and Methods) (Table S3).
Supplementary Material
Acknowledgments
We thank Dr. Robb Krumlauf and Dr. Marco Blanchette of the Stowers Institute for their critical comments on the manuscript. K.S. is supported by Searle Scholars Program, March of Dimes Basil O'Connor Starter Scholar Award, The Esther A. & Joseph Klingenstein Fund, and The McKnight Endowment Fund for Neuroscience.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequence reported in this paper has been deposited in the ArrayExpress database (accession no. E-TABM-527).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1004433107/-/DCSupplemental.
References
- 1.Casadio A, et al. A transient, neuron-wide form of CREB-mediated long-term facilitation can be stabilized at specific synapses by local protein synthesis. Cell. 1999;99:221–237. doi: 10.1016/s0092-8674(00)81653-0. [DOI] [PubMed] [Google Scholar]
- 2.Frey U, Morris RG. Synaptic tagging and long-term potentiation. Nature. 1997;385:533–536. doi: 10.1038/385533a0. [DOI] [PubMed] [Google Scholar]
- 3.Kang H, Schuman EM. A requirement for local protein synthesis in neurotrophin-induced hippocampal synaptic plasticity. Science. 1996;273:1402–1406. doi: 10.1126/science.273.5280.1402. [DOI] [PubMed] [Google Scholar]
- 4.Squire LR, Davis HP. Cerebral protein synthesis inhibition and discrimination training: Effect of extent and duration of inhibition. Behav Biol. 1975;13:49–57. doi: 10.1016/s0091-6773(75)90778-6. [DOI] [PubMed] [Google Scholar]
- 5.Steward O, Levy WB. Preferential localization of polyribosomes under the base of dendritic spines in granule cells of the dentate gyrus. J Neurosci. 1982;2:284–291. doi: 10.1523/JNEUROSCI.02-03-00284.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tanaka J, et al. Protein synthesis and neurotrophin-dependent structural plasticity of single dendritic spines. Science. 2008;319:1683–1687. doi: 10.1126/science.1152864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Si K, et al. A neuronal isoform of CPEB regulates local protein synthesis and stabilizes synapse-specific long-term facilitation in aplysia. Cell. 2003;115:893–904. doi: 10.1016/s0092-8674(03)01021-3. [DOI] [PubMed] [Google Scholar]
- 8.Richter JD. CPEB: A life in translation. Trends Biochem Sci. 2007;32:279–285. doi: 10.1016/j.tibs.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 9.Miniaci MC, et al. Sustained CPEB-dependent local protein synthesis is required to stabilize synaptic growth for persistence of long-term facilitation in Aplysia. Neuron. 2008;59:1024–1036. doi: 10.1016/j.neuron.2008.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Si K, Choi YB, White-Grindley E, Majumdar A, Kandel ER. Aplysia CPEB can form prion-like multimers in sensory neurons that contribute to long-term facilitation. Cell. 2010;140:421–435. doi: 10.1016/j.cell.2010.01.008. [DOI] [PubMed] [Google Scholar]
- 11.Si K, Lindquist S, Kandel ER. A neuronal isoform of the aplysia CPEB has prion-like properties. Cell. 2003;115:879–891. doi: 10.1016/s0092-8674(03)01020-1. [DOI] [PubMed] [Google Scholar]
- 12.Si K, Lindquist S, Kandel E. A possible epigenetic mechanism for the persistence of memory. Cold Spring Harb Symp Quant Biol. 2004;69:497–498. doi: 10.1101/sqb.2004.69.497. [DOI] [PubMed] [Google Scholar]
- 13.Keleman K, Krüttner S, Alenius M, Dickson BJ. Function of the Drosophila CPEB protein Orb2 in long-term courtship memory. Nat Neurosci. 2007;10:1587–1593. doi: 10.1038/nn1996. [DOI] [PubMed] [Google Scholar]
- 14.Dubnau J, et al. The staufen/pumilio pathway is involved in Drosophila long-term memory. Curr Biol. 2003;13:286–296. doi: 10.1016/s0960-9822(03)00064-2. [DOI] [PubMed] [Google Scholar]
- 15.Keene AC, Waddell S. Drosophila olfactory memory: Single genes to complex neural circuits. Nat Rev Neurosci. 2007;8:341–354. doi: 10.1038/nrn2098. [DOI] [PubMed] [Google Scholar]
- 16.Chang JS, Tan L, Schedl P. The Drosophila CPEB homolog, orb, is required for oskar protein expression in oocytes. Dev Biol. 1999;215:91–106. doi: 10.1006/dbio.1999.9444. [DOI] [PubMed] [Google Scholar]
- 17.Didelot G, et al. Tequila, a neurotrypsin ortholog, regulates long-term memory formation in Drosophila. Science. 2006;313:851–853. doi: 10.1126/science.1127215. [DOI] [PubMed] [Google Scholar]
- 18.Drier EA, et al. Memory enhancement and formation by atypical PKM activity in Drosophila melanogaster. Nat Neurosci. 2002;5:316–324. doi: 10.1038/nn820. [DOI] [PubMed] [Google Scholar]
- 19.Hake LE, Mendez R, Richter JD. Specificity of RNA binding by CPEB: Requirement for RNA recognition motifs and a novel zinc finger. Mol Cell Biol. 1998;18:685–693. doi: 10.1128/mcb.18.2.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Al-Shahrour F, et al. BABELOMICS: A systems biology perspective in the functional annotation of genome-scale experiments. Nucleic Acids Res. 2006;34:W472–W476. doi: 10.1093/nar/gkl172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gerber AP, Luschnig S, Krasnow MA, Brown PO, Herschlag D. Genome-wide identification of mRNAs associated with the translational regulator PUMILIO in Drosophila melanogaster. Proc Natl Acad Sci USA. 2006;103:4487–4492. doi: 10.1073/pnas.0509260103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.DiAntonio A, et al. Ubiquitination-dependent mechanisms regulate synaptic growth and function. Nature. 2001;412:449–452. doi: 10.1038/35086595. [DOI] [PubMed] [Google Scholar]
- 23.Huang YS, Kan MC, Lin CL, Richter JD. CPEB3 and CPEB4 in neurons: Analysis of RNA-binding specificity and translational control of AMPA receptor GluR2 mRNA. EMBO J. 2006;25:4865–4876. doi: 10.1038/sj.emboj.7601322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bingol B, Schuman EM. Activity-dependent dynamics and sequestration of proteasomes in dendritic spines. Nature. 2006;441:1144–1148. doi: 10.1038/nature04769. [DOI] [PubMed] [Google Scholar]
- 25.Ehlers MD. Activity level controls postsynaptic composition and signaling via the ubiquitin-proteasome system. Nat Neurosci. 2003;6:231–242. doi: 10.1038/nn1013. [DOI] [PubMed] [Google Scholar]
- 26.Fonseca R, Vabulas RM, Hartl FU, Bonhoeffer T, Nägerl UV. A balance of protein synthesis and proteasome-dependent degradation determines the maintenance of LTP. Neuron. 2006;52:239–245. doi: 10.1016/j.neuron.2006.08.015. [DOI] [PubMed] [Google Scholar]
- 27.Hegde AN, Goldberg AL, Schwartz JH. Regulatory subunits of cAMP-dependent protein kinases are degraded after conjugation to ubiquitin: A molecular mechanism underlying long-term synaptic plasticity. Proc Natl Acad Sci USA. 1993;90:7436–7440. doi: 10.1073/pnas.90.16.7436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee SH, et al. Synaptic protein degradation underlies destabilization of retrieved fear memory. Science. 2008;319:1253–1256. doi: 10.1126/science.1150541. [DOI] [PubMed] [Google Scholar]
- 29.Ruiz-Canada C, et al. New synaptic bouton formation is disrupted by misregulation of microtubule stability in aPKC mutants. Neuron. 2004;42:567–580. doi: 10.1016/s0896-6273(04)00255-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hannus M, Feiguin F, Heisenberg CP, Eaton S. Planar cell polarization requires Widerborst, a B’ regulatory subunit of protein phosphatase 2A. Development. 2002;129:3493–3503. doi: 10.1242/dev.129.14.3493. [DOI] [PubMed] [Google Scholar]
- 31.Dunlop J, Morin X, Corominas M, Serras F, Tear G. glaikit is essential for the formation of epithelial polarity and neuronal development. Curr Biol. 2004;14:2039–2045. doi: 10.1016/j.cub.2004.10.048. [DOI] [PubMed] [Google Scholar]
- 32.Shema R, Sacktor TC, Dudai Y. Rapid erasure of long-term memory associations in the cortex by an inhibitor of PKM zeta. Science. 2007;317:951–953. doi: 10.1126/science.1144334. [DOI] [PubMed] [Google Scholar]
- 33.Muslimov IA, et al. Dendritic transport and localization of protein kinase Mzeta mRNA: Implications for molecular memory consolidation. J Biol Chem. 2004;279:52613–52622. doi: 10.1074/jbc.M409240200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wills Z, et al. A Drosophila homolog of cyclase-associated proteins collaborates with the Abl tyrosine kinase to control midline axon pathfinding. Neuron. 2002;36:611–622. doi: 10.1016/s0896-6273(02)01022-x. [DOI] [PubMed] [Google Scholar]
- 35.Sone M, et al. Still life, a protein in synaptic terminals of Drosophila homologous to GDP-GTP exchangers. Science. 1997;275:543–547. doi: 10.1126/science.275.5299.543. [DOI] [PubMed] [Google Scholar]
- 36.Varoqueaux F, et al. Neuroligins determine synapse maturation and function. Neuron. 2006;51:741–754. doi: 10.1016/j.neuron.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 37.Sonoda J, Wharton RP. Drosophila Brain Tumor is a translational repressor. Genes Dev. 2001;15:762–773. doi: 10.1101/gad.870801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goberdhan DC, Meredith D, Boyd CA, Wilson C. PAT-related amino acid transporters regulate growth via a novel mechanism that does not require bulk transport of amino acids. Development. 2005;132:2365–2375. doi: 10.1242/dev.01821. [DOI] [PubMed] [Google Scholar]
- 39.Sutherland D, Samakovlis C, Krasnow MA. branchless encodes a Drosophila FGF homolog that controls tracheal cell migration and the pattern of branching. Cell. 1996;87:1091–1101. doi: 10.1016/s0092-8674(00)81803-6. [DOI] [PubMed] [Google Scholar]
- 40.Song JY, Ichtchenko K, Südhof TC, Brose N. Neuroligin 1 is a postsynaptic cell-adhesion molecule of excitatory synapses. Proc Natl Acad Sci USA. 1999;96:1100–1105. doi: 10.1073/pnas.96.3.1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.DiAntonio A, et al. Identification and characterization of Drosophila genes for synaptic vesicle proteins. J Neurosci. 1993;13:4924–4935. doi: 10.1523/JNEUROSCI.13-11-04924.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mohler J, et al. The embryonically active gene, unkempt, of Drosophila encodes a Cys3His finger protein. Genetics. 1992;131:377–388. doi: 10.1093/genetics/131.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang N, Wilkinson K, Bownes M. Cloning and analysis of expression of a ubiquitin carboxyl terminal hydrolase expressed during oogenesis in Drosophila melanogaster. Dev Biol. 1993;157:214–223. doi: 10.1006/dbio.1993.1125. [DOI] [PubMed] [Google Scholar]
- 44.Hölzl H, et al. The regulatory complex of Drosophila melanogaster 26S proteasomes. Subunit composition and localization of a deubiquitylating enzyme. J Cell Biol. 2000;150:119–130. doi: 10.1083/jcb.150.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.