Abstract
Previous studies demonstrated that cytokines regulate opioid and opioid receptor gene expression in neuronal and immune cells. The gene sequence analysis of opioid receptors revealed that μ-, δ- and κ-opioid receptor promoter regions contain potential cytokine response elements (NF-IL6 binding sites). It was postulated that the response elements present in opioid receptor promoter regions may have a role in the cytokine effects on opioid receptor gene expression through ciS-trans interaction. The present sandy investigated whether cytokines have an effect on opioid receptor gene expression by cytokine-induced transcription factor, NF-IL6, using a number of immune cell lines which respond to cytokines. Further investigation was made to determine whether the potential cytokine response element DNA sequences on opioid receptor promoter region have functional significance which may be affected by the DNA context of the opioid receptor promoter in immune cell lines. Tandem repeats of conserved cytokine response elements of IL-6 gene fused to a heteropromoter SV40 were utilized as a positive control and expressed 2-fold increased promoter activity after cytokine stimulation. Transient transfection studies in time courses (24–72 h) and different dose treatments (100–500 U/ml for IL-6 and 50–200 U/ml for IL-1 α + β) were also carried out to investigate the possibility that the upregulated gene expression may be transient or cytokine-dose-dependent. Our data demonstrated that there was no significant cytokine-stimulated opioid receptor gene expression in immune cells tested. In addition, the cytokine response elements (NF -IL6 binding sites) in opioid receptor genes are not functional. These results contradict the previous reports in which cytokines modulated the expression of opioid and opioid receptors in neuronal and immune cells. The possible reasons regarding the different results were discussed.
Keywords: Opioid receptor, Cytokine response element, NF-IL6 binding site
Previous studies have demonstrated that immune factors such as cytokines modulate the expression of opioid genes and opioid receptors in neuronal as well as immune cells [13]. Negro et al. [12] reported that interleukin-1β (IL-1β) stimulates proenkephalin gene expression in astrocytes cultured from rat cortex. In a later study, Ruzicka et al. [14] and Ruzicka and Akil [15] characterized more completely the astrocytic response of the opioid receptor to cytokine stimulation by examining the effect of IL-1β on μ-opioid receptor mRNA expression in primary astrocyte-enriched cultures derived from rat cortex, striatum, cerebellum, hippocampus, and hypothalamus. They reported that IL-1β treatment produced a 70–80% increase in μ-opioid receptor mRNA expression in striatal, cerebellar, and hippocampal cultures, but had no effect on this expression in cortical and hypothalamic cultures. More recently, Erich et al. [4] investigated the effect of interleukin-1 (IL-1) on opioid receptor gene expression in neural microvascular endothelial cells cultured from human brain. They observed that there was virtually no μ-opioid receptor expression at basal levels and no increase after either IL-lα or IL-1β treatment. However, simultaneous treatment with both IL-lα and IL-1β increased μ-opioid receptor gene expression.
Cytokines are known as the principal communication signals of the immune system. The synthesis of classical immune mediators such as IL-1, IL-1 receptor antagonist, and IL-6 and their receptors by cells in the brain and endocrine organs, implies a mechanism for the host’s immune system to communicate changes in immune status to the brain and associated neuroendocrine organs [2,19]. Although a clear signaling transcription pathway for these cytokines in neuronal or immune cells has yet to be elucidated, these cytokines have been shown to activate or induce several transcription factors, including nuclear factor-interleukin 6 (NF-IL6). Transcription factors provide the molecular link between post-receptor signaling events and changes in gene expression. Therefore, the expression of genes containing binding sites for these transcription factors would be expected to be modulated by these cytokines. Several studies showed that the cytokines exert their effects through cytokine response elements, such as the NF-IL6 binding sites of target genes in neuron and immune cells [1,6,19,21].
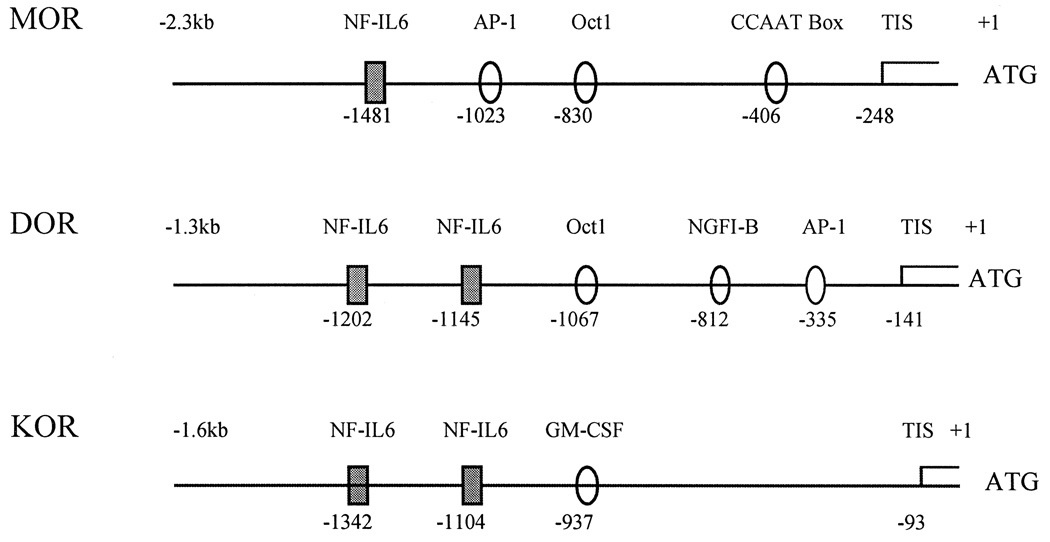
Min et al. [10] have shown that the promoter sequence of the μ-opioid receptor gene contains cytokine response elements, such as the NF-IL6 binding site. Sequence analysis of both δ- [8] and κ- [17] opioid receptor promoter regions revealed that these opioid receptor promoters also contain potential cytokine response elements with 1–2 bp mismatches as compared with those of already known sequences. Therefore, it has been postulated that the transcription factor, NF-IL6, may modulate the expression of the opioid receptor gene through specific DNA binding of cytokine response elements in the opioid receptor promoter region. The potential binding sites for transcription factors including cytokine response elements in opioid receptor promoter regions are shown in Fig. 1.
Fig 1.
The potential binding sites for transcription factors on μ-, δ- and κ-opioid receptor promoter regions. Translation start point (ATG) was marked as +1. All three opioid receptor promoter regions contain cytokine response elements (NF-IL6) as well as other potential transcription factor binding sites. Source: Refs. [8,10,17].
The NF-IL6 binding site is known as IL-1 response element or IL-6 response element as the transactivation activity of NF-IL6 is drastically induced by the cytokines such as IL-1 and IL-6 governing the expression of the target genes at the transcription level through cis-, trans-acting element interactions. Among the cytokines, IL-6 has been shown to be the most powerful inducer of NF-IL6 [5,9,11].
The present study investigates the role of cytokine response element in the increased opioid receptor promoter activity after cytokine stimulation. In these studies, the human myeloma cell line, U266, and macrophage Raw264.7, were used for transient transfection studies because they are known to be responsive to both cytokines, IL-1 and IL-6 as well as to express endogenous opioid receptors.
A series of 5′-deletion constructs of the opioid receptor promoters, μ, δ and κ, was transiently transfected into U266 and Raw264.7 along with an internal control plasmid pCH110 which contained a functional β-galactosidase gene by using SuperFect transfection methods (Qiagen). The transfected cells were incubated for 48–72 h in the presence or absence of mouse IL-6 (100–500 U/ml) or human IL-1 α + β (50–200 U/ml) purchased from Boehringer Mannheim. The reference plasmid p4 × NF-IL6Luc, containing four copies of tandemly repeated cytokine response elements of the IL-6 promoter (5′-ACA TTG CAC AAT CT-3′), was constructed as a cytokine-responsive positive control. This was achieved by placing the cytokine response elements upstream of a SV40 promoter at the SacI site of the pGL3-promoter vector, which contained a luciferase reporter gene system (Promega). Cell lysates were prepared and analyzed for luciferase and β-galactosidase activities. The relative luciferase units were calculated by normalizing the luciferase activities to those of β-galactosidase activities. In order to verify the functional significance of these potential cytokine response elements of the opioid receptor genes, four tandemly repeated core potential cytokine response elements of the μ-opioid receptor gene (5′-TGC TGA AAT G-3′) and δ-opioid receptor gene (5′-TCT TCC CTA A-3′ and 5′-TTT AGG AAT A-3′) were fused to a heteropromoter, SV40, in a similar way to the construction of the cytokine-responsive positive control, p4 × NF-IL6Luc. This resulted in the construction of μNF-IL6Luc, δNF-IL6Lucl and δNF-IL6Luc2, respectively. All the constructs were confirmed by sequencing. These constructs were then transiently transfected into U266 and Raw264.7 cells, and the cells were treated with IL-6 or IL-1 α + β. Transient transfection studies in time courses (24, 36, 48, 60, 72 h) and different dose treatments (100, 200, 300, 500 U/ml for lL-6 and 50, 100, 200 U/ml for IL-1 α + β) were also carried out to investigate the possibility that the upregulated gene expression may be transient or cytokine-dose-dependent. Several other immune cell lines as well as brain cell lines, such as, MolT-4, HSB2, CEM3, Raji, R1.1, NMB, NS20Y and PC12h, which express endogenous opioid receptors, were also examined in the transient transfection studies. Although the presence of the cytokine receptors on those cell lines has not been confirmed in this experiment, it has been reported that those immune cell lines may express cytokine receptors [7,16,18,20].
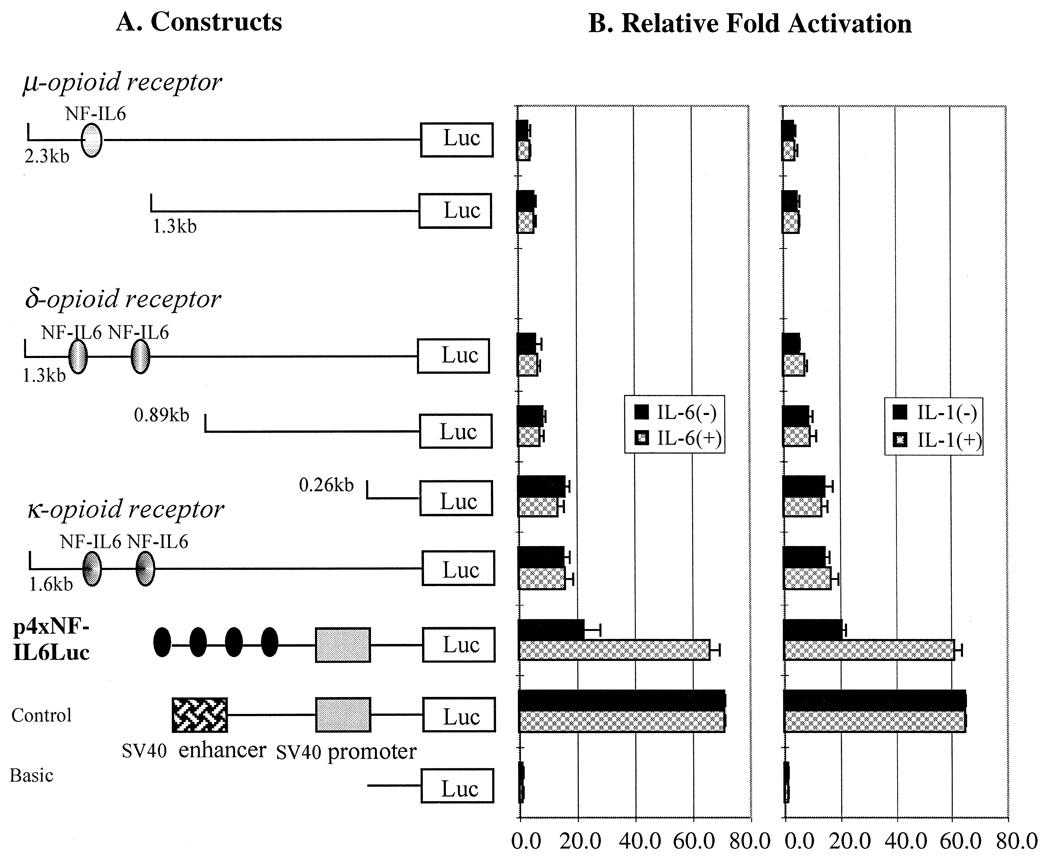
In contrast to the previous data from other groups using primary cultures of astrocytes and endothelial cells, the results obtained from our experiments demonstrated that neither IL-6 nor IL-1 α + β stimulated the promoter activity of μ-, δ- and κ-opioid receptors with either 48 or 72 h incubation after transfected into both cell lines, U266 and Raw264.7. The positive control, however, displayed more than 2-fold cytokine-enhanced promoter activity (Fig. 2). The increased amount of IL-6 (500 U/ml) or IL-1 α + β (200 U/ml) also provoked no significant differences in the luciferase activity when compared to those with less amount of cytokines9 (100 U/ml for IL-6 and 50 U/ml for IL-1 α + β (data not shown). Also, no increased promoter activity was observed when several other immune cell lines as well as the brain cell lines listed previously were tested (data not shown).
Fig 2.
A series of 5′-deletion constructs of μ- and δ-opioid receptor promoter (A) and the fold activation measured by luciferese activities after transfected into U266 and Raw264.7 with (+) and without (−) cytokine treatment (B). The cytokines used for this experiment are IL-1 α + β (200 U/ml) and IL-6 (500 U/ml) and incubated for 72 h. A reference plasmid, p4 × NF-IL6Luc which contains four copies of NF-IL6 binding sites of IL-6 gene, was used as a positive control for cytokine responses. There was no significant difference in luciferase activity between two different cell lines, U266 and Raw2647 in terms of the response to the cytokines as well as the promoter activity pattern. Therefore, one graphic bar represents both cell line, U266 and Raw264.7. Time course and dose-dependency experiments showed no different promoter activity from shown in this figure. (·) Indicates tandem repeats of NF-IL6 binding sites. Three times of individual experiments were performed with duplicates each time.
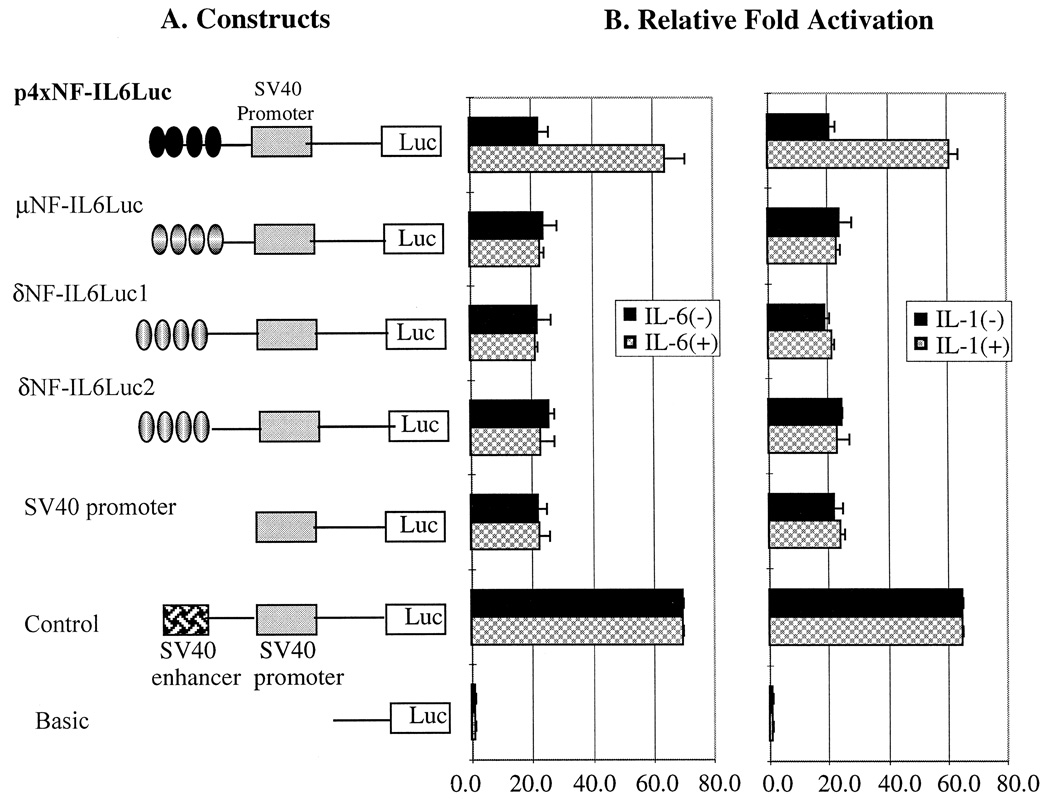
Even though all the deletion constructs, regardless of whether they contained NF-IL6 binding sites or not, showed no upregulated gene expression after cytokine stimulation (Fig. 2), the upregulated gene expression of the cytokine-responsive positive control encouraged us to focus on the potential cytokine response element DNA sequences in the opioid receptor genes to determine whether these potential cis-acting elements really have a functional significance through a cytokine-induced NF-IL6 pathway. If the potential cytokine response DNA sequences are functionally significant for gene expression, it is anticipated that the cytokines should be able to modulate the transcriptional level of these constructs that connected to the luciferase reporter gene system. The luciferase activities of these constructs containing tandemly repeated potential cytokine response elements from μ- and δ-opioid receptor gene and cytokine-responsive positive control, p4 × NF-IL6Luc, are shown in Fig. 3. As expected, the construct referred to as p4 × NF-IL6Luc demonstrated both basal and cytokine-inducible activities when the construct was transiently transfected to both U266 and Raw264.7 cells. However, the constructs containing the four copies of the potential cytokine response elements of the opioid receptor promoter region demonstrated basal activity only, even after stimulation with IL-6 (100–500 U/ml) or IL-1 α + β (50–200 U/ml). The 2-fold cytokine-enhanced promoter activity of the positive control demonstrated that the cell lines tested in this study respond to cytokine stimulation and, if these response elements are important for opioid receptor gene expression as a cis-acting transcriptional regulatory element, they should show cytokine effects. Therefore, these negative results demonstrate that potential cytokine response element DNA sequences in the opioid receptor genes have no functional significance. Our data contradict the previous hypothesis that the potential cytokine response elements of the opioid receptor promoter region may have a role for the cytokine effect on the opioid receptor gene expression through a direct DNA–protein interaction.
Fig 3.
Construction of four tandemly repeated core sequences of the cytokine response elements (NF-IL6 binding sites) from opioid receptor promoter region (A) and the fold activation measured by luciferase activities after transfected into U266 and Raw264.7 with (+) and without (−) cytokine treatment (B). The cytokines used for this experiment are IL-1 α + β (200 U/ml) and IL-6 (500 U/ml) and incubated for 72 h. A reference plasmid, p4 × NF-IL6Luc, which contains four copies of NF-IL6 binding sites of IL-6 gene, was used as a positive control for cytokine responses. There was no significant difference in luciferase activity between two different cell lines, U266 and Raw264.7 in terms of the represents both cell line, U266 and Raw264.7. Time course and dose-dependency experiments showed no different promoter activity from shown in this figure. (· · · ·) Indicates four tandem repeats of NF-IL6 binding sites. Three times of individual experiments were performed with duplicates each time.
There are several points to be considered as possible explanations for the differences between our results and the previous data regarding the upregulated-opioid receptor gene expression by cytokine stimulation. One possible explanation may be related to the differences between primary cell cultures that have shown a response to cytokines in previous studies and tumor cell lines used for these studies. In general, primary cell cultures have been shown to be much more sensitive in their response to external stimulation than tumor cells which require stronger stimulation for the same response or show no response at all. For example, bacterial lipopolysaccharide (LPS)-induced expression of the IL-6 or TNF-α gene was evident in primary cell culture but not in tumor cells, suggesting the different sensitivities to the stimulation (Dr. Roy, personal communication). However, working with primary cell culture for transient transfection studies is not practical at this moment because of the technical difficulties. Also, this experiment required a cell line which responds to both cytokines, IL-6 and IL-1, and preferably, expresses endogenous opioid receptors.
Another possible explanation for the different observations could be raised from the two different gene expression detection methods. Our results were based on data from transiently transfected promoter–reporter constructs, whereas those of other groups were based on data from reverse transcription polymerase chain reaction (RT-PCR) analysis from mRNA levels. Previous report suggested that regulation of levels of particular mRNA transcript could result from changes in transcription rates, altered stability of the transcript (mRNA half-life), or a combination of both mechanisms [3].
Different cell environments—immune and neuronal cells—could be considered as one of the reasons why our results contradict those of other groups. Roy et al. [13] reported that cytokine stimulation increased morphine binding sites on thymocytes. However, the low-affinity opioid receptors which are classified as non-classical opioid receptors present on immune cells appeared to contribute to this cytokine effect. The comparison of gene structure and sequence analysis between non-classical and classical opioid receptors isolated from immune cells has not been reported. In this study, opioid receptor promoters (classical opioid receptors) fused to reporter gene system were examined. Considering the positive control, p4 × NF-IL6Luc, which stimulated the promoter activity in immune cells upon cytokine addition, it would be possible that the cytokine response elements present in the opioid receptors may not be able to stimulate the classical opioid receptor promoter activity in immune cell environment or the opioid receptor genes stimulated by IL-1 or IL-6 may be subject to combinatorial regulation by NF-IL6 and other transcription factors induced by these cytokines such as AP-1 site binding factors. As shown in Fig. 1, μ- and δ-opioid receptor promoter regions contained potential AP-1 binding sites and AP-1 binding transcription factors; c-fos and c-jun are also induced by cytokines [14].
Even though opioid receptors do not respond to cytokine stimulation and the cytokine response elements in the opioid receptor genes appear not to be functional in a number of immune as well as neuronal cell lines from this study, there are several possible reasons why our results are different from those of previous reports as already mentioned. Therefore, it may be necessary to confirm these results by using primary astrocyte or endothelial cell cultures for the transient transfection study in order to better understand the mechanism of cellular effects on opioid receptor gene expression by cytokine stimulation which still remains to be elucidated.
Acknowledgements
This research was supported by NIH research grants DA-00546, DA-01583, DA-05695, K05-DA-70554, and A&F Stark fund of the Minnesota Medical Foundation. The authors would like to thank Dr. Sabita Roy for her invaluable discussions and kindness in providing IL-1β for this work.
References
- 1.Akira S, Isshiki H, Nakajima T, Kinoshita S, Nishio Y, Natsuka S, Kishimoto T. Regulation of expression of the interleukin 6 gene: structure and function of the transcription factor NF-IL6. Ciba Foundation Symposium. 1992;167:47–67. doi: 10.1002/9780470514269.ch4. [DOI] [PubMed] [Google Scholar]
- 2.Brown SL, Smith LR, Blalock JE. Interleukin 1 and interleukin 2 enhance proopiomelanocortin gene expression in pituitary cells. J. Immunol. 1987;139(10):3181–3183. [PubMed] [Google Scholar]
- 3.Elizabeth KG, Dawn MP, Carol RG, Kathleen MK, Robert AP. Regulation of mu opioid receptor mRNA levels by activation of protein kinase C in human SH-SY5Y neuroblastoma cells. Anesthesiology. 1997;87(5):1127–1138. doi: 10.1097/00000542-199711000-00017. [DOI] [PubMed] [Google Scholar]
- 4.Erich LV, Patel NA, Wu GD, Fiala M, Chant SL. Interleukin-1 induces the expression of μ-opioid receptor in endothelial cells. Immunopharmacology. 1998;38:261–266. doi: 10.1016/s0162-3109(97)00085-4. [DOI] [PubMed] [Google Scholar]
- 5.Geiger T, Andus T, Klapproth J, Hirano T, Kishimoto T, Heinrich PC. Induction of rat acute-phase proteins by interleukin 6 in vivo. Eur J. Immunol. 1998;18:717–721. doi: 10.1002/eji.1830180510. [DOI] [PubMed] [Google Scholar]
- 6.Hsu W, Kerppola TK, Chen PL, Curran T, Selina CK. Fos and Jun repress transcription activation by NF-IL6 through association at the basic zipper region. Mol. Cell. Biol. 1994;14(1):268–276. doi: 10.1128/mcb.14.1.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kobayashi N, Hamamoto Y, Koyanagi Y, Chen IS, Yamamoto N. Effect of interleukin-1 on the augmentation of human immunodeficiency virus gene expression. Biochem. Biophys. Res. Commun. 1989;165(2):715–721. doi: 10.1016/s0006-291x(89)80025-7. [DOI] [PubMed] [Google Scholar]
- 8.Lance BA, Felsheim RF, Min BH, Fuchs SM, Fuchs JA, Loh HH. Genomic structure of the mouse δ-opioid receptor gene. Biochem. Biophys. Res. Commun. 1995;207(1):111–119. doi: 10.1006/bbrc.1995.1160. [DOI] [PubMed] [Google Scholar]
- 9.Majello B, Ancone R, Toniatti C, Ciliberto G. Constitutive and IL-6-induced nuclear factors that interact with the human c-reactive protein promoter. IMBO J. 1990;9:457–465. doi: 10.1002/j.1460-2075.1990.tb08131.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Min BH, Lance BA, Roderick FF, James AF, Loh HH. Genomic structure and analysis of promoter sequence of a mouse μ-opioid receptor gene. Proc. Natl. Acad. Sci. USA. 1994;91:9081–9085. doi: 10.1073/pnas.91.19.9081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morrone G, Ciliberto G, Oliviero S, Arcone R, Dente L, Content J, Cortese R. Recombinant interleukin 6 regulates the transcriptional activation of a set of human acute phase genes. J. Biol. Chem. 1998;236:12554–12558. [PubMed] [Google Scholar]
- 12.Negro A, Tavella A, Facci L, Callegaro L, Skaper SD. Interleukin-1β regulates proenkephalin gene expression in astrocytes cultured from hat cortex. Glia. 1992;6:206–212. doi: 10.1002/glia.440060308. [DOI] [PubMed] [Google Scholar]
- 13.Roy S, Ge BL, Loh HH, Lee NM. Characterization of [3H]morphine binding to interleukin-1-activated thymocytes. J. Pharmacol. Exp. Ther. 1992;263:263–451. [PubMed] [Google Scholar]
- 14.Ruzicka BB, Thompson RC, Watson SJ, Akil H. Interleukin-1β-mediated regulation of μ-opioid receptor mRNA in primary astrocyte-enriched culture. J. Neurochem. 1996;66:425–428. doi: 10.1046/j.1471-4159.1996.66010425.x. [DOI] [PubMed] [Google Scholar]
- 15.Ruzicka BB, Akil H. The interleukin-1 beta-mediated regulation of proenkephalin and opioid receptor messenger RNA in primary astrocyte-enriched cultures. Neuroscience. 1997;79(2):517–524. doi: 10.1016/s0306-4522(96)00669-0. [DOI] [PubMed] [Google Scholar]
- 16.Sharma V, Knobloch TJ, Benjamin D. Differential expression of cytokine genes in HIV-1 tat transfected T and B cell lines. Biochem. Biophys. Res. Commun. 1995;208(2):704–713. doi: 10.1006/bbrc.1995.1395. [DOI] [PubMed] [Google Scholar]
- 17.Shengli L, Loh HH, Wei LN. Studies of dual promoters of mouse κ-opioid receptor gene. Mol. Pharmacol. 1997;52:1–6. [Google Scholar]
- 18.Teraoka H, Mikoshiba M, Takase K, Yamamoto K, Tsukada K. Reversible G1 arrest induced by dimethyl sulfoxide in human lymphoid cell lines: dimethyl sulfoxide inhibits IL-6-induced differentiation of SKW6-CL4 into IgM-secreting plasma cells. Exp. Cell Res. 1996;222(1):218–224. doi: 10.1006/excr.1996.0027. [DOI] [PubMed] [Google Scholar]
- 19.Wilder RL. Neuroendocrine–immune system interactions and autoimmunity. Annu. Rev. Immunol. 1995;13:307–338. doi: 10.1146/annurev.iy.13.040195.001515. [DOI] [PubMed] [Google Scholar]
- 20.Zeng L, An S, Goetzl EJ. EP4/EP2 receptor-specific prostaglandin E2 regulation of interleukin-6 generation by human HSB.2 early T cells. J. Pharmacol. Exp. Ther. 1998;286(3):1420–1426. [PubMed] [Google Scholar]
- 21.Zhang Y, Rom WN. Regulation of the Interleukin-1β (IL-1β) gene by mycobacterial components and lipopolysaccharide is mediated by two nuclear factor-IL6 motifs. Mol. Cell. Biol. 1993;13(6):3831–3837. doi: 10.1128/mcb.13.6.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]