Abstract
AIM: To investigate let-7a expression and analyze the correlation between let-7a and progression of gastric mucosa cancerization.
METHODS: The tissue microarray was constructed previously in 52 cases of human gastric carcinoma, 17 cases of chronic atrophic gastritis (atypical hyperplasia) and 11 cases of normal gastric tissue, and tissue microarrays combined with in situ hybridization were used to detect the expression of let-7a.
RESULTS: The positive rates of let-7a in normal gastric tissue, chronic atrophic gastritis and gastric carcinoma were 90.9%, 88.2% and 86.5%, respectively, without significant differences among the groups (P > 0.05). However, an intense signal of let-7a was observed in gastric epithelial cells, whereas a less intense signal was found in gastric atypical hyperplasia epithelial cells and a weak signal in gastric carcinoma epithelial cells. The expression of let-7a decreased along with the progression of gastric mucosa cancerization (P < 0.05). In the group of gastric carcinoma, the expression of let-7a was even significantly lower in gastric carcinomas with lymph node metastasis than in those without metastasis (P < 0.05).
CONCLUSION: Gastric carcinoma has relatively lower expression of let-7a. Reduced let-7a may be a fundamental factor in the formation and lymph node metastasis of gastric carcinoma.
Keywords: Tissue microarray, In situ hybridization, MicroRNA, Gastric carcinoma, Precancerous lesions
INTRODUCTION
MicroRNAs (miRNAs) are a recently discovered class of short noncoding RNA genes that act post-transcriptionally as negative regulators of gene expression[1,2]. A large body of research shows that animal miRNAs play a fundamental role in many biological processes, including cell growth and apoptosis, hematopoietic lineage differentiation, insulin secretion, brain morphogenesis, and muscle cell proliferation and differentiation[3]. About 1000 miRNA genes are thought to be encoded in the human genome[4,5]. Although it is still difficult to identify accurately individual miRNA/target interactions[6], computational predictions of miRNA target genes indicate that as many as one-third of all human protein-encoding genes may be regulated by miRNAs[7]. This suggests that miRNAs could be involved in a wide variety of human diseases, including cancer. As a result of in-depth researches over the last 5 years, miRNA-mediated regulation of tumorigenesis is emerging as a new paradigm in the field of cancer biology[8,9]. It has also been reported that these miRNA signatures may be a more robust tool than expression patterns of protein-encoding genes for distinguishing normal from tumor tissues[10,11]. One particular miRNA, let-7, was discovered in the early 1990s in humans and mice. It has been reported that let-7 as a novel candidate of tumor suppressor gene is involved in several cancers such as lung carcinoma and colon carcinoma.
Gastric carcinoma is one of the most frequent cancers and one of the most common causes of cancer-related mortality in China, with an incidence of 0.4 million new cases and 0.3 million deaths annually. Gastric carcinoma is also an inadequately understood, and often fatal disease when not detected at early stages. Atypical hyperplasia is generally accepted as a precancerous lesion. Cancerization from normal gastric mucosa, chronic atrophic gastritis to gastric carcinoma has been well acknowledged. In this study, the expression of let-7a in normal gastric tissue, chronic atrophic gastritis (atypical hyperplasia) and gastric carcinoma was detected by in situ hybridization (ISH) using a locked nucleic acid (LNA) probe and gastric mucosa cancerizing tissue microarrays; the relation between expression level of let-7a and the progression of gastric mucosa cancerization was discussed.
MATERIALS AND METHODS
Tissue microarray and probe
The tissue bank at Shanxi Chaoying Biotechnology Company maintains a series of formalin-fixed, paraffin-embedded (FFPE) tissue blocks of patients with gastric carcinoma, chronic atrophic gastritis (atypical hyperplasia) and normal gastric tissue. Information on tumor histologic grade, patient’s age and sex was collected and anonymized in a secure database. We generated a tissue microarray (TMA) from archived FFPE blocks of 80 cases, including 11 cases of normal gastric tissue, 17 cases of chronic atrophic gastritis with atypical hyperplasia (incomplete intestinal metaplasia) and 52 cases of gastric carcinoma (intestinal type). In the gastric carcinoma group, there were 10 cases with lymph node metastasis and 42 cases without lymph node metastasis. This study complied with the rules and regulations of the Ethics Committee of the hospital. Informed consent was obtained from all subjects. All FFPE blocks were reviewed and tissue cores were selected by a board-certified pathologist. The 5’Digoxin-labeled, LNA-modified oligonucleotide probe (sequence 5’-3’: aactatacaacctactacctca) complementary to the entire mature sequence of let-7a, was synthesized by Exiqon DNA technologies.
Detection of miRNAs by ISH in FFPE sections
The experiment was conducted at a laboratory of Shanxi Chaoying Biotechnology Company in November 2009. Sections (5 μm) of archived paraffin-embedded specimens were deparaffinized in xylenes and then rehydrated through an ethanol dilution series (from 100% to 25%). Slides were submerged in diethyl pyrocarbonate-treated water and subjected to proteinase K digestion (5-10 μg/mL) and 0.2% glycine treatment, refixed in 4% paraformaldehyde, and treated with acetylation solution [66 mmol/L HCl, 0.66% (v/v) acetic anhydride, and 1.5% (v/v) triethanolamine]; slides were rinsed thrice with 1 × PBS between the treatment. Slides were prehybridized in hybridization solution (50% formamide, 5 × SSC, 500 Ag/mL yeast tRNA, and 1 × Denhardt’s solution) at 50°C for 30 min. Then 5-10 pmol of Digoxin-labeled, LNA-modified probe, was added to 150 μL hybridization solution and hybridized for 18 h at a temperature of 42°C below the calculated Tm of the LNA probe. Subsequently, the samples were incubated at 37°C for 90 min with 50 μL biotin-labeled anti-digoxin antibody, rinsed with 0.5 × SSC four times for 5 min, incubated at 37°C for 30 min with 50 μL SABC solution, and rinsed with 0.5 × SSC solution three times for 5 min. After added 50 μL biotin-labeled peroxidase and incubated at 37°C for 30 min, the samples were rinsed five times with 0.5 × SSC for 5 min, then stained with Nitro-Blue-Tetrazolium/5-bromo-4-chloro-3-indolyl-phosphate. Finally, the sections were restained with eosin before covering. For a negative control, the hybridization solution was replaced with pre-hybridization solution. Likewise, β-actin was used as the probe of positive control.
Determination of results
Signals were visually quantified by a pathologist according to the reference of Frantz et al[12]. A total of 100 × 10 cells were counted randomly. Then, tissue sections were blindly examined by a second individual for an agreement with the initial quantifications. If there was no positive cell, the sample was scored as 0 point. If the percentage of positive cells was less than or equal to 30%, the sample was scored as 1 point; less than or equal to 70% as 2 points; and more than 70% as 3 points. The staining intensity was recorded as negative, light, moderate, or strong, and scored as 0, 1, 2 or 3 points, respectively. The two values were multiplied, and a sum of 0 point represented negative expression, and 1-9 points represented positive expression. Points 1-2 were marked ±, 3-4 marked +, 6 marked ++, and 9 marked +++.
Statistical analysis
Statistical analysis was performed using SPSS 11.5 Package. The χ2 and correlation tests were used in this study. A difference was considered significant when P value was less than 0.05.
RESULTS
Expression of let-7a was determined on 5-μm sections obtained from each of the TMA blocks. Generally, let-7a appeared as blue-purple particles in the cytoplasm or nucleus after staining, and predominantly expressed in normal gastric mucosa epithelia and its expression was down-regulated in atypical hyperplasia epithelia, especially in malignant cells. The let-7a probe revealed an intense cytoplasmic signal in gastric epithelial cells, whereas a less intense signal was observed in gastric atypical hyperplasia epithelial cells and a weak signal in gastric carcinoma epithelial cells (Figure 1). Let-7a was also detected in endothelial cells lining blood vessels and in fibroblasts (data not shown). The classification of let-7a expression grade in each group is shown in Table 1. There was no significant difference in the positive rates of let-7a among gastric carcinoma, chronic atrophic gastritis and normal gastric tissues (P > 0.05). However, by analysis of the signal levels of let-7a in each group, the levels of let-7a expression were reversely related to the progression of gastric mucosa cancerization (P = 0.008). The expression of let-7a decreased along with the progression of gastric mucosa cancerization. In the gastric carcinoma group, the expression of let-7a in the 10 cases with lymph node metastasis was even lower than in the 42 cases without lymph node metastasis (P = 0.043) as shown in Table 2. In this study, β-actin showed an intense cytoplasmic signal in each of the TMA blocks in positive control while no blue-purple staining was found in blocks of negative control.
Figure 1.
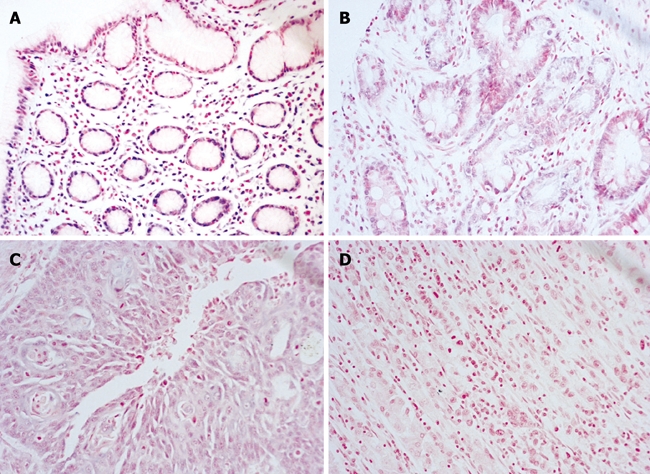
Expression let-7a in stomach cancer progression tissue array (in situ hybridization, Nitro-Blue-Tetrazolium/5-bromo-4-chloro-3-indolyl-phosphate staining, original magnification × 200). A: Positive expression (++) of let-7a in gastric tissue; B: Positive expression (+) of let-7a in chronic atrophic gastritis; C: Positive expression (±) of let-7a in gastric carcinoma; D: Negative expression of let-7a in gastric carcinoma.
Table 1.
Expression of let-7a in gastric mucosa cancerization tissue array
| Cases (n) |
Results |
Positive rate (%) | |||||
| - | ± | + | ++ | +++ | |||
| Normal gastric mucosa | 11 | 1 | 0 | 2 | 8 | 0 | 90.9 |
| Chronic atrophic gastritis | 17 | 2 | 2 | 7 | 5 | 1 | 88.2 |
| Gastric carcinoma | 52 | 7 | 18 | 18 | 7 | 2 | 86.5 |
Table 2.
Expression of let-7a in gastric carcinomas
| Lymph node metastasis | Cases (n) |
Results |
P value | ||||
| - | ± | + | ++ | +++ | 0.043 | ||
| Negative | 42 | 4 | 11 | 18 | 7 | 2 | |
| Positive | 10 | 3 | 6 | 1 | 0 | 0 | |
DISCUSSION
The let-7 family consists of 12 genes encoding for nine distinct miRNAs (let-7a to let-7i). Historically, let-7 was found to bind to the 3’ UTR of lin-41 and hbl-1 (lin-57) to inhibit their translation in nematodes[13], and control the developmental transition from the L4 stage into the adult stage[14-16]. An evolutionarily conserved regulation of the RAS family of growth control proteins by let-7 from nematodes to human cancer cells has also been demonstrated. Recently, let-7 implication in cancerogenesis has been extended to the repression of high mobility group A2, thus preventing oncogenic transformation in many tumors[17]. Johnson et al[18] showed that let-7 miRNA expression was lower in lung tumors than in normal lung tissues. The detection of mature miRNAs by ISH is technically challenging because of their small size. LNAs are a class of bicyclic high-affinity RNA analogues, in which the furanose ring in the sugar-phosphate backbone is chemically locked in a conformation mimicking the North-type (C3’-endo) conformation of RNA[19,20]. This results in an unprecedented hybridization affinity of LNA toward complementary single-stranded RNA molecules. A limitation to identifying and validating potential biomarkers makes it difficult to conduct retrospective studies with archived tumor specimens due in part to protein and RNA degradation[21]. Hence, the very small size of miRNAs offers a unique advantage because short RNA molecules are substantially less susceptible than mRNAs to enzymatic and mechanical degradation. In this study, we used a method for detection of miRNAs in gastric carcinoma specimens, LNA-ISH, which offers a spatial resolution of miRNA expression unsurpassed by other available techniques. Zhang at al[22] established an accurate and rapid stem-loop reverse transcriptional real-time PCR (RT-PCR) method to quantify human let-7a miRNA in gastric cancer. It was found that the expression level of let-7a miRNA in gastric tumor tissues was significantly lower than in normal tissues of 14 samples from 32 patients. There might be a discordance between ISH and RT-PCR results, which could be explained by the overrepresentation of epithelial cells in tumor lesions, resulting in a high let-7 signal in gross tissue biopsies. However, let-7a expression is diminished within individual cancer cells, which can be determined only by the ISH approach. We noticed that long fixation periods in 10% formalin (more than 12-18 h) can affect negatively the intensity of the miRNA in situ signal. There is no significant difference of the positive rates of let-7a expression among gastric carcinoma, chronic atrophic gastritis and normal gastric tissue, which may be partly due to the limitation of normal gastric tissue cases in the TMA. Decrease of let-7a expression is already apparent in hyperplasia (possibly preneoplastic). Its further decrease in gastric carcinoma is reminiscent of reduction of let-7a related to the gastric mucosa cancerization progression. Hence, this suggests that reduction of let-7a is an early event during intestinal-type gastric carcinoma formation. The most essential behavior of the malignant tumor is metastasis. Lymph nodes metastasis is an important index for clinical stages of gastric carcinoma before surgery and it is also an important prognosis factor for gastric carcinoma patients. Our study showed that the expression of let-7a was even lower in gastric carcinoma with lymph nodes metastasis than in those without lymph nodes metastasis. Decreased expression or absence of let-7a may determine initially the occurrence, development, metastasis and prognosis of gastric carcinoma. Using combined LNA-ISH and RT-PCR, quantitative detection of let-7a may have a potential clinical application as a novel biomarker for early diagnosis and prognosis of gastric carcinoma. Infecting breast tumor-initiating cells with let-7-lentivirus reduced proliferation, mammosphere formation, and the proportion of undifferentiated cells in vitro and tumor formation and metastasis in NOD/SCID mice[23]. Thus, the reason why let-7a in human gastric carcinoma has generated great interest is that it not only has a potency in diagnosis, but also can shed light on the molecular mechanisms of the tumorigenic process and has a high potential as novel targets for therapeutic intervention using synthetic oligonucleotide technologies. Further studies are being carried out on the relationship between gastric carcinoma and let-7a in our laboratory.
COMMENTS
Background
Gastric carcinoma is one of the malignancies with highest incidence and mortality rates and it is of great significance to investigate the mechanisms behind the occurrence and development of gastric carcinoma. Down-regulated expression let-7a has been shown to play an important role in the tumorigenesis and cancer progression. Therefore, the exploration into the expression of let-7a in gastric carcinoma and precancerous lesions may shed a new light on early detection and therapy of gastric carcinoma.
Research frontiers
Let-7a has been shown to exert tumor suppressor gene effects in various human neoplasms. In this study, the author investigated the expression of let-7a in normal gastric mucosa, chronic atrophic gastritis and gastric carcinoma to explore its correlation with gastric carcinogenesis and metastasis. They found that reduced expression of let-7a may take part in early carcinogenesis and metastasis of gastric carcinoma.
Innovations and breakthroughs
Let-7a is down-regulated in a wide range of human cancers such as gastric, colorectal and breast carcinomas, and may play a key role in cancer pathogenesis. Down-regulated let-7a increases cancer cell proliferation, migration and invasion. This is the first study to report that expression of let-7a has a reverse correlation with gastric cancerization.
Applications
The expression level of let-7a was significantly lower in gastric carcinoma than in chronic atrophic gastritis and normal gastric mucosa. In the malignant tumor group, the expression level of let-7a was even significantly lower in gastric carcinoma with lymph node metastasis than in those without metastasis. Detection of let-7a expression might be helpful in early detection and prognosis judgment of gastric carcinoma patients. In addition, let-7a may serve as a potential therapeutic tool for gastric carcinoma.
Peer review
In this descriptive study, let-7a miRNA expression is analyzed in human gastric mucosa, chronic atrophic gastritis, and gastric cancer with in situ hybridization technique on tissue microarray. The study confirms the results by Zhang et al demonstrating a decrease of let-7a in gastric carcinogenesis.
Footnotes
Supported by Research Grants from the Key Science and Technology Projects of Guangxi Zhuang Autonomous Region
Peer reviewers: Nikolaus Gassler, Professor, Institute of Pathology, University Hospital RWTH Aachen, Pauwelsstrasse 30, 52074 Aachen, Germany; Wei Tang, MD, EngD, Assistant Professor, H-B-P Surgery Division, Artificial Organ and Transplantation Division, Department of Surgery, Graduate School of Medicine, The University of Tokyo, Tokyo 113-8655, Japan
S- Editor Tian L L- Editor Ma JY E- Editor Ma WH
References
- 1.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 2.Bartel DP, Chen CZ. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat Rev Genet. 2004;5:396–400. doi: 10.1038/nrg1328. [DOI] [PubMed] [Google Scholar]
- 3.Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Dev Cell. 2006;11:441–450. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 4.Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O, Barzilai A, Einat P, Einav U, Meiri E, et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet. 2005;37:766–770. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- 5.Berezikov E, van Tetering G, Verheul M, van de Belt J, van Laake L, Vos J, Verloop R, van de Wetering M, Guryev V, Takada S, et al. Many novel mammalian microRNA candidates identified by extensive cloning and RAKE analysis. Genome Res. 2006;16:1289–1298. doi: 10.1101/gr.5159906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Didiano D, Hobert O. Perfect seed pairing is not a generally reliable predictor for miRNA-target interactions. Nat Struct Mol Biol. 2006;13:849–851. doi: 10.1038/nsmb1138. [DOI] [PubMed] [Google Scholar]
- 7.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 8.Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302:1–12. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 9.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 10.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 12.Frantz GD, Pham TQ, Peale FV Jr, Hillan KJ. Detection of novel gene expression in paraffin-embedded tissues by isotopic in situ hybridization in tissue microarrays. J Pathol. 2001;195:87–96. doi: 10.1002/1096-9896(200109)195:1<87::AID-PATH932>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 13.Jérôme T, Laurie P, Louis B, Pierre C. Enjoy the Silence: The Story of let-7 MicroRNA and Cancer. Curr Genomics. 2007;8:229–233. doi: 10.2174/138920207781386933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 15.Abrahante JE, Daul AL, Li M, Volk ML, Tennessen JM, Miller EA, Rougvie AE. The Caenorhabditis elegans hunchback-like gene lin-57/hbl-1 controls developmental time and is regulated by microRNAs. Dev Cell. 2003;4:625–637. doi: 10.1016/s1534-5807(03)00127-8. [DOI] [PubMed] [Google Scholar]
- 16.Lin SY, Johnson SM, Abraham M, Vella MC, Pasquinelli A, Gamberi C, Gottlieb E, Slack FJ. The C elegans hunchback homolog, hbl-1, controls temporal patterning and is a probable microRNA target. Dev Cell. 2003;4:639–650. doi: 10.1016/s1534-5807(03)00124-2. [DOI] [PubMed] [Google Scholar]
- 17.Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science. 2007;315:1576–1579. doi: 10.1126/science.1137999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 19.Kauppinen S, Vester B, Wengel J. Locked nucleic acid: high-affinity targeting of complementary RNA for RNomics. Handb Exp Pharmacol. 2006:405–422. doi: 10.1007/3-540-27262-3_21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koshkin AA, Wengel J. Synthesis of Novel 2’,3’-Linked Bicyclic Thymine Ribonucleosides. J Org Chem. 1998;63:2778–2781. doi: 10.1021/jo972239c. [DOI] [PubMed] [Google Scholar]
- 21.Ludwig JA, Weinstein JN. Biomarkers in cancer staging, prognosis and treatment selection. Nat Rev Cancer. 2005;5:845–856. doi: 10.1038/nrc1739. [DOI] [PubMed] [Google Scholar]
- 22.Zhang HH, Wang XJ, Li GX, Yang E, Yang NM. Detection of let-7a microRNA by real-time PCR in gastric carcinoma. World J Gastroenterol. 2007;13:2883–2888. doi: 10.3748/wjg.v13.i20.2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong C, Huang Y, Hu X, Su F, Lieberman J, et al. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007;131:1109–1123. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]


