Abstract
The pituitary-specific homeodomain protein Pit-1 cooperates with other transcription factors, in cluding CCAAT/enhancer binding protein α (C/ EBPα), in the regulation of pituitary lactotrope gene transcription. Here, we correlate cooperative activation of prolactin (PRL) gene transcription by Pit-1 and C/EBPα with changes in the subnuclear localization of these factors in living pituitary cells. Transiently expressed C/EBPα induced PRL gene transcription in pituitary GHFT1–5 cells, whereas the coexpression of Pit-1 and C/EBPα in HeLa cells demonstrated their cooperativity at the PRL promoter. Individually expressed Pit-1 or C/EBPα, fused to color variants of fluorescent proteins, occupied different subnuclear compartments in living pituitary cells. When coexpressed, Pit-1 recruited C/EBPα from regions of transcriptionally quiescent centromeric heterochromatin to the nuclear regions occupied by Pit-1. The homeodomain region of Pit-1 was necessary for the recruitment of C/EBPα. A point mutation in the Pit-1 homeodomain associated with the syndrome of combined pituitary hormone deficiency in humans also failed to recruit C/EBPα. This Pit-1 mutant functioned as a dominant inhibitor of PRL gene transcription and, instead of recruiting C/EBPα, was itself recruited by C/EBPα to centromeric heterochromatin. Together our results suggest that the intranuclear positioning of these factors determines whether they activate or silence PRL promoter activity.
THE TISSUE-SELECTIVE TRANSCRIPTION of genes requires the combinatorial interactions of specific transcription factors with other coregulatory proteins. Through these interactions, cell-specific combinations of factors modify chromatin structure, recruit the general transcription apparatus, and regulate RNA polymerase activity at those genes. It is generally assumed that the cooperating transcription factors have similar access to all potential gene targets. However, many observations indicate that transcription factors and coregulatory proteins may be assembled at particular intranuclear sites (1–5). The restriction of transcription factors to particular intranuclear sites would be expected to affect the combinatorial interactions available for gene transcription. Thus, the location of gene-regulatory complexes within the nucleus may represent a key step in the control of tissue-specific gene transcription.
We have investigated how the subnuclear localization of transcription factors in anterior pituitary cells was correlated with their cooperative interactions in the regulation of prolactin (PRL) gene expression. The transcription of the PRL gene is restricted to lactotropes, making it an excellent model for cell type specific gene regulation. The pituitary-specific homeodomain (HD) protein Pit-1, which is required for the development of the lactotrope, somatotrope, and thyrotrope cell lineages, is also necessary for the transcriptional regulation of the genes encoding the hormone products of these cell types. Because of its central role in the genesis of these pituitary cell types, patients that have inactivating mutations in Pit-1 develop combined pituitary hormone deficiency (CPHD), a disease characterized by the lack of the hormones produced by these cells (6, 7).
The promoter and enhancer regions of the PRL gene direct its pituitary cell-specific expression and contain multiple binding sites for Pit-1 (8). Pit-1, alone, is necessary, but not sufficient, for PRL gene transcription, and it is the interplay between Pit-1 and other generegulatory proteins that controls the expression of PRL (9). In this regard, several other transcription factors, including the CCAAT/enhancer binding protein-α (C/ EBPα), are known to participate in the regulation of PRL gene expression. C/EBPα is a member of the basic region-leucine zipper (B-ZIP) family of transcription factors (10) and controls the expression of genes involved in terminal differentiation and energy metabolism (10, 11). C/EBPα controls the transcription of both the PRL and GH genes by binding to promoter elements adjacent to critical binding sites for Pit-1 (12–14).
Here we investigated the role of intranuclear compartmentalization of Pit-1 and C/EBPα in their cooperative activation of PRL gene transcription. Fluorescence imaging of pituitary cells expressing either C/EBPα or Pit-1 as fusions to green fluorescent protein (GFP) revealed that each had a distinct pattern of distribution relative to other nuclear markers. How ever, when coexpressed in the same cells as fusions to FP color variants, we observed that C/EBPα was recruited to the intranuclear sites occupied by Pit-1. Mutational studies indicated that the HD of Pit-1 was required for this recruitment activity. Further, we found that a point mutation in the Pit-1 HD also failed to recruit C/EBPα. This Pit-1 mutant, which acts as a dominant inhibitor of PRL gene expression and is associated with CPHD syndrome in humans, became associated with C/EBPα in regions of centromeric heterochromatin. Together these studies provide evidence for an organizational role of Pit-1 in directing other cooperating factors to particular intranuclear sites. The disruption of the organizational function of Pit-1 by the CPHD point mutation suggests that intranuclear location may be a critical determinant of transcriptional outcome. The ability of Pit-1, C/EBPα, and other factors involved in pituitary-specific gene expression to assemble at particular subnuclear sites may constitute an overlooked epigenetic component of the combinatorial code responsible for cell-specific gene transcription.
RESULTS
C/EBPα Induces Transcription from the PRL Promoter
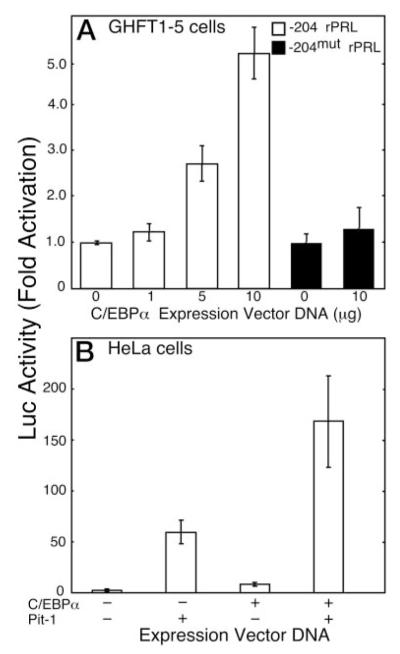
Mouse pituitary GHFT1–5 and human cervical carcinoma HeLa cells were used to examine the cooperative interactions between C/EBPα and Pit-1 in the regulation of PRL gene transcription. GHFT1–5 cells have the characteristics of the progenitor for the anterior pituitary somatotrope and lactotrope cell lineages and do not express the PRL gene (15). GHFT1–5 cells also do not express C/EBPα, but C/EBPα is found in other differentiated pituitary cell lines that do express the PRL gene (12). The expression of exogenous C/EBPα in GHFT1–5 cells induced the transcription of a cotransfected luciferase reporter gene coupled to the rat PRL gene promoter [−204 to +34, −204 rat PRL (rPRL)] in a dose-dependent manner (Fig. 1A). A C/EBPα-response element was identified previously at position −101 to −92 (relative to the transcription start site) of the rat PRL promoter (13). We found that mutation of −97 to −91 bp (−204mut rPRL) abolished the promoter responsiveness to C/EBPα in GHFT1–5 cells (Fig. 1A).
Fig. 1.
Transcription from the −204 rPRL Promoter Is Induced by C/EBPα
A, GHFT1–5 cells were transfected with a plasmid containing either the −204 rPRL promoter (open bars) or −204mut rPRL promoter (black bars) linked to the Luc reporter gene and the indicated amount of C/EBPα expression vector. After 24 h, cell extracts were prepared, and luciferase activity, corrected for total cellular protein, was determined and normalized to the activity of the reporter alone. The error is the SEM from three independent experiments, each done in triplicate. B, HeLa cells were transfected with the −204 rPRL luc reporter gene alone or with expression vectors for either Pit-1 (5 μg), C/EBPα (10 μg), or both. Luciferase activity was corrected for total cellular protein and normalized to the activity of the reporter alone. The error is the SEM from six independent experiments, each done in triplicate.
We extended these observations to show that the activity of C/EBPα at the PRL promoter was integrated with that of Pit-1. Because mouse GHFT1–5 cells express a low level of Pit-1 (Refs. 12 and 15; also see Fig. 2A), we assessed the functional interactions involving C/EBPα and Pit-1 at the PRL promoter in nonpituitary, human HeLa cells. Transfection of the HeLa cells with the expression vector for Pit-1 alone induced −204 rPRL reporter gene expression 50-fold, whereas expression of C/EBPα alone resulted in approximately 5-fold induction (Fig. 1B). When combined, the expression of C/EBPα and Pit-1 resulted in more than additive induction of reporter gene expression (Fig. 1B).
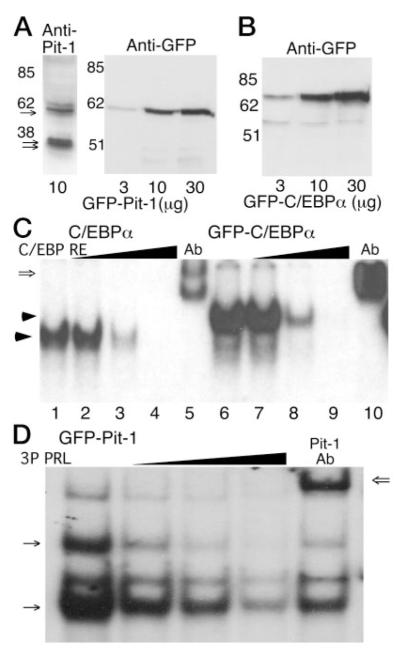
Fig. 2.
Analysis of the Expression and the DNA Binding Characteristics of GFP-Pit-1 and GFP-C/EBPα
A, Western blot analysis of proteins extracted from GHFT1–5 cells transfected with the indicated amount of the GFP-Pit-1 expression plasmid. The transferred proteins were probed with either an antibody against Pit-1 (left panel) or an anti-GFP antibody (right panel). The anti-Pit-1 antibody detected both the doublet for endogenous 31- and 33-kDa Pit-1 proteins (double arrows) and the expressed 60 kDa GFP-Pit-1 (arrow), whereas the GFP antibody detected the 60-kDa GFP-Pit-1. B, Cell extracts from GHFT1–5 cells transfected with the indicated amount of the GFP-C/EBPα expression plasmid were subjected to Western blot analysis using the anti-GFP antibody. The GFP antibody detected the 70-kDa GFP-C/EBPα fusion protein. C, EMSA showing that untagged C/EBPα (lanes 1–5) and GFP-C/ EBPα (lanes 6–10) have similar DNA-binding characteristics. Cell extracts were prepared from GHFT1–5 cells expressing the indicated protein, and samples were incubated with a labeled C/EBPα response element (16) as described in Materials and Methods. After gel electrophoresis a single DNA-protein complex was observed (arrow). Binding specificity was demonstrated by competition with an excess of unlabeled oligonucleotide (lanes 2–4 and 7–9), and the presence of C/EBPα in the complex was verified by a shift in mobility resulting from the addition of an antibody specific for C/EBPα (open arrow, lanes 5 and 10). D, Cell extracts were prepared from 3T3-L1 cells expressing GFP-Pit-1, and samples were incubated with a labeled PRL 3P Pit-1 response element as described in Materials and Methods. GFP-Pit-1 formed two distinct complexes (arrows) with the PRL 3P DNA-element. Probe specificity was demonstrated using 3- to 100-fold excess unlabeled oligonucleotide (wedge) and mobility shift with addition of a Pit-1-specific antibody. Other DNA-protein complexes relate to ETS-protein binding to the 3P DNA element (18).
Characterization of GFP-Tagged C/EBPα and Pit-1
Because C/EBPα and Pit-1 interacted cooperatively to induce PRL promoter activity, we determined whether these proteins also colocalized in nuclei of the living GHFT1–5 cells. This was accomplished by expression of either Pit-1 or C/EBPα fused to different color variants of the fluorescent proteins (FPs) in GHFT1–5 cells. Western blot analysis of extracts prepared from the transfected cells verified that the expressed GFP Pit-1 (Fig. 2A) and GFP-C/EBPα (Fig. 2B) fusion proteins were the appropriate size. In addition, the expressed GFP-Pit-1, detected with an antibody directed against Pit-1, was similar in abundance to the endogenous Pit-1 protein in the GHFT1–5 cell extracts (Fig. 2A). The amount of endogenous Pit-1 in GHFT1–5 cells is about 10–30% of that present in mature, PRL secreting cell types (12). Our fluorescence microscopy studies (see below) showed that the GFP-fusion proteins could be detected in at least 40% of the transfected cells. Although there is cell-to-cell variation in the expression level of the GFP-fusion proteins, these results shown in Fig. 2A that, on average, the fusion proteins were expressed at levels similar to other endogenous nuclear proteins.
To assess the function of the GFP-tagged C/EBPα and Pit-1, we measured their ability to bind to appropriate DNA-response elements. Extracts were prepared from GHFT1–5 cells expressing C/EBPα and GFP-C/EBPα or from 3T3 -L1 cells expressing GFP Pit-1. These protein extracts were then incubated with the indicated DNA-binding sites and subjected to EMSA. The results shown in Fig. 2C demonstrated that C/EBPα (lane 1) or GFP-C/EBPα (lane 6) each formed a single DNA-protein complex with the consensus C/EBPα DNA-response element (16). Competition by 3- to 100-fold excess unlabeled oligonucleotide demonstrated that the DNA-protein complexes were probe-specific (Fig. 2C, wedges). Incubation with an antibody directed against C/EBPα resulted in decreased mobility of the DNA-protein complex, confirming that C/EBPα was present in these complexes (Fig. 2C, lanes 5 and 10). Similarly, GFP-Pit-1 was found to bind specifically to the PRL gene promoter 3P Pit-1 DNA-response element (Fig. 2D). Consistent with previous studies of Pit-1 DNA-binding activity (8), we observed two distinct DNA-protein complexes formed by GFP-Pit-1 (arrows, Fig. 2D), as well as a complex containing ETS-family proteins that interact with the 3P DNA element in conjunction with Pit-1 (17, 18). Competition with an excess of unlabeled 3P DNA (wedge, lanes 2–4) and an antibody directed against Pit-1 showed these complexes each contained GFP Pit-1. These results demonstrated that the GFP tag did not alter the DNA binding characteristics of either C/EBPα or Pit-1.
Coincident Subnuclear Localization of Endogenous and GFP-Tagged C/EBPα and Pit-1
C/EBPα is present in rat pituitary cell lines that secrete GH and PRL, but is absent from the mouse pituitary progenitor GHFT1–5 cells, which do not express these hormones (14). We showed that the expression of exogenous C/EBPα in GHFT1–5 cells leads to activation of a cotransfected GH gene promoter (14) and PRL gene promoter (Fig. 1A), and results in inhibition of cell proliferation (19). Because the GHFT1–5 cells do not express C/EBPα, we characterized the intranuclear positioning of endogenous C/EBPα in a different mouse cell line. Earlier studies showed that C/EBP was expressed in mouse 3T3-L1 cells that were induced to differentiate into adipocytes, where it plays a key role in the adipogenic cascade (20, 21). Here, the endogenous C/EBPα was detected by immunohisto chemical staining in the nuclei of 3T3-L1 cells that had differentiated to adipocytes by hormone treatment (Fig. 3A). To provide a marker for chromatin structure, the cells were stained with the blue fluorescent DNA dye, Hoechst 33342 (H33342). The H33342 preferentially binds to the tracts of satellite DNA repeats located at centromere regions of interphase chromosomes in mouse cells (14, 22, 23). The results showed that the endogenous C/EBPα was highly concentrated in large foci in the nuclei of the induced 3T3-L1 cells (Fig. 3A). Overlaying the red fluorescent C/EBPα image with the blue fluorescent H33342 image resulted in a purple-colored image (Fig. 3A, merge) indicating that the distributions of C/EBPα and H33342-stained DNA were identical. No C/EBPα was detected in undifferentiated 3T3-L1 cells (data not shown). A very similar pattern of intranuclear distribution was observed for GFP-C/EBPα when expressed in the 3T3-L1 preadipocyte cells (Fig. 3A, right panel).
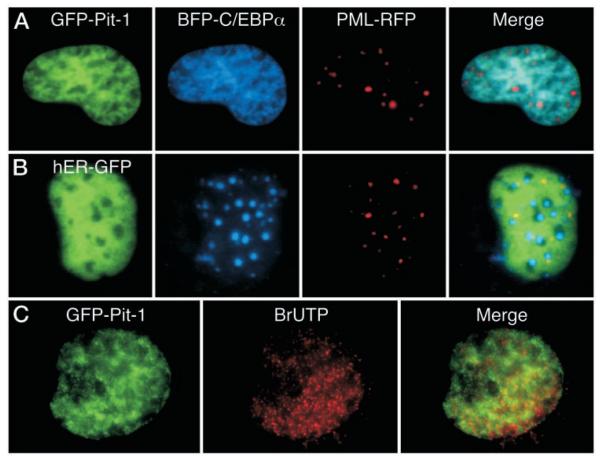
Fig. 3.
The Expressed GFP-Pit-1 and GFP-C/EBPα Have the Same Intranuclear Distribution as Their Endogenous Counterparts
A, Immunohistochemical staining of C/EBPα in mouse 3T3-L1 cells induced to differentiate to adipocytes. Endogenous C/EBPα stained with an anti-C/EBPα antibody and a rhodamine-linked secondary antibody. Overlay of the red C/EBPα fluorescence and blue H33342 fluorescence images appears purple (merge) demonstrating that endogenous, adipocyte C/EBPα was colocalized with the H33342-stained chromatin. In a separate experiment, 3T3-L1 cells were then transfected with the plasmid encoding GFP-C/EBPα, and GFP fluorescence was detected directly by fluorescence microscopy (right panel). B, Immunohistochemical staining of Pit-1 in mouse GHFT1–5 cells. The endogenous Pit-1 was detected with antisera to Pit-1 anda Texas red-linked secondary antibody. Merger of the red Pit-1 fluorescence and blue H33342 fluorescence images show that Pit-1 is not concentrated in regions of heterochromatin preferentially stained by H33342. An image of a GHFT1–5 cell nucleus expressing GFP-Pit-1 is shown for comparison (right panel). GHFT1–5 cells were then transfected with plasmids encoding either GFP-C/EBPα (panel C), GFP-C/EBPΔ244 (panel D), or GFP-Pit1 (panel E) and grown on cover glass in 35-mm culture dishes. After 24 h the living cells were stained for 20 min with the cell-permeable DNA dye H33342. Sequential images were acquired of the GFP fusion protein and the stained DNA in the same focal plane as described in Materials and Methods. The calibration bar indicates 10 μm. The images were merged to show regions of overlap, which appear as cyan color in the merged image. An intensity profile was obtained for both GFP emission (green line) and H33342 fluorescence (blue line) at the position indicted by the line in each merged image, and the results were plotted (right panels).
The mouse GHFT1–5 cells express low levels of endogenous Pit-1 (Fig. 2A) that is approximately 10–30% of that present in mature, somatolactotrope cell types (12, 14). The endogenous Pit-1 protein was detected by immunohistochemical staining using an antibody against Pit-1 (Fig. 3B), and its intranuclear distribution relative to chromatin was determined by staining with H33342. The endogenous Pit-1 protein had a reticular pattern throughout the nucleus, and there were distinct regions where the protein was not concentrated (Fig. 3B). Chromatin staining with H33342 revealed that centromeric heterochromatin was localized to these same regions of low Pit-1 concentration (Fig. 3B, merge). There was no nuclear staining observed for cells incubated with secondary antibody alone (data not shown). The pattern of intranuclear distribution observed for GFP-Pit-1 expressed in a different GHFT1–5 cell was very similar to that observed for the endogenous protein (Fig. 3A, right panel). Together these results showed that the expressed GFP-fusion proteins adopt the same intranuclear distributions as their endogenous counterparts.
We then used fluorescence microscopy to visualize the distribution of these GFP-fusion proteins relative to chromatin in the nucleus of living GHFT1–5. As we reported previously (14, 24), GFP-C/EBPα was concentrated in regions of centromeric chromatin that were preferentially stained with the H33342 dye (Fig. 3C). There was also a significant amount of GFP-C/ EBPα localized to regions outside of the heterochromatin foci stained with H33342 (Fig. 3C, intensity profile). In contrast, when only the carboxy-terminal B-ZIP region of C/EBPα fused to GFP (GFP-C/EBPΔ244) was expressed in GHFT1–5 cells, it was predominately localized to the heterochromatin foci (Fig. 3D, intensity profile). This result suggests that activities associated with the amino-terminal transactivation domain could function to direct C/EBPα into other nuclear domains. As was shown for the endogenous Pit-1 protein (Fig. 3B), the expressed GFP-Pit-1 adopted a web-like pattern of nuclear distribution and did not concentrate in the regions of heterochromatin that were preferentially stained by H33342 (Fig. 3E). Therefore, the intranuclear locations adopted by the GFP-tagged C/EBPα and Pit-1 fusion proteins faithfully mimicked the intranuclear locations of their endogenous counterparts.
Pit-1 Coexpression Dispersed C/EBPα Away from Peri-Centromeric Heterochromatin
We then determined whether the coexpression of Pit-1 and C/EBPα in the same cell might influence their individual patterns of intranuclear distribution. For these experiments we used the promyelocytic leukemia (PML) protein as a marker for nuclear structure. PML is a member of a family of proteins that form well defined 0.5-μm subnuclear structures called nuclear bodies (25). The sequence encoding PML was fused in-frame to that of the Discosoma sp. red fluorescent protein (RFP) (26). We demonstrated previously that the expressed PML-RFP localized to discrete nuclear bodies (24) and that it colocalized with endogenous PML in HeLa cells (not shown).
When GHFT1–5 cells were cotransfected with plasmids encoding GFP-Pit-1, blue fluorescent protein (BFP)-C/EBPα, and PML-RFP, we observed that the C/EBPα was no longer localized to regions of centromeric heterochromatin, but was instead localized in a reticular pattern that was coincident with GFP-Pit-1 (Fig. 4A). PML-RFP remained localized to the well defined nuclear bodies, arguing against a restructuring of the nucleus in these transfected cells. Importantly, not all factors that were coexpressed with BFP-C/ EBPα influenced its subnuclear localization. When BFP-C/EBPα was coexpressed with a functional estrogen receptorα (ERα)-GFP fusion protein (human ER-GFP; Ref. 27), the two fusion proteins distributed independently, with little overlap in their subnuclear localization (Fig. 4B). Again, the PML-RFP was localized to nuclear bodies, and these were distinct from the foci occupied by BFP-C/EBPα. Together, these results provide evidence for the specific recruitment of C/EBPα to the intranuclear sites occupied by the coexpressed Pit-1.
Fig. 4.
Pit-1 Recruits the Coexpressed C/EBPα
GHFT1–5 cells were cotransfected with BFP-C/EBPα, PML-RFP, and either GFP-Pit1 (panel A) or GFP-ER (panel B). Sequential images from the same focal plane were acquired using suitable filters as described in Materials and Methods. The images were merged to show regions of overlap, with blue and green overlap indicated by cyan color, and regions of red and green overlap indicated by yellow color in the merged image. C, GHFT1–5 cells expressing GFP-Pit1 were permeabilized and exposed to BrUTP for 20 min. BrUTP was then immunohistochemically detected in fixed cells with a Texas red-conjugated secondary antibody as described in Materials and Methods. Dual color images were obtained in the same focal plane, and the images were merged to show regions of overlap, which appear yellow in the merged image.
Because Pit-1 and C/EBPα interacted cooperatively to activate PRL gene expression (Fig. 1), we next examined the relationship of the nuclear regions occupied by GFP-Pit-1 to sites of active transcription. Nascent mRNA transcripts in GHFT1–5 cells transfected with GFP-Pit-1 were detected by labeling with bromouridine (BrUTP). The results shown in Fig. 4C demonstrated that the distribution of nascent mRNAs and GFP-Pit-1 partially overlapped. This was in contrast to the observation that the foci occupied by C/EBPα in transfected GHFT1–5 cells were relatively devoid of nascent mRNA transcripts (14).
The Pit-1 HD Is Required for the Recruitment of C/EBPα from Heterochromatin
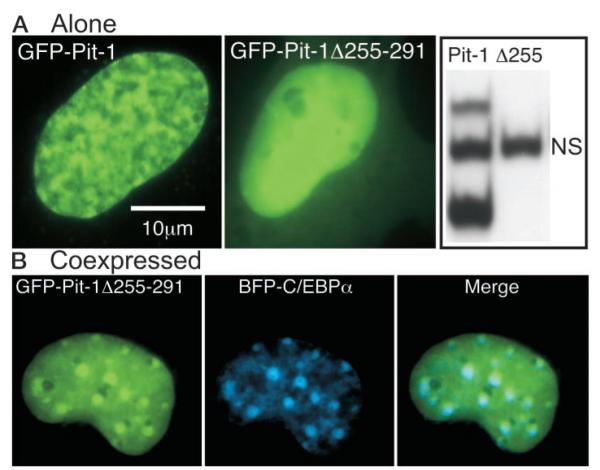
Because the conserved Pit-1 HD was shown to mediate interactions with other proteins (28, 29), we examined what effect a deletion within the HD had on recruitment of C/EBPα. Deletion of the carboxy-terminal portion of the Pit-1 HD (amino acid residues 255–291, Pit-1Δ255–291) abolished the ability of the protein to bind to a Pit-1 DNA element from the PRL promoter (Fig. 5A, right panel). Imaging cells expressing GFP Pit-1Δ255–291 revealed a diffuse pattern of intranuclear distribution distinct from the reticular pattern observed for the intact GFP-Pit-1 (Fig. 5A). When GFP Pit-1Δ255–291 was coexpressed with BFP-C/EBPα, the BFP-C/EBPα was not relocalized by the truncated Pit-1 protein (Fig. 5B). Instead, the GFP-Pit-1Δ255–291 tended to accumulate in the sites occupied by BFP-C/EBPα (Fig. 5B, merge). This result indicates that domains other than the Pit-1 HD may also interact with C/EBPα or other associated coregulatory factors, but that the recruitment activity of Pit-1 for C/EBPα required the intact HD.
Fig. 5.
The HD of Pit-1 Is Necessary for Specific Subnuclear Interactions with C/EBPα
A, Fluorescence images of living GHFT1–5 cells expressing either GFP-Pit-1 (left panel) or GFP-Pit1 lacking the carboxyterminal portion of the HD (GFP-Pit-1Δ255–291, middle panel). Cell extracts were prepared from HeLa cells expressing either GFP-Pit-1 or GFP-Pit-1Δ255–291 and samples were incubated with a labeled PRL 1P Pit-1 response element as described in Materials and Methods. EMSA showed that GFP-Pit-1 (lane 1) bound to the Pit-1 DNA element resolved as two distinct complexes, whereas there was no detectable binding of GFP-Pit1Δ255 (lane 2) to the 1P Pit-1 DNA element (NS, nonspecific, see Fig. 2). B, Dual color fluorescence images of living GHFT1–5 cells coexpressing GFP-Pit1Δ255 and BFP-CEBPα were acquired from the same focal plane. The images were merged to show regions of overlap, which appear as cyan color in the merged image.
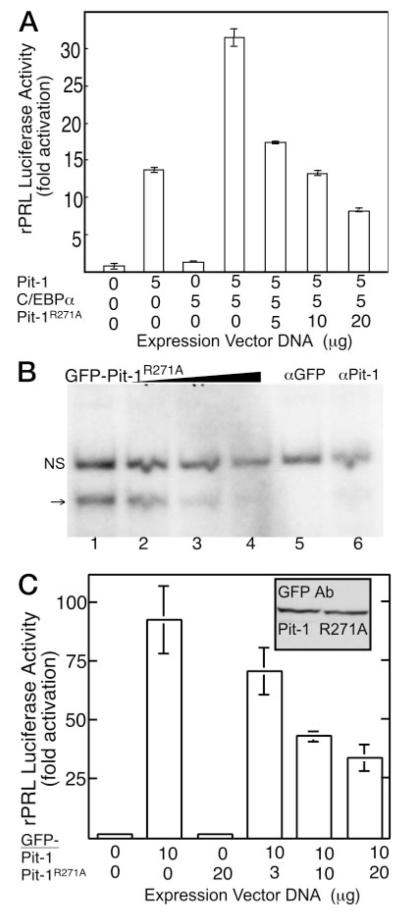
The Pit-1 HD Mutant, R271A, Fails to Redistribute C/EBPα and Blocks PRL Promoter Activation
Because the Pit-1 HD is necessary for site-specific DNA binding, the loss of DNA binding could explain the failure of Pit-1Δ255–291 to recruit C/EBPα. Several different Pit-1 mutations linked to the syndrome of CPHD that do not interfere with DNA binding have been reported (6, 7). Specifically, mutations altering the arginine residue at position 271 of the Pit-1 HD produce a dominant inhibitor of Pit-1-dependent transcription that binds with high affinity and specificity to Pit-1 DNA elements (30, 31). This mutant Pit-1 protein allowed us to examine how changes in the Pit-1 HD that do not disrupt specific DNA binding might effect the ability of Pit-1 to interact with other nuclear proteins.
A mutant Pit-1 protein with substitution of alanine for arginine 271 (Pit-1R271A) was prepared, and the dominant inhibitory activity of this protein was determined. The results in Fig. 6 demonstrate the ability of Pit-1R271A to block the cooperative actions of Pit-1 with C/EBPα at the rPRL promoter in transfected HeLa cells (Fig. 6A). A GFP fusion to Pit-1R271A was made to examine its intranuclear distribution and ability to interact with C/EBPα. The DNA-binding activity of this fusion protein was assessed by EMSA using extracts prepared from transfected HeLa cells. The results shown in Fig. 6B demonstrated that GFP-Pit-1R271A mutant bound with high specificity to the PRL gene 1P Pit-1 DNA element, but only a single, specific DNA-protein complex was observed (arrow, Fig. 6B). This complex was cleared from the reaction by incubation with an antibody that disrupts Pit-1 binding to DNA. These results are in accord with an earlier study indicating that the CPHD Pit-1R271W bound to Pit-1 DNA elements predominately as a monomer whereas wildtype Pit-1 bound both as a monomer and a dimer (31). As a further measure of function, reporter gene analysis demonstrated that GFP-Pit-1R271A retained the dominant inhibitory activity. When expressed in HeLa cells, GFP-Pit-1 strongly induced the luciferase reporter gene linked to the −204 rPRL promoter, and increasing concentrations of the plasmid encoding the GFP-Pit-1R271A inhibited this activity (Fig. 6C). Western blot analysis (inset, Fig. 6C) showed these GFP tagged proteins were expressed equivalently after transfection of identical amounts of expression vector. Together, these results confirmed the dominant negative attributes of Pit-1 mutated at amino acid residue 271 and showed that mutation at this residue blocked the cooperative activation of the PRL promoter by Pit-1 and C/EBPα.
Fig. 6.
The Pit-1 Mutant, R271A, Binds Specifically to DNA and Functions as a Dominant Inhibitor of PRL Gene Expression
A, HeLa cells were transfected with the rPRL luc reporter and the indicated amounts of the expression plasmids encoding C/EBPα, Pit-1, the Pit-1R271A mutant, or the indicated combination. Luciferase activity was determined after 24 h and was corrected for total cellular protein. The error is the SEM from three independent experiments, each done in triplicate and normalized to reporter alone. B, GFP-Pit1R271A was bound specifically to the PRL promoter 1P DNA element, but formed only a monomeric complex. Probe specificity was demonstrated using 3- to 100-fold excess unlabeled oligonucleotide (wedge, lanes 2–4), and immunoclearing was observed with either a GFP-specific (lane 5) or Pit-1 specific (lane 6) antibody; NS, Nonspecific complex. C, HeLa cells were transfected with the rPRL luc reporter and the indicated amount of GFP-Pit-1 and GFP-Pit1mut. Inset, Western blot demonstrating that the GFP-Pit1 and GFP-Pit-1R271A were expressed at equivalent levels. Error is SEM from three independent experiments, each done in triplicate and normalized to reporter alone.
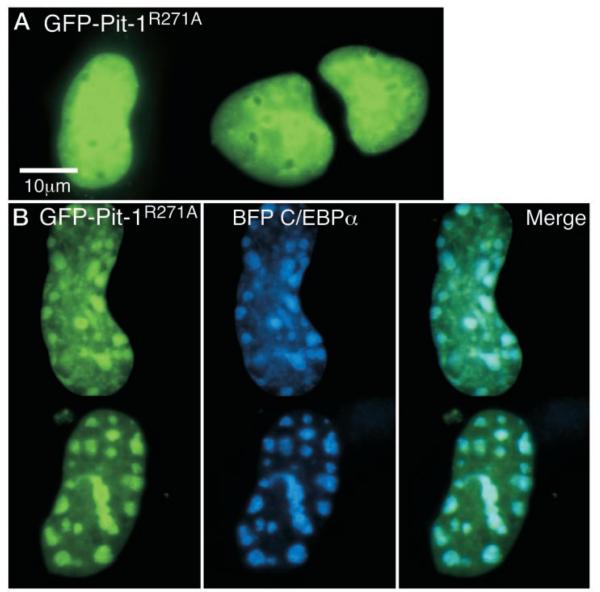
When expressed in GHFT1–5 cells, the GFP-tagged Pit-1R271A adopted a pattern of subnuclear distribution that was distinct from GFP-Pit-1 and very similar to that observed for Pit-1Δ255–291 deletion, having a uniform distribution throughout the nucleus (Fig. 7A; compare with Fig. 5). Therefore, the ability of Pit-1R271A to bind to specific Pit-1 DNA response elements (Fig. 6B) was insufficient to direct the subnuclear targeting of the protein. Further, when the GFP-Pit-1R271A was coexpressed with BFP-C/EBPα, the Pit-1 mutant failed to influence the distribution of the BFP-C/EBPα (Fig. 7B). This result demonstrated that the carboxy-terminal end of the HD played a critical role in the recruitment activity of Pit-1. Interestingly, as was observed for the Pit-1 HD deletion (Fig. 5), there was a strong tendency for GFP Pit-1R271A to accumulate in the sites occupied by BFP-C/EBPα (Fig. 7B). Thus, the CPHD Pit-1 mutation, which inhibited both Pit-1- and C/EBPα-regulated PRL gene expression, localized to centromeric heterochromatin when coexpressed with C/EBPα.
Fig. 7.
FP-Pit1R271A Fails to Recruit C/EBPα
A, GHFT1–5 cells were transfected with the expression plasmid encoding GFP-Pit1mut and grown on cover glasses in 35-mm culture dishes. After 24 h, images were acquired of the GFP-fusion protein. Calibration bar indicates 10 μm. B, GHFT1–5 cells were cotransfected with plasmids encoding BFP-C/EBPα and GFP-Pit-1R271A. Sequential blue and green fluorescent images from the same focal plane were acquired using suitable filters as described in Materials and Methods. The images were merged to show regions of overlap, with blue and green overlap indicated by cyan color.
DISCUSSION
The transcription factor Pit-1 is essential for the establishment of pituitary somatotrope, lactotrope, and thyrotrope cells (9, 32–34). The central role of Pit-1 in pituitary cell development is exemplified by various inactivating mutations that cause the syndrome of CPHD (6, 7, 32). Although some of these mutants prevent Pit-1 activity by disrupting DNA binding, other mutations that do not alter DNA-binding still function as dominant inhibitors by mechanisms that remain unclear. The results described here supply a possible explanation for the dominant inhibitory activity of one CPHD Pit-1 mutant and provided evidence for a general mechanism by which the intranuclear location of transcriptional complexes contributes to either gene activation or silencing.
Cooperative Interactions Between Pit-1 and C/EBPα Induce PRL Transcription
Pit-1 cooperates with other transcription factors to regulate both the basal expression and hormonal regulation of the pituitary-specific PRL gene (9, 13, 17, 18, 28, 29, 35–38). We previously demonstrated that Pit-1 and C/EBPα cooperate in the regulation of GH transcription (12, 14). Similarly, Jacob and Stanley (13) observed the interaction of Pit-1 and C/EBPα at the PRL gene promoter. Here, we showed that expression of C/EBPα in pituitary GHFT1–5 cells, which have low levels of Pit-1 and are devoid of endogenous C/EBPα, could induce rPRL promoter activity (Fig. 1A). This activity required the PRL promoter element between −97 to −91 bp (Fig. 1A), a site implicated in earlier studies as necessary for both basal and hormonestimulated promoter activities (13, 36–38). A functional interaction between C/EBPα and Pit-1 was shown in the nonpituitary human HeLa cells, where the coexpressed proteins cooperated in the activation of PRL transcription (Fig. 1B). Together, our results suggest a cooperative interaction between C/EBPα and Pit-1, which may involve the physical association of these transcription factors. Our recent studies using fluorescence resonance energy transfer microscopy supports this view (39).
The Intranuclear Positioning of C/EBPα
Many recent studies suggest that transcription factors and other regulatory proteins assemble in higher order transcriptional complexes at specific subnuclear sites (39a–43). We observed here that the endogenous C/EBPα in differentiated 3T3-L1cells was preferentially localized to regions of centromeric heterochromatin (Fig. 3). Similarly, we found GFP-C/EBPα localized to these same subnuclear sites when expressed in either mouse 3T3-L1 preadipocyte cells or mouse GHFT1–5 pituitary progenitor cells (Fig. 3 and Ref. 14). Our earlier studies demonstrated that the subnuclear distribution of coregulatory proteins, including the CREB binding protein, the TATA binding protein, and acetylated histone H3, was altered upon C/EBPα expression such that they relocalized to the centromeric heterochromatin domains occupied by C/EBPα (14, 43a). In addition, we have found that point mutants of C/EBPα, which fail to concentrate at the sites of centromeric heterochromatin, are more transcriptionally active (Bo Wu, R. N. D., and F. Schaufele, unpublished data). This observation favors a model in which C/EBPα may be sequestered at transcriptionally inactive heterochromatin (14). The C/EBP family of proteins are known to regulate cell proliferation and differentiation (10, 11). For example, Tang and Lane (20) showed that the centromeric localization of the endogenous C/EBPα in mouse adipocytes functioned to control cell proliferation during adipocyte terminal differentiation. The GHFT1–5 cells used in our studies have the phenotype of the progenitor of the somatolactotrope cell lineage (15), and the targeting of GFP C/EBPα to centromeres may be indicative of their stage in pituitary differentiation. These results indicate that the targeting of C/EBPα to regions of centromeric heterochromatin may be intrinsic to its biological functions.
The centromeric heterochromatin in mammalian cells consists of arrays of tandemly repeated satellite DNA that is assembled into higher order chromatin structure (22). These long arrays of repeated satellite DNA are present in all mammalian cells, but mouse cells possess large blocks of highly condensed centromeric heterochromatin that is readily visualized by staining with Hoechst DNA dyes (22, 23). Our ability to detect the positioning of C/EBPα at these subnuclear sites was possible because these regions are particularly well defined in mouse cells (44, 45). In this regard our observations of the positioning of C/EBPα relative to centromeric heterochromatin could be a property that is unique to mouse cells. However, recent studies indicate that the structure of the centromeres is highly conserved across species, particularly at the level of the protein components of the kinetochore (46, 47). For example, the evolutionarily conserved histone-like centromere protein A is involved in the assembly of centromeric DNA into higher order structure in a variety of different organisms (47). Furthermore, C/EBPα was shown to be a strong inhibitor of cell proliferation in both mouse and human cell lines (20, 49), and we observed previously that the expression of C/EBPα in pituitary GHFT1–5 cells also inhibited cell growth (19). This similar activity in cell lines from different species argues against a mouse cell-specific function of C/EBPα. The well defined centromeric regions that are visible in the mouse cell nucleus, however, have enabled us to study the role of C/EBPα subnuclear distribution in its transcriptional and antiproliferative activities (14, 19, 24).
C/EBPα Is Recruited to the Intranuclear Sites Occupied by Pit-1
These earlier findings prompted us to investigate whether the discrete intranuclear positioning of C/EBPα in the mouse pituitary cells could be influenced by Pit-1. We found that the low level of endogenous Pit-1 protein in GHFT1–5 cells was distributed in a reticular pattern throughout the nucleus, but was not concentrated at sites of centromeric heterochromatin (Fig. 3). This same pattern of subnuclear distribution was observed for GFP-Pit-1 expressed in GHFT1–5 cells (Fig. 3). We showed that when GFP-C/ EBPα was expressed in the pituitary GHFT1–5 cells, it was incompletely localized to regions centromeric heterochromatin (Fig. 3A). In contrast, we found that a truncated GFP-C/EBPΔ244 protein, containing only the B-ZIP DNA-binding domain, was almost exclusively localized to these sites (Fig. 3B). This could indicate that C/EBPΔ244 has a higher affinity for heterochromatin. However, studies by others demonstrated that the DNA binding specificity and affinity for the B-ZIP domain alone were very similar to those of the full-length protein (50). Alternatively, our results might indicate that activities associated with the amino-terminal transactivation domain function to direct C/EBPα to regions outside the heterochromatin foci.
In this regard, we found that when GFP-Pit-1 and BFP-C/EBPα were coexpressed in the same pituitary cells, C/EBPα adopted a pattern of intranuclear distribution that completely overlapped that of Pit-1 (Fig. 4). The colocalization of C/EBPα with Pit-1 did not result from nonspecific interactions of the FPs, because a third coexpressed protein, PML-RFP, remained localized to separate nuclear bodies in these same cells (Fig. 4). Moreover, when BFP-C/EBPα was coexpressed with the ERα-GFP, C/EBPα remained localized to foci independent of the ERα-GFP. We also observed in fixed cells that GFP-Pit-1 partially overlapped sites of active gene transcription marked by BrUTP-labeled nascent mRNAs. In contrast, we previously demonstrated that C/EBPα accumulated at heterochromatin foci that were relatively devoid of nascent mRNA transcripts. Together, these results suggest that the recruitment activities of Pit-1 for C/EBPα may play an organizational role that is essential for their cooperative activation of pituitary-specific PRL and GH gene transcription, and the developmental progression of the somatolactotrope progenitor cells.
The HD of Pit-1and the Recruitment of C/EBPα
The specific organizational activity of Pit-1 for C/EBPα was demonstrated by the effect of deletions and mutations in Pit-1. A truncation of Pit-1 that removed the HD and abolished its activity at both the GH (51) and PRL (52) promoters failed to redistribute C/EBPα from the regions of centromeric heterochromatin (Fig. 5). Instead, when coexpressed with C/EBPα, there was a tendency for the truncated Pit-1 protein to associate with C/EBPα in regions of centromeric heterochromatin. This suggests that the Pit-1 HD deletion retained some ability to interact with C/EBPα, but that the recruitment activities of Pit-1 required the intact HD.
The HD forms half of a conserved bipartite DNAbinding motif (8, 31), and binding to specific DNA sites may be prerequisite for the recruitment of other interacting protein partners (35). We tested this hypothesis by using a Pit-1 protein mutated at the R271 residue, which lies outside the DNA-binding domain. The dominant inhibitory activity previously reported for Pit-1 R271 mutants is causative for the syndrome of CPHD (6, 7, 30). We found that the Pit-1R271A mutant acted as a dominant inhibitor of PRL gene transcription, preventing the cooperation between Pit-1 and C/EBPα (Fig. 7). The Pit-1R271A mutant bound specifically to Pit-1 DNA response elements predominately as a monomer. The intranuclear distribution of GFP Pit-1R271A, however, was diffuse like that of the Pit-1 HD deletion, which did not bind to Pit-1 DNA elements (compare Figs. 5 and 7). This indicated that binding to specific DNA elements alone was not sufficient to direct the subnuclear targeting of Pit-1. Further, when coexpressed with C/EBPα, the Pit-1 point mutant failed to recruit C/EBPα and instead became associated with C/EBPα in regions of centromeric heterochromatin. It is possible that disruption of the Pit-1 dimerization interface by mutation of R271 (31), which prevents the protein from assuming dimer conformations, also prevents the formation of other important protein-protein contacts, such as that observed here for C/EBPα. These observations suggest that the dominant inhibitory actions of the CPHD Pit-1 mutant could be related to its redirection into the transcriptionally inactive centromeric heterochromatin. This raises the possibility that other proteins associated in complexes with Pit-1 would similarly be directed to these transcriptionally silent subnuclear domains.
Nuclear Compartmentalization as a Regulator of Gene Expression Patterns
Together our results indicate that the location of transcription factors within the nucleus may be a critical epigenetic determinant for the formation of transcriptional complexes necessary for the regulation of specific genes. In this case, the positioning of transcription factor complexes in nuclear compartments associated with centromeric heterochromatin may function to silence specific genes. The interphase centromere is thought to position chromosome territories (53), and the silencing of transcriptional activity can occur when genes are located close to the heterochromatin (54). A recent analysis of the human β-globin locus control region demonstrated that a transcriptional enhancer functions to maintain the gene at a distance from centromeric heterochromatin, thus preventing its silencing (55).
The pituitary cell-specific expression of the GH gene requires a locus control region positioned 15 kb up stream of the promoter, and Pit-1 binds to an array of DNA-elements within this region (56). It is possible that the location of Pit-1 and its cooperating factors within the nucleus plays a critical role in the positioning of this locus control region. Thus, the Pit-1 CPHD mutant shown here, which localized to centromeric chromatin when coexpressed with C/EBPα, could function to position genes with suitable DNA elements near these regions of silencing. Indeed, other transcription factors are known to function as repressors through this type of mechanism. For example, during B cell development the zinc-finger protein Ikaros/Lyf-1 becomes localized to centromeres where it functions to recruit genes to be silenced during lymphocyte activation (57–60). In addition, proteins with the Krüppel-associated box also function as transcriptional repressors, exerting their silencing activity by recruitment of other factors and target gene loci to the transcriptionally inert centromeric heterochromatin (61). Further studies using techniques such as fluorescence in situ hybridization and chromatin immunoprecipitation assay will be necessary to determine how the distribution of these protein complexes is related to the positioning and the transcriptional state of specific endogenous gene loci in pituitary cells.
Our observations of the colocalization of Pit-1 and C/EBPα in the pituitary cell nucleus using conventional fluorescence microscopy are limited by the diffraction of light to a resolution of approximately 200 nm. Although these observations imply that C/EBPα and Pit-1 are associated, they do not conclusively establish this point. Our failure to detect a physical interaction between these proteins using more traditional biochemical approaches suggested either an indirect interaction or an interaction dependent upon conditions only present in the environment within the intact living cell. Significantly, we have used fluorescence resonance energy transfer microscopy to demonstrate that Pit-1 and C/EBPα are in close physical association in the living pituitary cell nucleus (39). Together with the studies reported here, we provide striking evidence that some transcription factors can specifically interact with, and direct, cooperating factors to particular sites in the nucleus, and that a mutation associated with human disease can dramatically alter the intranuclear targeting of these factors.
MATERIALS AND METHODS
Construction of Expression Vectors, Transfection of Cell Lines, and Reporter Gene Assays
The cDNAs encoding either rat C/EBPα, rat Pit-1 or the mutants were each inserted into the pCDNA His3.1 expression vector (Invitrogen, San Diego, CA). These sequences and the sequence encoding either the human estrogen receptor α (ERα) or human PML protein were also fused in frame to sequences encoding each of the indicated fluorescent proteins (CLONTECH Laboratories, Inc., Palo Alto, CA) as described previously (14, 24). For transfection, GHFT1–5, HeLa, or 3T3 -L1 cells were maintained as a monolayer in DMEM containing 10% fetal calf serum (FCS), harvested, and transfected with the indicated plasmid DNA(s) by electroporation as described previously (14, 24). The total amount of DNA was kept constant using the pCDNA His3.1 expression vector. For the reporter gene experiments, the rat PRL (rPRL) promoter −204 to +34) or the same promoter containing cluster point mutations at positions −97 to −91 (36) were coupled to the luciferase (Luc) reporter gene. Cell extracts were prepared 24 h after transfection for determination of Luc activity as described by the manufacturer (Promega Corp., Madison WI). Luc activity for each sample was determined in duplicate and corrected for total protein, and the combined results from at least three independent transfections are shown.
Western Blotting and EMSA
For Western blotting, GHFT1–5 cells were transfected with the indicated protein expression vector, and detergent lysates were prepared after 24 h as described previously (63). After electrophoresis and transfer to nitrocellulose membranes, the proteins were detected by incubation with anti body against C/EBPα (sc-61, Santa Cruz Biotechnology, Inc., Santa Cruz, CA; 1:200 final dilution) or Pit-1 (sc-442; 1:5000 final dilution). This was followed by incubation with secondary antibody (horseradish peroxidase-conjugated goat antirabbit, Pierce Chemical Co., Rockford, IL) at a final dilution of 1:50,000. The membranes were then washed and incubated in enhanced chemiluminescence substrate (Amersham Pharmacia Biotech, Arlington Heights, IL) for 1 min and then exposed to film for 10 min as described previously (62).
EMSAs were performed on whole-cell extracts prepared from transiently transfected GHFT1–5 cells as described previously (18). Duplex oligonucleotides probes corresponding to either a consensus C/EBP binding site (16) or Pit-1 sites were (18):
CEBP RE 5−-GATCGAGCCCCATTGCGCAATCTATATTCG
PRL 1P 5−-CCTGATTACATGAATATTCATGAAGGTG
PRL 3P 5−-GGCTTCCTGAATATGAATAAGA
Each probe was prepared by end-labeling using [γ-32P] ATP and T4 polynucleotide kinase. Samples of cell extracts (10 μg) were added to the reaction mixtures assembled on ice. For competition studies, unlabeled duplex oligonucleotide was added in 3-, 30-, or 100-fold excess. Where indicated, antisera against the expressed protein (0.2 μl) was added to the reaction mixtures and incubated for 20 min at 4 C. Antibodies specific for Pit-1 that either super-shifted the DNA-protein complex (Geneka Biotechnology, Inc., Montréal, Canada) or disrupted DNA binding (63) were used in these studies. The reaction mixtures were transferred to tubes containing 25,000–50,000 cpm of the end-labeled probe and incubated for 20 min at room temperature. The samples were then fractionated on prerun 6.0% polyacrylamide gels, followed by autoradiography as described previously (18).
Immunohistochemistry and Labeling of Nascent Transcripts
To induce differentiation to adipocytes, the 3T3-L1 cells were incubated in medium containing 10% FCS that was supplemented with 1 μg/ml insulin, 1 μM dexamethasone, and 0.5 mM 3-isobutyl-1-methylxanthine for 2 d, followed by incubation in medium containing 10% FCS with 1 μg/ml insulin (20). The nontransfected mouse pituitary GHFT1–5 cells and adipocyte cells were cultured on glass cover slips. Cells were maintained in culture 24–48 h, and then fixed by incubation in 1.5% formaldehyde in PBS, and then processed for immunohistochemical detection. Endogenous C/EBP was detected in fixed adipocytes by incubation with a rabbit polyclonal C/EBPα primary antibody (1:100 dilution of sc-61, Santa Cruz Biotechnology, Inc.) followed by incubation with an antirabbit rhodamine-conjugated secondary antibody. The endogenous Pit-1 was detected in fixed GHFT1–5 cells by incubation with a rabbit polyclonal Pit-1 antiserum (1:500 dilution), followed by incubation with antirabbit Texas redconjugated secondary antibody. The antiserum to Pit-1 was described previously (63). The cover slips were washed and the fixed cells were stained with 0.2 μg/ml H33342 for 5 min and then rinsed. The cover slips were then mounted on slides using Prolong Antifade mounting media (Molecular Probes, Inc., Eugene, OR) and viewed by fluorescence microscopy using the appropriate filter sets.
Labeling of nascent mRNA transcripts was performed as previously described (4) except cells were exposed to BrUTP for 20 min. Briefly, transfected cells were grown on cover glasses for 24 h and then permeabilized with saponin. The cells were exposed to BrUTP for 20 min at 33 C to label nascent mRNA and then fixed in paraformaldehyde. After fixation, cells were washed and incubated overnight at 4 C with antibromouracil antibody (BMC9318, Roche Molecular Biochemicals, Indianapolis, IN; 1:100 final dilution). The next day cells were washed followed by detection with a Texas Red-conjugated secondary antibody. Cells were washed again and stained with H33342 at a concentration of 0.2 μg/ml, and the cover glasses were mounted using Vectashield (Vector Laboratories, Inc., Burlingame, CA). Fluorescent images were captured and processed as above for the live cell imaging, with the gray-level Texas red signal being assigned to the red channel of the red-green-blue image.
Microscopy and Image Analysis
Pituitary GHFT1–5 cells were transfected with between 3 and 30 μg of expression plasmid DNA encoding the proteins of interest fused to the fluorescent proteins, and inoculated into culture dishes containing 25-mm cover glasses. The cells were maintained in culture for 24 h as described above and then subjected to fluorescence microscopy as described previously (24). For experiments involving staining with H33342, the stain was added to a final concentration of 0.5 μg/ml approximately 20 min before imaging of living cells or at 0.2 μg/ml for 5 min to image fixed cells. The fluorescence images were acquired using an inverted IX-70 (Olympus Corp., Lake Success, NY) equipped with a 60× aqueous-immersion objective lens. The filter combinations were 485/22 nm excitation and 535/50 nm emission for GFP; 365/15 nm excitation and 460/50 nm emission for H33342 or BFP images, and a tetramethyl rhodamine isothiocyanate filter set for RFP and Texas red imaging (Chroma Technology Corp., Brattelboro, VT). Grayscale images with no saturated pixels were obtained using a cooled digital interline camera (Orca-200, Hamamatsu, Bridgewater, NJ). All images were collected at a similar gray-level intensity by controlling the excitation intensity using neutral density filtration, and by varying the oncamera integration time. ISEE software (Inovision Corp., Raleigh, NC) was used to background subtract and then convert the digital images to red-green-blue images. All image files were processed for presentation using Canvas 7.0 (Deneba Systems, Miami, FL).
Acknowledgments
We thank Cindy Booker, Molly Gangopadhyay, and Phat Tran for expert assistance, and Ammasi Periasamy of the W.M. Keck Center for Cellular Imaging for microscopy advice. We also thank Dr. Sue Moenter for helpful discussions.
This work was supported by NIH Grant DK-47301 and the National Science Foundation Center for Biological Timing (to R.N.D.) and NIH Grant F32DK-10093 (to J.F.E.).
Abbreviations
- BFP
Blue fluorescent protein
- BrUTP
bromouridine
- C/EBPα
CCAAT/enhancer binding protein-α
- CPHD
combined pituitary hormone deficiency
- B-ZIP
basic region-leucine zipper
- ER
estrogen receptor
- FCS
fetal calf serum
- FP
fluorescent protein
- GFP
green fluorescent protein
- H33342
Hoechst 33342
- HD
homeodomain
- PML
promyelocytic leukemia protein
- PRL
prolactin
- RFP
red fluorescent protein
- rPRL
rat prolactin
REFERENCES
- 1.FJ Iborra, DA Jackson, PR Cook. The path of transcripts from extra-nucleolar synthetic sites to nuclear pores: transcripts in transit are concentrated in discrete structures containing SR proteins. J Cell Sci. 1996;111:2269–2282. doi: 10.1242/jcs.111.15.2269. [DOI] [PubMed] [Google Scholar]
- 2.TA Nardozza, MM Quigley, RH Getzenberg. Association of transcription factors with the nuclear matrix. J Cell Biochem. 1996;61:467–477. doi: 10.1002/(SICI)1097-4644(19960601)61:3%3C467::AID-JCB14%3E3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 3.AI Lamond, WC Earnshaw. Structure and function in the nucleus. Science. 1998;280:547–553. doi: 10.1126/science.280.5363.547. [DOI] [PubMed] [Google Scholar]
- 4.A Pombo, P Cuello, W Schul, JB Yoon, RG Roeder, PR Cook, S Murphy. Regional and temporal specialization in the nucleus: a transcriptionally-active nuclear domain rich in PTF, Oct1 and PIKA antigens associates with specific chromosomes early in the cell cycle. EMBO J. 1998;17:1768–1778. doi: 10.1093/emboj/17.6.1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.T Misteli. Protein dynamics: implications for nuclear architecture and gene expression. Science. 2001;291:843–847. doi: 10.1126/science.291.5505.843. [DOI] [PubMed] [Google Scholar]
- 6.S Radovick, LE Cohen, FE Wondisford. The molecular basis of hypopituitarism. Horm Res. 1998;49(Suppl 1):30–36. doi: 10.1159/000053065. [DOI] [PubMed] [Google Scholar]
- 7.JS Parks, MR Brown, DL Hurley, CJ Phelps, MP Wajnrajch. Heritable disorders of pituitary development. J Clin Endocrinol Metab. 1999;84:4362–4370. doi: 10.1210/jcem.84.12.6209. [DOI] [PubMed] [Google Scholar]
- 8.HA Ingraham, SE Flynn, JW Voss, VR Albert, MS Kapiloff, L Wilson, MG Rosenfeld. The POU-specific domain of Pit-1 is essential for sequence-specific, high affinity DNA binding and DNA-dependent Pit-1-Pit-1 interactions. Cell. 1990;61:1021–1033. doi: 10.1016/0092-8674(90)90067-o. [DOI] [PubMed] [Google Scholar]
- 9.JS Dasen, MG Rosenfeld. Combinatorial codes in signaling and synergy: lessons from pituitary development. Curr Opin Genet Dev. 1999;9:566–574. doi: 10.1016/s0959-437x(99)00015-5. [DOI] [PubMed] [Google Scholar]
- 10.SL McKnight, MD Lane, S Gluecksohn-Waelsch. Is CCAAT/enhancer-binding protein a central regulator of energy metabolism? Genes Dev. 1989;3:2021–2024. doi: 10.1101/gad.3.12b.2021. [DOI] [PubMed] [Google Scholar]
- 11.GJ Darlington, SE Ross, OA MacDougald. The role of C/EBP genes in adipocyte differentiation. J Biol Chem. 1998;273:30057–30060. doi: 10.1074/jbc.273.46.30057. [DOI] [PubMed] [Google Scholar]
- 12.F Schaufele. CCAAT/enhancer-binding protein α activation of the rat growth hormone promoter in pituitary progenitor GHFT1–5 cells. J Biol Chem. 1996;271:21484–21489. doi: 10.1074/jbc.271.35.21484. [DOI] [PubMed] [Google Scholar]
- 13.KK Jacob, FM Stanley. CCAAT/enhancer-binding protein α is a physiological regulator of prolactin gene expression. Endocrinology. 1999;140:4542–4550. doi: 10.1210/endo.140.10.7076. [DOI] [PubMed] [Google Scholar]
- 14.F Schaufele, JF Enwright, III, X Wang, C Teoh, R Srihari, R Erickson, OA MacDougald, RN Day. CCAAT/enhancer binding protein α assembles essential cooperating factors in common subnuclear domains. Mol Endocrinol. 2001;15:1665–1676. doi: 10.1210/mend.15.10.0716. [DOI] [PubMed] [Google Scholar]
- 15.D Lew, H Brady, K Klausing, K Yaginuma, LE Theill, C Stauber, M Karin, PL Mellon. GHF-1-promoter-targeted immortalization of a somatotropic progenitor cell results in dwarfism in transgenic mice. Genes Dev. 1993;7:683–693. doi: 10.1101/gad.7.4.683. [DOI] [PubMed] [Google Scholar]
- 16.WH Landschulz, PF Johnson, EY Adashi, BJ Graves, SL McKnight. Isolation of a recombinant copy of the gene encoding C/EBP. Genes Dev. 1988;2:786–800. doi: 10.1101/gad.2.7.786. [DOI] [PubMed] [Google Scholar]
- 17.AP Bradford, C Wasylyk, B Wasylyk, A Gutierrez-Hartmann. Interaction of Ets-1 and the POU-homeodomain protein GHF-1/Pit-1 reconstitutes pituitary-specific gene expression. Mol Cell Biol. 1997;17:1065–1074. doi: 10.1128/mcb.17.3.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.RN Day, J Liu, V Sundmark, M Kawecki, D Berry, HP Elsholtz. Selective inhibition of prolactin gene transcription by the ETS-2 repressor factor. J Biol Chem. 1998;273:31909–31915. doi: 10.1074/jbc.273.48.31909. [DOI] [PubMed] [Google Scholar]
- 19.W Liu, JF Enwright, W Hyun, RN Day, F Schaufele. CCAAT/enhancer binding protein α uses distinct domains to prolong pituitary cells in the growth 1 and DNA synthesis phases of the cell cycle. BMC Cell Biol. 2002;3:6–17. doi: 10.1186/1471-2121-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.QQ Tang, MD Lane. Activation and centromeric localization of CCAAT/enhancer-binding proteins during the mitotic clonal expansion of adipocyte differentiation. Genes Dev. 1999;13:2231–2241. doi: 10.1101/gad.13.17.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.RL Erickson, N Hemati, SE Ross, OA MacDougald. p300 Coactivates the adipogenic transcription factor CCAAT/enhancer-binding protein. J Biol Chem. 2001;276:16348–16355. doi: 10.1074/jbc.m100128200. [DOI] [PubMed] [Google Scholar]
- 22.JM Craig, WC Earnshaw, P Vagnarelli. Mammalian centromeres: DNA sequence, protein composition, and role in cell cycle progression. Exp Cell Res. 1999;246:249–262. doi: 10.1006/excr.1998.4278. [DOI] [PubMed] [Google Scholar]
- 23.BK Vig, M Willcourt. Decondensation of pericentric heterchromatin alters the sequence of centromere separation in mouse cells. Chromosoma. 1998;107:417–423. doi: 10.1007/s004120050325. [DOI] [PubMed] [Google Scholar]
- 24.RN Day, A Periasamy, F Schaufele. Fluorescence resonance energy transfer microscopy of localized protein interactions in the living cell nucleus. Methods. 2001;25:4–18. doi: 10.1006/meth.2001.1211. [DOI] [PubMed] [Google Scholar]
- 25.FM Boisvert, MJ Hendzel, DP Bazett-Jones. Promyelocytic leukemia (PML) nuclear bodies are protein structures that do not accumulate RNA. J Cell Biol. 2000;48:283–292. doi: 10.1083/jcb.148.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.GS Baird, DA Zacharias, RY Tsien. Biochemistry, mutagenesis, and oligomerization of DsRed, a red fluorescent protein from coral. Proc Natl Acad Sci USA. 1989;97:11984–11991. doi: 10.1073/pnas.97.22.11984. 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.F Schaufele, CY Chang, W Liu, JD Baxter, SK Nordeen, Y Wan, RN Day, DP McDonnell. Temporally distinct and ligand-specific recruitment of nuclear receptor-interacting peptides and cofactors to subnuclear domains containing the estrogen receptor. Mol Endocrinol. 2000;14:2024–2039. doi: 10.1210/mend.14.12.0572. [DOI] [PubMed] [Google Scholar]
- 28.L Xu, RM Lavinsky, JS Dasen, SE Flynn, EM McInerney, TM Mullen, T Heinzel, D Szeto, E Korzus, R Kurokawa, AK Aggarwal, DW Rose, CK Glass, MG Rosenfeld. Signal-specific coactivator domain requirements for Pit-1 activation. Nature. 1998;395:301–306. doi: 10.1038/26270. [DOI] [PubMed] [Google Scholar]
- 29.AP Bradford, KS Brodsky, SE Diamond, LC Kuhn, Y Liu, A Gutierrez-Hartmann. The Pit-1 homeodomain and β-domain interact with Ets-1 and modulate synergistic activation of the rat prolactin promoter. J Biol Chem. 2000;275:3100–3106. doi: 10.1074/jbc.275.5.3100. [DOI] [PubMed] [Google Scholar]
- 30.S Radovick, M Nations, Y Du, LA Berg, BD Weintraub, FE Wondisford. A mutation in the POU-homeodomain of Pit-1 responsible for combined pituitary hormone deficiency. Science. 1992;257:1115–1118. doi: 10.1126/science.257.5073.1115. [DOI] [PubMed] [Google Scholar]
- 31.EM Jacobson, P Li, A Leon-del-Rio, MG Rosenfeld, AK Aggarwal. Structure of Pit-1 POU domain bound to DNA as a dimer: unexpected arrangement and flexibility. Genes Dev. 1997;11:198–212. doi: 10.1101/gad.11.2.198. [DOI] [PubMed] [Google Scholar]
- 32.SA Camper, TL Saunders, RW Katz, RH Reeves. The Pit-1 transcription factor gene is a candidate for the murine Snell dwarf mutation. Genomics. 1990;8:586–590. doi: 10.1016/0888-7543(90)90050-5. [DOI] [PubMed] [Google Scholar]
- 33.S Li, EB Crenshaw, III, EJ Rawson, DM Simmons, LW Swanson, MG Rosenfeld. Dwarf locus mutants lacking three pituitary cell types result from mutations in the POU-domain gene pit-1. Nature. 1990;347:528–533. doi: 10.1038/347528a0. [DOI] [PubMed] [Google Scholar]
- 34.DM Simmons, JW Voss, HA Ingraham, JM Holloway, RS Broide, MG Rosenfeld, LW Swanson. Pituitary cell phenotypes involve cell-specific Pit-1 mRNA translation and synergistic interactions with other classes of transcription factors. Genes Dev. 1990;4:695–711. doi: 10.1101/gad.4.5.695. [DOI] [PubMed] [Google Scholar]
- 35.KM Scully, EM Jacobson, K Jepsen, V Lunyak, H Viadiu, C Carriere, DW Rose, F Hooshmand, AK Aggarwal, MG Rosenfeld. Allosteric effects of Pit-1 DNA sites on long-term repression in cell type specification. Science. 2000;290:1127–1131. doi: 10.1126/science.290.5494.1127. [DOI] [PubMed] [Google Scholar]
- 36.RA Iverson, KH Day, M d’Emden, RN Day, RA Maurer. Clustered point mutation analysis of the rat prolactin promoter. Mol Endocrinol. 1990;4:1564–1571. doi: 10.1210/mend-4-10-1564. [DOI] [PubMed] [Google Scholar]
- 37.SM Jackson, CA Keech, DJ Williamson, A Gutierrez-Hartmann. Interaction of basal positive and negative transcription elements controls repression of the proximal rat prolactin promoter in nonpituitary cells. Mol Cell Biol. 1992;12:2708–2719. doi: 10.1128/mcb.12.6.2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.RE Schweppe, A Gutierrez-Hartmann. Pituitary Ets-1 and GABP bind to the growth factor regulatory sites of the rat prolactin promoter. Nucleic Acids Res. 2001;29:1251–1260. doi: 10.1093/nar/29.5.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.RN Day, JF Enwright, III, CF Booker, TC Voss, F Schaufele, A Periasamy. Imaging the localized protein interactions between Pit-1 and the CCAAT/enhancer binding protein alpha (C/EBPα) in the living pituitary cell nucleus. Mol Endocrinol. doi: 10.1210/me.2002-0136. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39a.C Zeng, S McNeil, S Pockwinse, J Nickerson, L Shopland, JB Lawrence, S Penman, S Hiebert, JB Lian, AJ van Wijnen, JL Stein, GS Stein. Intranuclear targeting of AML/CBFα regulatory factors to nuclear matrix-associated transcriptional domains. Proc Natl Acad Sci USA. 1998;95:1585–1589. doi: 10.1073/pnas.95.4.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.D Thanos, T Maniatis. Virus induction of human IFN β gene expression requires the assembly of an enhanceosome. Cell. 1995;83:1091–1100. doi: 10.1016/0092-8674(95)90136-1. [DOI] [PubMed] [Google Scholar]
- 41.TK Kim, T Maniatis. The mechanism of transcriptional synergy of an in vitro assembled interferon-β enhanceosome. Mol Cell. 1997;1:119–129. doi: 10.1016/s1097-2765(00)80013-1. [DOI] [PubMed] [Google Scholar]
- 42.M Merika, AJ Williams, G Chen, T Collins, D Thanos. Recruitment of CBP/p300 by the IFNβ enhanceosome is required for synergistic activation of transcription. Mol Cell. 1998;1:277–287. doi: 10.1016/s1097-2765(00)80028-3. [DOI] [PubMed] [Google Scholar]
- 43.N Munshi, M Merika, J Yie, K Senger, G Chen, D Thanos. Acetylation of HMGI(Y) by CBP turns off IFNα expression by disrupting the enhanceosome. Mol Cell. 1998;1:457–467. doi: 10.1016/s1097-2765(00)80145-8. [DOI] [PubMed] [Google Scholar]
- 43a.W-H Zhang, R Srihari, RN Day, F Schaufele. CCAAT/enhancer binding protein α alters histone H3 acetylation at large subnuclear domains. J Biol Chem. 2001;276:40373–40376. doi: 10.1074/jbc.C100505200. [DOI] [PubMed] [Google Scholar]
- 44.OJ Miller, W Schnedl, J Allen, BF Erlanger. 5-Methylcytosine localised in mammalian constitutive heterochromatin. Nature. 1974;251:636–637. doi: 10.1038/251636a0. [DOI] [PubMed] [Google Scholar]
- 45.LM Lica, S Narayanswami, BA Hamkalo. Mouse satellite DNA, centromere structure, and sister chromatid pairing. J Cell Biol. 1986;103:1145–1151. doi: 10.1083/jcb.103.4.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.BA Sullivan, MD Blower, GH Karpen. Determining centromere identity: cyclical stories and forking paths. Nat Rev Genet. 2001;2:584–596. doi: 10.1038/35084512. [DOI] [PubMed] [Google Scholar]
- 47.MD Blower, BA Sullivan, GH Karpen. Conserved organization of centromeric chromatin in flies and humans. Dev Cell. 2002;2:319–330. doi: 10.1016/s1534-5807(02)00135-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Deleted in proof
- 49.H Wang, P Iakova, M Wilde, A Welm, T Goode, WJ Roesler, NA Timchenko. C/EBPα arrests cell proliferation through direct inhibition of Cdk2 and Cdk4. Mol Cell. 2001;8:817–828. doi: 10.1016/s1097-2765(01)00366-5. [DOI] [PubMed] [Google Scholar]
- 50.M Olive, SC Williams, C Dezan, PF Johnson, C Vinson. Design of a C/EBP-specific, dominant-negative bZIP protein with both inhibitory and gain-of-function properties. J Biol Chem. 1996;271:2040–2047. doi: 10.1074/jbc.271.4.2040. [DOI] [PubMed] [Google Scholar]
- 51.LE Theill, JL Castrillo, D Wu, M Karin. Dissection of functional domains of the pituitary-specific transcription factor GHF-1. Nature. 1989;342:945–948. doi: 10.1038/342945a0. [DOI] [PubMed] [Google Scholar]
- 52.SE Diamond, M Chiono, A Gutierrez-Hartmann. Reconstitution of the protein kinase A response of the rat prolactin promoter: differential effects of distinct Pit-1 isoforms and functional interaction with Oct-1. Mol Endocrinol. 1999;13:228–238. doi: 10.1210/mend.13.2.0227. [DOI] [PubMed] [Google Scholar]
- 53.D He, BR Brinkley. Structure and dynamic organization of centromeres/prekinetochores in the nucleus of mammalian cells. J Cell Sci. 1996;109:2693–2704. doi: 10.1242/jcs.109.11.2693. [DOI] [PubMed] [Google Scholar]
- 54.M Cockell, SM Gasser. Nuclear compartments and gene regulation. Curr Opin Genet Dev. 1999;9:199–205. doi: 10.1016/S0959-437X(99)80030-6. [DOI] [PubMed] [Google Scholar]
- 55.C Francastel, MC Walters, M Groudine, DI Martin. A functional enhancer suppresses silencing of a transgene and prevents its localization close to centrometric heterochromatin. Cell. 1999;99:259–269. doi: 10.1016/s0092-8674(00)81657-8. [DOI] [PubMed] [Google Scholar]
- 56.BM Shewchuk, SL Asa, NE Cooke, SA Liebhaber. Pit-1 binding sites at the somatotrope-specific DNase I hypersensitive sites I, II of the human growth hormone locus control region are essential for in vivo hGH-N gene activation. J Biol Chem. 1999;274:35725–35733. doi: 10.1074/jbc.274.50.35725. [DOI] [PubMed] [Google Scholar]
- 57.KE Brown, SS Guest, ST Smale, K Hahm, M Merkenschlager, AG Fisher. Association of transcriptionally silent genes with Ikaros complexes at centromeric heterochromatin. Cell. 1997;91:845–854. doi: 10.1016/s0092-8674(00)80472-9. [DOI] [PubMed] [Google Scholar]
- 58.KE Brown, J Baxter, D Graf, M Merkenschlager, AG Fisher. Dynamic repositioning of genes in the nucleus of lymphocytes preparing for cell division. Mol Cell. 1999;3:207–217. doi: 10.1016/s1097-2765(00)80311-1. [DOI] [PubMed] [Google Scholar]
- 59.K Hahm, BS Cobb, AS McCarty, KE Brown, CA Klug, R Lee, K Akashi, IL Weissman, AG Fisher, ST Smale. Helios, a T cell-restricted Ikaros family member that quantitatively associates with Ikaros at centromeric het erochromatin. Genes Dev. 1998;12:782–796. doi: 10.1101/gad.12.6.782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.J Kim, S Sif, B Jones, A Jackson, J Koipally, E Heller, S Winandy, A Viel, A Sawyer, T Ikeda, R Kingston, K Georgopoulos. Ikaros DNA-binding proteins direct formation of chromatin remodeling complexes in lymphocytes. Immunity. 1999;10:345–355. doi: 10.1016/s1074-7613(00)80034-5. [DOI] [PubMed] [Google Scholar]
- 61.E Matsuda, Y Agata, M Sugai, T Katakai, H Gonda, A Shimizu. Targeting of Kruppel-associated box containing zinc finger proteins to centromeric heterochromatin. Implication for the gene silencing mechanisms. J Biol Chem. 2001;276:14222–14229. doi: 10.1074/jbc.M010663200. [DOI] [PubMed] [Google Scholar]
- 62.RN Day, KH Day. An alternatively spliced form of Pit-1 represses prolactin gene expression. Mol Endocrinol. 1994;8:374–381. doi: 10.1210/mend.8.3.8015554. [DOI] [PubMed] [Google Scholar]
- 63.PW Howard, RA Maurer. Thyrotropin releasing hormone stimulates transient phosphorylation of the tissue specific transcription factor, Pit-1. J Biol Chem. 1994;269:28662–28669. [PubMed] [Google Scholar]