Abstract
Background
Quantitative spectral analysis of the radio-frequency (RF) signals that underlie grayscale EUS images can be used to provide additional, objective information about tissue state.
Objective
Our purpose was to validate RF spectral analysis as a method to distinguish between (1) benign and malignant lymph nodes and (2) normal pancreas (NP), chronic pancreatitis (CP) and pancreatic cancer (PC).
Design & Setting
A prospective validation study of eligible patients was conducted to compare with pilot study RF data.
Patients
Forty-three patients underwent EUS of the esophagus, stomach, pancreas, and surrounding intra-abdominal and mediastinal lymph nodes (19 from previous pilot study and 24 additional patients).
Main Outcome Measurements
Midband fit, slope, intercept, and correlation coefficient from a linear regression of the calibrated RF power spectra were determined.
Results
Discriminant analysis of mean pilot-study parameters was then performed to classify validation-study parameters. For benign vs. malignant lymph nodes, midband-fit and intercept (both with t-test p < 0.058) provided classification with 67% accuracy and area under ROC curve (AUC) of 0.86. For diseased vs. NP, midband-fit and correlation coefficient (both with ANOVA p < 0.001) provided 93% accuracy and AUC of 0.98. For PC vs. CP, the same parameters provided 77% accuracy and AUC of 0.89. Results improved further when classification was performed with all data.
Limitations
Moderate sample size and spatial averaging inherent to the technique.
Conclusions
This study confirms that mean spectral parameters provide a non-invasive method to quantitatively discriminate benign and malignant lymph nodes as well as normal and diseased pancreas.
Keywords: Endoscopic ultrasound, Spectrum analysis, Ultrasound backscatter, Pancreatic cancer, Chronic pancreatitis, Lymph nodes, Linear discriminant analysis, Receiver-operating characteristic curve, Computer aided diagnosis
Endoscopic ultrasound (EUS) imaging is integral to the staging of most gastrointestinal cancers. During examination, EUS assesses key factors such as depth of tumor invasion and spread to regional lymph nodes. Typically, diagnosis using conventional EUS grayscale imaging is based on judgements regarding the sonographic characteristics of the imaged areas of interest, including size, morphology, relative echogenicity, and level of homo- or heterogeneity. However, deciphering these characteristics in real-time is dependent on the stage, traversability of the tumor, and operator/center experience.1-6 Differentiation of pancreatic cancer from chronic pancreatitis7, 8 as well as benign from malignant lymph nodes9-12 continue to be challenging. Although EUS guided fine needle aspiration (FNA) can obtain diagnostic cytologic material, imaging characteristics are still vital to guiding the target for FNA. As such, additional and more objective means of differentiating benign or inflammatory tissue from neoplastic or malignant tissue could increase the diagnostic accuracy of EUS, improve the yield of FNA, and significantly impact patient care.
Spectrum analysis of backscattered radio-frequency (RF) ultrasound13-15 has proven to be an effective method for identifying changes in tissue state in the contexts of prostate cancer,16-20 breast cancer21 and associated lymph node metastases,22 ocular cancer,23 liver disease,24 intravascular plaque,25-28 and hyperthermic lesions,29 and has even been implement to perform real-time tissue-type imaging17. Ultrasound is backscattered due to local inhomogeneity in tissue or other imbedded acoustical scatterers. The characteristics of the backscattered signals depend on the effective size and concentration of the scatterers, and are also a spatial function of the acoustic impedance (density and sound speed) of the tissue. For example, a malignant tumor scatters ultrasound differently than normal tissue because of its different microstructure.30 Analysis of the backscattered RF signals may allow different tissue types to be distinguished.31 The assessment by spectral parameters is quantitative and, with proper calibration, is independent of the system and user.
In the context of gastrointestinal cancer, ex vivo studies of lymph node metastases of colorectal cancer have shown that ultrasound backscatter analysis performed better than B-mode ultrasound32 even when multiple B-mode sonographic parameters were considered.33 Our previous in vivo pilot study34 showed that mean spectral parameters computed from EUS RF data can provide a non-invasive method to discriminate normal pancreas from diseased pancreas and benign from malignant lymph nodes. The mean intercept, slope, and midband fit of the spectra differed significantly among normal pancreas, pancreatic cancer, and chronic pancreatitis when all were compared to each other, and, on direct comparison, mean midband fit for pancreatic cancer differed significantly from chronic pancreatitis. For lymph nodes, mean midband fit and intercept differed significantly among benign and malignant lymph nodes.
In this in vivo validation study, our aim was to test spectral analysis of EUS backscattered signals using parameters derived from our pilot study (Study 1) with a new set of patients to distinguish between (i) benign and malignant lymph nodes and (ii) normal pancreas and diseased pancreas.
Methods
Patients and clinical protocol
A total of 24 patients (9 men and 15 women; mean age 66, range 40–90) already scheduled for EGD/EUS were enrolled in the validation study. As the data acquisition process does not affect the procedure itself, a waiver of patient consent for this study was granted by our institutional review board. The indications for the procedures included suspected disease in or around the esophagus (2 cases), stomach (4 cases), bile duct (2 cases), lymph nodes (2 cases) and pancreas (14 cases). In some cases images were also taken in areas outside of the organ of primary interest, to act as normal controls.
Data acquisition and analysis
As the procedure for data acquisition and analysis has been previously described in detail,34 only a summary is provided here. A commercially available clinical ultrasound system (Model Exera EU-M60, Olympus America, Center Valley, Pa) was used with an ultrasonic gastrovideoscope (Model GF-UM160, Olympus America, Center Valley, Pa). The endoscope contains a single-element transducer that spins about the axis of the scope in the lumen, thereby creating B-scan cross-sectional images, consisting of 256 A-scan lines. All data were acquired when the EUS system was operated in the C5 mode (transducer center frequency 6 MHz, transducer focal distance 20 mm, pulse repetition frequency 3.415 kHz). The RF data were obtained via a specially equipped output port (provided by Olympus America) using a digital oscilloscope (Model LT372, LeCroy Corporation, Chestnut Ridge, NY) in 8-bit mode at a sampling rate of 100 million samples per second. In each case, the B-scan image generated by the Olympus system (system image) was saved shortly before the RF data were acquired. The acquisition could not be achieved simultaneously because of technical limitations of the data transfer speed but were taken as close together as possible, with a time delay less than a few seconds. No complications were encountered during the data acquisition of any procedure.
The RF data were then imported into our custom-designed, MATLAB-based (MATLAB 2006b, Mathworks, Natick, Mass) analysis software for image reconstruction and data processing. Our analysis software first re-created images from the acquired RF data (reconstructed image). Prior to RF data analysis, regions of interest (ROIs) were identified and manually segmented by endosonographers on the system image according to the evaluation criteria described below. The ROIs were then independently translated onto the reconstructed image to select corresponding segments of RF data. Each sector-shaped area was sized to be maximum within the designated ROIs. The power spectrum was calculated for the signals of each A-scan RF data within the ROI gated by a series of sliding Hamming windows35, 36 of 1.5 μs, each offset by 0.1 μs.
Spectrum calibration
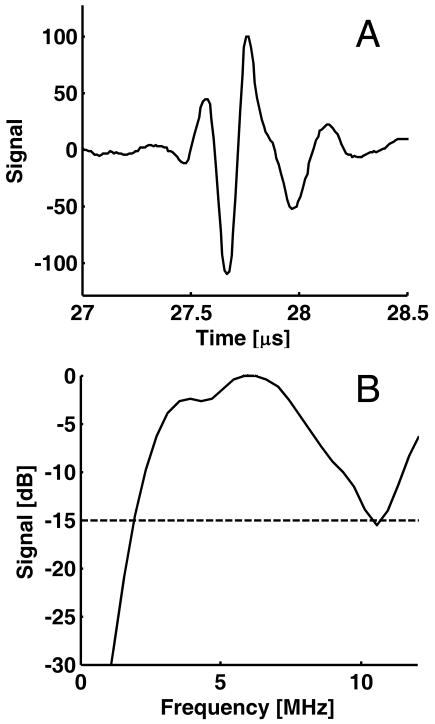
To remove artifacts associated with the composite transfer function of the electronic transmitter/receiver and transducer, calibration was performed by dividing the power spectrum by the spectrum of a perfect reflector. Our previous study34 used a glass cylinder filled with water as the reflector, but repeated measurements showed that this method was prone to signal saturation from the receiver electronics of the EUS system due to the large amplitude of the specular reflection. To prevent this problem, an acrylic plastic cylinder (5.08 cm inner diameter, 6.35 cm outer diameter) was filled with a cylindrical annulus (0.635 cm inner diameter, 5.08 cm outer diameter) composed of a custom-made gelatin phantom (consisting of Type A porcine skin gelatin, 10–20 μm Amberlite particles, bleach, and water) with attenuation of 0.7 dB/MHz/cm. The central hole in the gelatin phantom was filled with water prior to the insertion of the endoscope transducer, and the endoscope's balloon was inflated with water until the balloon was in full contact with the phantom. The orientation of the endoscope tip was then manually adjusted to ensure that the endoscope was centered and parallel to the axis of the cylinder. Figure 1A shows the pulse reflected from the annulus, and Fig. 1B shows the corresponding attenuation-corrected, normalized power spectrum in dB. The spectrum is broad, fairly uniform (15 dB bandwidth 2–10 MHz), and generally consistent with the frequency characteristics specified by the manufacturer. This spectrum was used for calibration of the validation data and the re-analysis of the pilot study data.
Figure 1.
Radio-frequency (RF) signal used for spectral calibration. A, Ultrasound pulse for the C5 mode of EUS system. B, Corresponding power spectrum. The usable bandwidth, defined by the range within −15 dB from the maximum of the spectrum, is 2 to 10.6 MHz or 8.6 MHz.
To perform the calibration, the tissue spectrum is divided by the calibration spectrum. The primary effect of the new calibration as compared to the previous calibration34 was the reduction of the midband fit and intercept by a nearly constant value, while the slope and R2 remained relatively unchanged (data not shown). Because the calibrated spectra are typically quasi-linear in shape over the ultrasound frequency range used,34 they can be effectively characterized by linear regression using their slope, intercept, and their midband fit, which is the value of the linear function evaluated at the midpoint of the −15dB bandwidth. The square of the correlation coefficient R2 for each fit was also recorded as a simple measure of the deviation of the calibrated spectrum from linearity.
Diagnostic criteria
Lymph node cases were classified as benign or malignant according to the following definitions. Benign lymph node cases met all of the following criteria: (1) Patient was referred for EUS for reasons other than cancer staging. (2) Patient had no prior or current diagnosis of malignancy. (3) Patient had no identification of mass lesions at EUS. (4) Patient had at least 1 EUS feature suggestive of benign nodes: (a) draping configuration (b) hyperechoic (c) < 1 cm in diameter. (5) No diagnosis of cancer in the year following EUS examination as determined by follow-up telephone questionnaire and/or review of medical records. Malignant lymph node cases met all of the following criteria: (1) Patient had lymph node identified at EUS that exhibited at least three established EUS features of malignant lymphadenopathy9, 37: (a) diameter > 1 cm, (b) round or oval shape, (c) diffusely hypoechoic, (d) sharp edges. (2) FNA cytology positive for carcinoma.
Pancreas cases were classified according to the following definitions. Normal pancreas (NP) cases met all of the following criteria: (1) Patient was referred for EGD/EUS for a non-pancreas indication (e.g. evaluation of submucosal nodules). (2) Patient had no history of alcohol abuse as defined by habitual consumption of > 40 g ethanol weekly. (3) Patient had no prior personal history of pancreatitis (4) Patient had no family history of pancreatitis. (5) Patient had no diagnosis of pancreatitis or pancreatic cancer in the year following EUS examination, as determined by follow-up telephone questionnaire and/or review of medical records. Chronic pancreatitis (CP) cases met all of the following criteria: (1) Patient was referred for EGD/EUS for evaluation of pancreas. (2) Patient had EUS examination with greater than or equal to 5 of the following established EUS criteria38: (a) hyperechoic foci (b) hyperechoic stranding (c) lobularity (d) cyst (e) calcification (f) ductal dilation (g) side branch dilation (h) duct irregularity (i) hyperechoic duct margins (j) atrophy (k) inhomogeneous echo pattern. (3) Patient had no diagnosis of pancreatic cancer in the year following EUS examination, as determined by follow-up telephone questionnaire and/or review of medical records. Pancreatic cancer (PC) cases met all of the following criteria: (1) Patient had a pancreatic mass lesion identified on EUS and (2) Patient had (a) an FNA cytology positive for adenocarcinoma or (b) positive ERCP brush cytology for adenocarcinoma or (c) positive mucosal biopsy for adenocarcinoma or (d) surgical pathology positive for adenocarcinoma.
Statistical analysis
The spectral parameters generated from each window were averaged over each ROI. The resulting values were then analyzed using Student's T-test for the two tissue types in the lymph-node data and a one-way ANOVA for the three tissue types in the pancreas data. Once parameters were identified that provided statistically significant differences between group means, linear discriminant analysis (LDA) was performed to classify the data using equal prior probabilities for each group and the within-groups covariance matrix. For LDA where the same data was used for training and testing, the leave-one-out approach was used for cross-validation. All statistical calculations were performed using SPSS (Version 16, SPSS, Chicago, Ill). A binormal receiver operating characteristic (ROC) curve was then fit to the resulting discriminant scores using the maximum likelihood estimation routine of ROCKIT39 (Version 1.1B2, University of Chicago, Chicago, Ill). Classification performance was assessed by computing the area under the curve (AUC) from ROCKIT.
Combined studies
The RF data from the pilot study was re-analyzed using the new calibration spectrum (Fig. 1). The combined data set resulting from both the re-analyzed pilot study (Study 1) and validation study (Study 2) was derived from a total of 43 subjects (16 men and 27 women; mean age 66, range 36–90). For reference, the pilot-study data34 had 8 cases of benign lymph nodes and 6 cases of malignant lymph nodes and also had 25 cases of normal pancreas, 3 cases of pancreatic cancer, and 4 cases of chronic pancreatitis.
Sample size calculations
For the validation study, sample size calculations were performed a priori using G*Power40 (Version 3, Heinrich-Heine-University, Dusseldorf, Germany) based on the results of our previous pilot study, assuming a significance level of 0.05, power of 0.80, and equal group sizes. These calculations estimated that 12–18 lymph-node cases, and a total of 12–18 pancreas cases with 8–14 cases of diseased pancreas were needed. Post hoc power calculations using the same software for the data from the combined studies indicated that the actual power achieved was greater than 0.93 for all reported cases of significant differences between groups based on the computed spectral parameters.
Results
Lymph nodes
In the validation study, RF data from 9 subjects (4 men and 5 women, mean age 66, range 48–90) were examined, yielding a total of 12 ROIs in the resulting images. The first three rows of Table 1 lists the mean and standard deviation of all the spectral parameters for the benign and malignant lymph nodes along with the results of Student's T-test between groups. The benign nodes have higher midband fit, lower (more negative) slope, higher intercept, and higher correlation coefficient than the malignant nodes, which is consistent with the results in the pilot study.34 The midband fit shows a statistically significant difference between the groups at the p = 0.05 levels, while the intercept is almost significantly different at this level.
Table 1. Descriptive statistics and hypothesis testing results for lymph node data.
Rows 1–2: Means and standard deviations of spectral parameters for benign and malignant lymph node cases from Study 2 (validation study). Row 3: The p-values resulting from Student's T-test assuming equal variance between groups. Rows 4–5: Means and standard deviations of spectral parameters for benign and malignant lymph node cases from both studies. Row 6: The p-values resulting from Student's T-test assuming equal variance between groups for slope and R2 and unequal variance between groups for midband fit and intercept.
| Data Set | B-mode appearance | Midband Fit [dB] | Slope [dB/MHz] | Intercept [dB] | Correlation coeff. R2 |
|---|---|---|---|---|---|
| Study 2 (Validation Study) | Benign (n = 7) | −42.2±8.6 | −0.71±0.36 | −37.9±9.1 | 0.16±0.03 |
| Malignant (n = 5) | −51.8±4.6 | −0.60±0.38 | −48.1±6.7 | 0.13±0.06 | |
| p-value | < 0.05 | 0.61 | 0.058 | 0.37 | |
| Study 1 + Study 2 | Benign (n = 15) | −43.8±6.6 | −1.20±0.80 | −36.5±6.8 | 0.24±0.12 |
| Malignant (n = 11) | −51.8±3.2 | −0.90±0.81 | −46.3±7.1 | 0.17±0.09 | |
| p-value | < 0.001 | 0.34 | < 0.002 | 0.13 | |
In the combined data set, RF data from 16 subjects (7 men and 9 women, mean age 66, range 48–90) were examined, yielding a total of 26 ROIs in the resulting images. The second three rows of Table 1 list the mean and standard deviation of all the spectral parameters for the benign and malignant lymph node cases. The midband fit and intercept show statistically significant differences between the two groups at the p = 0.05 level. These two parameters were subsequently used for the linear discriminant analysis.
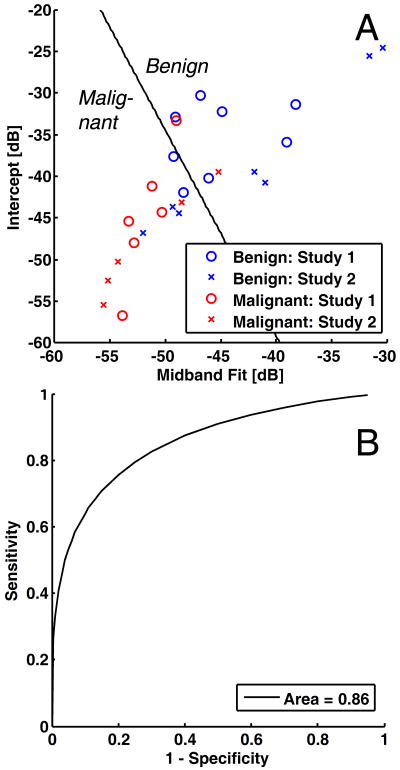
Table 2 shows the results of the LDA classification using various training and testing sets. When only the data from Study 1 (training set) were used to classify only the data from Study 2 (testing set), the classification had sensitivity of 57%, specificity of 80%, positive predictive value of 80%, negative predictive value of 57%, and overall accuracy of 67% (Table 2, Rows 1–2). Figure 2A shows the corresponding scatterplot of the data using midband fit and intercept. The solid line shows the approximate dividing line between benign and malignant cases according to the LDA derived from Study 1 data only. Figure 2B shows the corresponding binormal ROC curve with AUC of 0.86. When the combined data were used to classify the combined data using leave-one-out cross-validation, the classification had sensitivity of 67%, specificity of 82%, positive predictive value of 83%, negative predictive value of 75%, and overall accuracy of 73% (Table 2, Rows 3–4). The resulting ROC curve from this classification (not shown) has a somewhat higher AUC of 0.90.
Table 2. Classification results from linear discriminant analysis of lymph node data.
Classification was performed using midband fit and intercept as independent variables. Rows 1–2: Classification results for Study 2 data only based on training with Study 1 data only. Rows 3–4: Classification results using leave-one-out cross-validation for all data based on training with all data. (See Figure 2 for the corresponding scatterplot and ROC curve.)
| Training Data | Testing Data | Tissue State | Predicted State | ||
|---|---|---|---|---|---|
| Malignant | Benign | Total | |||
| Study 1 (n = 14) | Study 2 (n = 12) | Malignant | 4 | 1 | 5 |
| Benign | 3 | 4 | 7 | ||
| Study 1 + Study 2 (n = 26) | Study 1 + Study 2 (n = 26) | Malignant | 9 | 2 | 11 |
| Benign | 5 | 10 | 15 | ||
Figure 2.
Linear discriminant analysis and receiver operating characteristics (ROC) curves for the lymph node data. A, Scatterplot of data from Study 1 (pilot study) and Study 2 (validation study) with coordinates given by midband fit and intercept. The dividing line between benign and malignant classification is based on linear discriminant analysis of Study 1 data only (training set). B, Corresponding binormal maximum likelihood estimate of the ROC curve with discriminant scores from Study 2 only (testing set). The area the curve is 0.86. (See Table 2 for the corresponding classification matrix.)
Pancreas
In the validation study, RF data from 21 subjects (8 men and 13 women, mean age 64, range 40–78) were acquired for total of 28 ROIs in the resulting images. Rows 1–3 of Table 3 provide a summary of the mean values over each ROI for each spectral parameter in all the examined regions. The results are again consistent with those of the pilot study.34 The midband fit, intercept, and correlation coefficient were higher on average for NP than PC and CP, while the slope was lower (more negative) on average. ANOVA calculations indicated that at least one of the tissue types is distinguishable from the others with p < 0.01 (Table 3, Row 4). Post hoc multiple comparisons performed using the Bonferroni criterion (Table 3, Rows 5–7) indicated that NP differed significantly from PC for all parameters and from CP for midband fit and intercept, but there was not a significant difference between PC and CP at the p = 0.05 level.
Table 3. Descriptive statistics and hypothesis testing results for pancreas data.
Rows 1–3: Means and standard deviations of spectral parameters for normal pancreas (NP), pancreatic cancer (PC), and chronic pancreatitis (CP) cases from Study 2 (validation study). Row 4: The p-values resulting from ANOVA calculations assuming equal variance between groups. Rows 5–7: The p-values resulting from multiple comparisons using Bonferroni's criterion. Rows 8–10: Means and standard deviations of spectral parameters normal pancreas (NP), pancreatic cancer (PC), and chronic pancreatitis (CP) cases from both studies. Row 11: The p-values resulting from ANOVA calculations assuming equal variance between groups. Rows 12–14: The p-values resulting from multiple comparisons. Bonferroni's criterion was used for slope and intercept, whereas Tamhane's T2 criterion was used for midband fit and R2.
| Data Set | B-mode appearance | Midband Fit (dB) | Slope (dB/MHz) | Intercept (dB) | Correlation coeff. R2 |
|---|---|---|---|---|---|
| Study 2 (Validation Study) | Normal Pancreas (n = 15) | −40.9±3.8 | −1.44±0.31 | −32.2±4.7 | 0.31±0.08 |
| Pancreatic Cancer (n = 10) | −53.3±1.2 | −0.94±0.33 | −47.6±2.8 | 0.17±0.05 | |
| Chronic Pancreatitis (n = 3) | −49.5±3.3 | −1.28±0.54 | −41.8±6.5 | 0.25±0.10 | |
| ANOVA p-value | < 0.001 | < 0.01 | < 0.001 | < 0.001 | |
| NP vs. PC p-value | < 0.01 | < 0.01 | < 0.001 | < 0.001 | |
| NP vs. CP p-value | < 0.001 | 1 | < 0.01 | 0.59 | |
| PC vs. CP p-value | 0.21 | 0.43 | 0.14 | 0.27 | |
| Study 1 + Study 2 | Normal Pancreas (n = 40) | −41.5±3.4 | −1.86±0.76 | −30.3±5.4 | 0.40±0.12 |
| Pancreatic Cancer (n = 13) | −53.2±1.1 | −0.98±0.31 | −47.3±2.6 | 0.16±0.04 | |
| Chronic Pancreatitis (n = 7) | −50.2±2.1 | −1.21±0.34 | −42.8±4.0 | 0.26±0.07 | |
| ANOVA p-value | < 0.001 | < 0.01 | < 0.001 | < 0.001 | |
| NP vs. PC p-value | < 0.001 | < 0.01 | < 0.001 | < 0.001 | |
| NP vs. CP p-value | < 0.001 | 0.055 | < 0.001 | < 0.01 | |
| PC vs. CP p-value | < 0.05 | 1 | 0.14 | < 0.05 | |
When the data from both studies were combined, RF data from 41 subjects (14 men and 27 women, mean age 66, range 36–85) were acquired for total of 60 ROIs in the resulting images. Rows 8–11 of Table 3 show a summary of the mean values over each ROI for each spectral parameter in all the examined regions. NP is significantly different from PC for all parameters and from CP for all parameters except slope (Table 3, Rows 13–14). When PC and CP are compared, midband fit and correlation coefficient R2 both had significant differences between groups (Table 3, Row 15). As a result, these two parameters were used subsequently for the linear discriminant analysis.
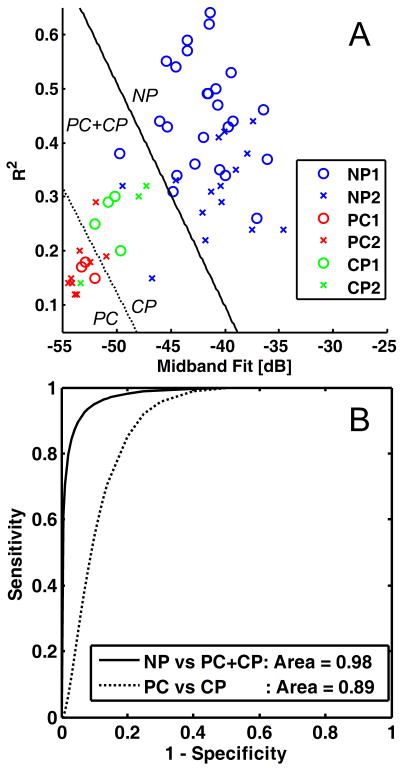
First, a LDA was performed to classify the data between normal pancreas (NP) and diseased pancreas (PC and CP grouped together). Table 4 shows the results of the LDA classification using various training and testing sets. When only the data from Study 1 (training set) were used to classify only the data from Study 2 (testing set), the classification had sensitivity of 100%, specificity of 87%, positive predictive value of 87%, negative predictive value of 100%, and overall accuracy of 93% (Table 4, Rows 1–2). Figure 3A shows the scatterplot of the data along with the dividing line between the normal and diseased pancreas cases (solid line) according to the LDA derived from Study 1 data only, while Fig. 3B shows the corresponding binormal ROC curve (solid line) with AUC of 0.98. When the combined data were used to classify the combined data using leave-one-out cross-validation, the classification had sensitivity of 95%, specificity of 93%, positive predictive value of 86%, negative predictive value of 97%, and overall accuracy of 93% (Table 4, Rows 3–4). The resulting ROC curve from this classification (not shown) has a slightly higher AUC of 0.99.
Table 4. Classification results from linear discriminant analysis of pancreas data into normal pancreas (NP) and diseased pancreas (PC and CP) groups.
Classification was performed using midband fit and correlation coefficient R2 as independent variables. Rows 1–2: Classification results for Study 2 data only based on training with Study 1 data only. Rows 3–4: Classification results using leave-one-out cross-validation for all data based on training with all data. (See Figure 3 for the corresponding scatterplot and ROC curve.)
| Training Data | Testing Data | Tissue State | Predicted State | ||
|---|---|---|---|---|---|
| Diseased | Normal | Total | |||
| Study 1 (n = 32) | Study 2 (n = 28) | Diseased | 13 | 0 | 13 |
| Normal | 2 | 13 | 15 | ||
| Study 1 + Study 2 (n = 60) | Study 1 + Study 2 (n = 60) | Diseased | 19 | 1 | 20 |
| Normal | 3 | 37 | 40 | ||
Figure 3.
Linear discriminant analysis and receiver operating characteristics (ROC) curves for the pancreas data. A, Scatterplot of data from Study 1 (pilot study) and Study 2 (validation study) with coordinates given by midband fit and correlation coefficient R2. The dividing lines for the classification of normal pancreas (NP) vs diseased pancreas (PC and CP together) [solid] and pancreatic cancer (PC) vs. chronic pancreatitis (CP) excluding NP [dashed] are based on linear discriminant analysis of Study 1 data only (training set). The legend indicates which tissue state and study number corresponds to each point. B, Corresponding binormal maximum likelihood estimates of the ROC curves for each classification with discriminant scores from Study 2 only (testing set). The area under each curve is given in the legend. (See Tables 4 and 5 for the corresponding classification matrices.)
Given the excellent separation between normal and diseased pancreas, a LDA was next performed between PC and CP cases with NP cases excluded. The results are shown in Table 5 using various training and testing sets. When only the data from Study 1 (training set) were used to classify only the data from Study 2 (testing set), the classification had sensitivity of 80%, specificity of 67%, positive predictive value of 89%, negative predictive value of 50%, and overall accuracy of 77% (Table 5, Rows 1–2). Figure 3A shows the scatterplot of the data along with the dividing line between the normal and diseased pancreas cases (dashed line) according to the LDA derived from Study 1 data only, while Fig. 3B shows the corresponding binormal maximum likelihood estimate of the ROC curve (dashed line) with AUC of 0.89. When the combined data were used to classify the combined data using leave-one-out cross-validation, the classification had sensitivity of 85%, specificity of 71%, positive predictive value of 85%, negative predictive value of 86%, and overall accuracy of 85% (Table 5, Rows 3–4). The resulting ROC curve from this classification (not shown) improves slightly to an AUC of 0.91.
Table 5. Classification results from linear discriminant analysis of pancreas data into pancreatic cancer (PC) and chronic pancreatitis (CP) groups, excluding normal pancreas (NP) data.
Classification was performed using midband fit and correlation coefficient R2 as independent variables. Rows 1–2: Classification results for Study 2 data only based on training with Study 1 data only. Rows 3–4: Classification results using leave-one-out cross-validation for all data based on training with all data. (See Figure 3 for the corresponding scatterplot and ROC curve.)
| Training Data | Testing Data | Tissue State | Predicted State | ||
|---|---|---|---|---|---|
| PC | CP | Total | |||
| Study 1 (n = 7) | Study 2 (n = 13) | PC | 8 | 2 | 10 |
| CP | 1 | 2 | 3 | ||
| Study 1 + Study 2 (n = 20) | Study 1 + Study 2 (n = 20) | PC | 11 | 2 | 13 |
| CP | 2 | 5 | 7 | ||
Discussion
Computer-aided diagnosis is a current area of research that may improve diagnostic results. In the context of EUS of pancreas and lymph nodes, initial efforts have largely focused on computerized digital image analysis of grayscale images41-46. Irisawa et al.41, 42 showed that statistically different changes in the size of hyperechoic area computed from EUS images occurred between normal pancreas and chronic pancreatitis, where echogenicity was measured relative to a control area in the initial set of echoes (rings surrounding the transducer). Norton et al.43 employed four image texture parameters and a neural network classifier to differentiate chronic pancreatitis from pancreatic cancer with diagnostic accuracy up to 89%. Das et al.44 used eleven texture parameters (out of an initial test set of 288 parameters), mostly based on various statistical parameters of gray-level nonuniformity, with neural network classification to distinguish pancreatic cancer from chronic pancreatitis and normal tissue with ROC AUC of 0.93. For lymph nodes, Loren et al.45 performed analysis of images with three parameters and found that malignant nodes had statistically significant lower echogenicity, higher whole node heterogeneity, and more roundness (lower long to short axis ratio) as compared to benign lymph nodes as well as somewhat less inter-node variability.
Analyses of grayscale images have the advantage that they can be performed with all imaging systems, but the imaging conditions need to be highly controlled to ensure reproducibility (e.g. same gain and contrast setting, same transducer, etc.) and the derived metrics can be difficult to relate back to tissue histology or other tissue properties objectively and in a meaningful way. Recently, other approaches such as spectral analysis of EUS backscattered signals34 and elastography47-52 have been explored, both of which involve computerized analysis of the RF data underlying the grayscale images. These methods require special software and/or hardware that are not yet implemented or not yet widely available with commercial systems, but have the potential to provide additional useful information beyond what can be obtained with conventional systems.
Our studies demonstrate that spectral analysis of RF data from EUS in vivo is an effective method to differentiate malignant from benign lymph nodes and to differentiate between normal and diseased pancreas, based on quantitative and objective criteria. The spectral method draws upon frequency and phase information of the backscattered ultrasound signals which is already acquired by EUS systems but omitted in traditional B-mode image creation. As such, clinical implementation can be done with minimal modification to current equipment and procedures, including systems with digital signals and linear arrays.
Previous analyses of ultrasound backscatter spectra have shown their utility to extract potentially useful information about tissue microstructure which is characterized by effective acoustic scatterers in tissue.30, 31 The slope of the spectrum depends primarily on scatterer size, while the midband fit and intercept contain information about the scatterer concentration and relative acoustic impedance between the scatterers and surrounding tissue.14 Our results show that slope is not as effective as midband fit and intercept for differentiating between malignant and non-malignant tissue; this suggests that scatterer size is a less important factor than scatterer concentration and acoustic impedance for tissue differentiation in the current applications. For the pancreas data, the reduced midband fit between NP and PC is consistent with the typically reduced echogenicity of tumors relative to surrounding tissue. Interestingly, midband fit has also been shown to be the most important parameter for differentiation between cancerous and noncancerous prostate biopsies.16 For the lymph node data, the reduced midband fit and intercept of the lymph node data is consistent with the lower echogenicity of malignant nodes relative to benign nodes.45 Intercept has also been shown to be a useful parameter for the differentiation of ex vivo metastatic and nonmetastatic lymph nodes in the context of breast cancer22 and colon cancer.33
In this study, classification was performed based on a linear discriminant analysis. LDA is a relatively straightforward approach that typically works best when the groups occupy different but separable regions of parameter space. Other classification techniques like artificial neural networks, support vector machines, and classification and regression trees have been shown to provide more robust classification of more complex ultrasound data17, 20, 26, although this comes often at the cost of increased complexity (and potentially reduced understanding) of the diagnostic criteria. Our classification based on spectral parameters did not perform as well as some others reported in the literature43, 44 in terms of sensitivity or ROC AUC, but more parameters were used in these studies (4 and 11, respectively). Thus our approach might be further improved by other classification techniques or in combination with additional independent parameters. For example, the computer-aided analyses of EUS images described previously have examined morphological criteria and shown that chronic pancreatitis has increased area of hyperechogencity of relative to normal pancreas41, 42 and malignant nodes have increased roundness relative to benign nodes.45 It is possible that combination of spectral parameters with information about morphology, heterogeneity, or other complementary tissue properties (e.g., tissue stiffness from elastography) could further enhance diagnostic capabilities.
Our study has several limitations. First, the spatial averaging inherent in the ROI method limits the spatial resolution over which the spectral parameters can be computed. The use of higher frequencies could improve resolution, provided that the tissue to be imaged is close enough to the transducer so the signals are not decreased severely by attenuation. Second, the acoustical properties of the pancreas are known to vary at different locations in the pancreas (i.e., head, body, tail), and the location was not considered in the current analysis. Third, histology results were only available for cases of suspected malignancy (malignant lymph nodes, pancreatic cancer) with biopsy, and thus only relatively well defined cases of normal pancreas, chronic pancreatitis, and pancreatic cancer and well cases of defined benign malignant lymph nodes were studied to reduce the uncertainty of diagnosis. Fourth, the relatively small sample size precludes the sensible use of more sophisticated classification techniques. This issue could be addressed by sub-sampling the ROIs used in the current data, particularly of the pancreas ROIs, although at the risk of decreasing the independence of the individual data. Finally, these studies were performed with older mechanical radial scanning ultrasound endoscopes. In particular, the FNA biopsies had to be performed with a curvilinear array echoendoscope that was different from the endoscope used to acquire the RF data. This made it challenging to ensure that the biopsied region was identical to the spectrally-analyzed region, particularly for the lymph node cases. Many major EUS centers have transitioned to electronic scanning ultrasound endoscopes. In theory, the same principles should be applicable to electronic scanning but it will be necessary to repeat these studies with the newer equipment.
Additional work is necessary to fully demonstrate the effectiveness of the technique. Ex vivo characterization of resected tissue may be helpful to obtain a more thorough understanding of the relationship between the measured spectral parameters and the physical and histological characteristics of the tissue. Ideally, a classification scheme could be developed that would allow for parametric “tissue-type” images like those that have been developed in ultrasound imaging of the prostate19, 20 and coronary artery28, in which grayscale B-mode images are encoded or “digitally stained” with coloration corresponding to the probability of various normal or disease states. Co-registration of histological and “tissue type” images could then be performed. Larger-scale prospective studies are also required to determine the utility of this technique over a broader range of patients, conditions, and equipment and such studies using electronic scanning ultrasound endoscopes are already in progress, including a curvilinear array echoendoscope used to perform FNA biopsies. If reliable diagnostic criteria can be developed based on these studies, it is possible that spectral analysis method could lead to improved targeting of pancreatic lesions and lymph nodes for EUS guided fine needle aspiration, even in real-time.
Conclusion
This study shows that spectral analysis of the EUS RF backscatter signals in vivo can provide a method to discriminate between benign and malignant lymph nodes as well as between normal pancreas, chronic pancreatitis, and pancreatic cancer. The combination of mean midband fit and intercept performed best for differentiation between lymph node states (ROC area = 0.86–0.90). The combination of mean midband fit and correlation coefficient provided an excellent method for distinguishing normal pancreas from diseased pancreas (ROC area = 0.98–0.99) and a good method for distinguishing normal pancreas from diseased pancreas (ROC area = 0.89–0.91). However, additional work is needed to develop the method to be sufficiently robust and reliable for clinical practice.
Acknowledgments
The authors would like to acknowledge Olympus for making the RF data available, Brian Wolf for his technical assistance with the EUS system, and Jingping Xu for the construction of the phantom used during the calibration measurements.
Grant support: This work was supported in part by the Ohio Wright Center of Innovation/Biomedical Research and Technology Transfer Grant “Biomedical structural, functional and molecular imaging enterprise.” Amitabh Chak was supported by a K24 Midcareer Award in Patient Oriented Research, National Institutes of Health (Grant DK002800). Farees T. Farooq and Ronald E. Kumon were supported in part by an ASGE Research, Outcomes, and Effectiveness Award.
Acronyms
- ANOVA
Analysis of variance
- AUC
Area under the curve
- CP
Chronic pancreatitis
- EGD
Esophagogastroduodenoscopy
- EUS
Endoscopic ultrasound
- FNA
Fine need aspiration
- LDA
Linear discriminant analysis
- NP
Normal pancreas
- PC
Pancreatic cancer
- RF
Radio frequency
- ROC
Receiver operating characteristic
- ROI
Region of interest
Footnotes
Participating Institutions: Division of Gastroenterology, University Hospitals Case Medical Center, Case Western Reserve University, Cleveland, Ohio, USA, and Department of Biomedical Engineering, Case Western Reserve University, Cleveland, Ohio, USA
Conflict of interest disclosures: None
Author contributions: (A) Conception and design [A.L.F., A.C., C.X.D., R.E.K., F.T.F., M.P.]; (B) Analysis and interpretation of the data [R.E.K., V.K.C., K.O., F.T.F., M.P., Y.Z., R.C.K.W., G.A.I., M.V.S., A.C., C.X.D.]; (C) Drafting of the article [R.E.K., M.P., F.T.F.]; (D) Critical revision of the article for important intellectual content [all]; (E) Final approval of the article [all]
Capsule Summary
- Identifying pancreatic cancers in the setting of chronic pancreatitis and differentiating benign from malignant lymph nodes can be challenging using conventional grayscale EUS imaging.
- Spectral analysis of ultrasound backscatter has been previously demonstrated in both EUS and non-EUS contexts to be an objective, quantitative method for tissue characterization and uses information in the backscattered ultrasound signals otherwise discarded in grayscale EUS imaging.
- This validation study shows that spectral parameters such as midband fit, intercept, and correlation coefficient of the EUS backscatter spectra can quantitatively discriminate between normal pancreas, pancreatic cancer, and chronic pancreatitis, as well as between benign and malignant lymph nodes.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rifkin MD, Ehrlich SM, Marks G. Staging of rectal carcinoma: prospective comparison of endorectal US and CT. Radiology. 1989;170:319–22. doi: 10.1148/radiology.170.2.2643135. [DOI] [PubMed] [Google Scholar]
- 2.Botet JF, Lightdale CJ, Zauber AG, Gerdes H, Winawer SJ, Urmacher C, Brennan MF. Preoperative staging of gastric cancer: comparison of endoscopic US and dynamic CT. Radiology. 1991;181:426–32. doi: 10.1148/radiology.181.2.1924784. [DOI] [PubMed] [Google Scholar]
- 3.Meyenberger C, Huch Boni RA, Bertschinger P, Zala GF, Klotz HP, Krestin GP. Endoscopic ultrasound and endorectal magnetic resonance imaging: a prospective, comparative study for preoperative staging and follow-up of rectal cancer. Endoscopy. 1995;27:469–79. doi: 10.1055/s-2007-1005751. [DOI] [PubMed] [Google Scholar]
- 4.Willis S, Truong S, Gribnitz S, Fass J, Schumpelick V. Endoscopic ultrasonography in the preoperative staging of gastric cancer: accuracy and impact on surgical therapy. Surg Endosc. 2000;14:951–4. doi: 10.1007/s004640010040. [DOI] [PubMed] [Google Scholar]
- 5.van Vliet EP, Eijkemans MJ, Poley JW, Steyerberg EW, Kuipers EJ, Siersema PD. Staging of esophageal carcinoma in a low-volume EUS center compared with reported results from high-volume centers. Gastrointest Endosc. 2006;63:938–47. doi: 10.1016/j.gie.2006.01.053. [DOI] [PubMed] [Google Scholar]
- 6.Dewitt J, Devereaux BM, Lehman GA, Sherman S, Imperiale TF. Comparison of endoscopic ultrasound and computed tomography for the preoperative evaluation of pancreatic cancer: a systematic review. Clin Gastroenterol Hepatol. 2006;4:717–25. doi: 10.1016/j.cgh.2006.02.020. quiz 664. [DOI] [PubMed] [Google Scholar]
- 7.Bhutani MS, Gress FG, Giovannini M, Erickson RA, Catalano MF, Chak A, Deprez PH, Faigel DO, Nguyen CC. The No Endosonographic Detection of Tumor (NEST) Study: a case series of pancreatic cancers missed on endoscopic ultrasonography. Endoscopy. 2004;36:385–9. doi: 10.1055/s-2004-814320. [DOI] [PubMed] [Google Scholar]
- 8.Chong AK, Hawes RH, Hoffman BJ, Adams DB, Lewin DN, Romagnuolo J. Diagnostic performance of EUS for chronic pancreatitis: a comparison with histopathology. Gastrointest Endosc. 2007;65:808–14. doi: 10.1016/j.gie.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 9.Catalano MF, Alcocer E, Chak A, Nguyen CC, Raijman I, Geenen JE, Lahoti S, Sivak MV., Jr Evaluation of metastatic celiac axis lymph nodes in patients with esophageal carcinoma: accuracy of EUS. Gastrointest Endosc. 1999;50:352–6. doi: 10.1053/ge.1999.v50.98154. [DOI] [PubMed] [Google Scholar]
- 10.Kanamori A, Hirooka Y, Itoh A, Hashimoto S, Kawashima H, Hara K, Uchida H, Goto J, Ohmiya N, Niwa Y, Goto H. Usefulness of contrast-enhanced endoscopic ultrasonography in the differentiation between malignant and benign lymphadenopathy. Am J Gastroenterol. 2006;101:45–51. doi: 10.1111/j.1572-0241.2006.00394.x. [DOI] [PubMed] [Google Scholar]
- 11.Bhutani MS, Hawes RH, Hoffman BJ. A comparison of the accuracy of echo features during endoscopic ultrasound (EUS) and EUS-guided fine-needle aspiration for diagnosis of malignant lymph node invasion. Gastrointest Endosc. 1997;45:474–9. doi: 10.1016/s0016-5107(97)70176-7. [DOI] [PubMed] [Google Scholar]
- 12.Chen VK, Eloubeidi MA. Endoscopic ultrasound-guided fine needle aspiration is superior to lymph node echofeatures: a prospective evaluation of mediastinal and peri-intestinal lymphadenopathy. Am J Gastroenterol. 2004;99:628–33. doi: 10.1111/j.1572-0241.2004.04064.x. [DOI] [PubMed] [Google Scholar]
- 13.Lizzi FL, Greenebaum M, Feleppa EJ, Elbaum M, Coleman DJ. Theoretical framework for spectrum analysis in ultrasonic tissue characterization. J Acoust Soc Am. 1983;73:1366–73. doi: 10.1121/1.389241. [DOI] [PubMed] [Google Scholar]
- 14.Lizzi FL, Astor M, Kalisz A, Liu T, Coleman DJ, Silverman R, Ursea R, Rondeau M. Ultrasonic spectrum analysis for assays of different scatterer morphologies: Theory and very-high frequency results. 1996 IEEE Ultrasonics Symposium; Piscataway, NJ. 1996. [Google Scholar]
- 15.Lizzi FL, Feleppa EJ, Alam SK, Deng CX. Ultrasonic spectrum analysis for tissue evaluation. Pattern Recog Lett. 2003;24:637–658. [Google Scholar]
- 16.Feleppa EJ, Liu T, Kalisz A, Shao MC, Fleshner N, Reuter V, Fair WR. Ultrasonic Spectral-Parameter Imaging of the Prostate. Int J Imaging Sys Tech. 1997;8:11–25. [Google Scholar]
- 17.Feleppa EJ, Ennis RD, Schiff PB, Wuu CS, Kalisz A, Ketterling J, Urban S, Liu T, Fair WR, Porter CR, Gillespie JR. Spectrum-analysis and neural networks for imaging to detect and treat prostate cancer. Ultrason Imaging. 2001;23:135–46. doi: 10.1177/016173460102300301. [DOI] [PubMed] [Google Scholar]
- 18.Scheipers U, Ermert H, Sommerfeld HJ, Garcia-Schurmann M, Senge T, Philippou S. Ultrasonic multifeature tissue characterization for prostate diagnostics. Ultrasound Med Biol. 2003;29:1137–49. doi: 10.1016/s0301-5629(03)00062-0. [DOI] [PubMed] [Google Scholar]
- 19.Feleppa EJ, Porter CR, Ketterling J, Lee P, Dasgupta S, Urban S, Kalisz A. Recent developments in tissue-type imaging (TTI) for planning and monitoring treatment of prostate cancer. Ultrason Imaging. 2004;26:163–72. doi: 10.1177/016173460402600303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feleppa EJ. Ultrasonic tissue-type imaging of the prostate: Implications for biopsy and treatment guidance. Cancer Biomark. 2008;4:201–12. doi: 10.3233/cbm-2008-44-504. [DOI] [PubMed] [Google Scholar]
- 21.Golub RM, Parsons RE, Sigel B, Feleppa EJ, Justin J, Zaren HA, Rorke M, Sokil-Melgar J, Kimitsuki H. Differentiation of breast tumors by ultrasonic tissue characterization. J Ultrasound Med. 1993;12:601–8. doi: 10.7863/jum.1993.12.10.601. [DOI] [PubMed] [Google Scholar]
- 22.Tateishi T, Machi J, Feleppa EJ, Oishi R, Jucha J, Yanagihara E, McCarthy LJ, Noritomi T, Shirouzu K. In vitro diagnosis of axillary lymph node metastases in breast cancer by spectrum analysis of radio frequency echo signals. Ultrasound Med Biol. 1998;24:1151–9. doi: 10.1016/s0301-5629(98)00100-8. [DOI] [PubMed] [Google Scholar]
- 23.Silverman RH, Folberg R, Rondeau MJ, Boldt HC, Lloyd HO, Chen X, Lizzi FL, Weingeist TA, Coleman DJ. Spectral parameter imaging for detection of prognostically significant histologic features in uveal melanoma. Ultrasound Med Biol. 2003;29:951–9. doi: 10.1016/s0301-5629(03)00907-4. [DOI] [PubMed] [Google Scholar]
- 24.King DL, Lizzi FL, Feleppa EJ, Wai PM, Yaremko MM, Rorke MC, Herbst J. Focal and diffuse liver disease studied by quantitative microstructural sonography. Radiology. 1985;155:457–62. doi: 10.1148/radiology.155.2.2984720. [DOI] [PubMed] [Google Scholar]
- 25.Noritomi T, Sigel B, Swami V, Justin J, Gahtan V, Chen X, Feleppa EJ, Roberts AB, Shirouzu K. Carotid plaque typing by multiple-parameter ultrasonic tissue characterization. Ultrasound Med Biol. 1997;23:643–50. doi: 10.1016/s0301-5629(97)00013-6. [DOI] [PubMed] [Google Scholar]
- 26.Nair A, Kuban BD, Obuchowski N, Vince DG. Assessing spectral algorithms to predict atherosclerotic plaque composition with normalized and raw intravascular ultrasound data. Ultrasound Med Biol. 2001;27:1319–31. doi: 10.1016/s0301-5629(01)00436-7. [DOI] [PubMed] [Google Scholar]
- 27.Waters KR, Bridal SL, Cohen-Bacrie C, Levrier C, Fornes P, Laugier P. Parametric analysis of carotid plaque using a clinical ultrasound imaging system. Ultrasound Med Biol. 2003;29:1521–30. doi: 10.1016/j.ultrasmedbio.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 28.Nasu K, Tsuchikane E, Katoh O, Vince DG, Virmani R, Surmely JF, Murata A, Takeda Y, Ito T, Ehara M, Matsubara T, Terashima M, Suzuki T. Accuracy of in vivo coronary plaque morphology assessment: a validation study of in vivo virtual histology compared with in vitro histopathology. J Am Coll Cardiol. 2006;47:2405–12. doi: 10.1016/j.jacc.2006.02.044. [DOI] [PubMed] [Google Scholar]
- 29.Lizzi FL, Astor M, Liu T, Deng C, Coleman DJ, Silverman RH. Ultrasonic spectrum analysis for tissue assays and therapy evaluation. Int J Imaging Sys Tech. 1997;8:3–10. [Google Scholar]
- 30.Oelze ML, O'Brien WD, Jr, Blue JP, Zachary JF. Differentiation and characterization of rat mammary fibroadenomas and 4T1 mouse carcinomas using quantitative ultrasound imaging. IEEE Trans Med Imaging. 2004;23:764–71. doi: 10.1109/tmi.2004.826953. [DOI] [PubMed] [Google Scholar]
- 31.Lizzi FL, Ostromogilsky M, Feleppa EJ, Rorke MC, Yaremko MM. Relationship of ultrasonic spectral parameters to features of tissue microstructure. IEEE Trans Ultrason Ferroelectr Freq Control. 1987;34:319–29. doi: 10.1109/t-uffc.1987.26950. [DOI] [PubMed] [Google Scholar]
- 32.Noritomi T, Machi J, Feleppa EJ, Yanagihara E, Shirouzu K. In vitro investigation of lymph node metastasis of colorectal cancer using ultrasonic spectral parameters. Ultrasound Med Biol. 1998;24:235–43. doi: 10.1016/s0301-5629(97)00274-3. [DOI] [PubMed] [Google Scholar]
- 33.Tateishi T, Machi J, Feleppa EJ, Oishi AJ, Furumoto NL, Oishi RH, McCarthy LJ, Yanagihara E, Shirouzu K. In vitro investigation of detectability of colorectal lymph nodes and diagnosis of lymph node metastasis in colorectal cancer using B-mode sonography. J Clin Ultrasound. 2004;32:1–7. doi: 10.1002/jcu.10210. [DOI] [PubMed] [Google Scholar]
- 34.Kumon RE, Olowe K, Faulx AL, Farooq FT, Chen VK, Zhou Y, Wong RC, Isenberg GA, Sivak MV, Chak A, Deng CX. EUS spectrum analysis for in vivo characterization of pancreatic and lymph node tissue: a pilot study. Gastrointest Endosc. 2007;66:1096–106. doi: 10.1016/j.gie.2007.05.052. [DOI] [PubMed] [Google Scholar]
- 35.Harris FJ. Use of windows for harmonic-analysis with discrete Fourier-transform. Proceedings of the IEEE. 1978;66:51–83. [Google Scholar]
- 36.Press WH, Flannery BP, Teukolsky SA, Vetterling WT. Numerical Recipes in FORTRAN: The Art of Scientific Computing. New York, NY: Cambridge University Press; 1992. [Google Scholar]
- 37.Schmulewitz N, Wildi SM, Varadarajulu S, Roberts S, Hawes RH, Hoffman BJ, Durkalski V, Silvestri GA, Block MI, Reed C, Wallace MB. Accuracy of EUS criteria and primary tumor site for identification of mediastinal lymph node metastasis from non-small-cell lung cancer. Gastrointest Endosc. 2004;59:205–12. doi: 10.1016/s0016-5107(03)02692-0. [DOI] [PubMed] [Google Scholar]
- 38.Wallace MB, Hawes RH. Endoscopic ultrasound in the evaluation and treatment of chronic pancreatitis. Pancreas. 2001;23:26–35. doi: 10.1097/00006676-200107000-00004. [DOI] [PubMed] [Google Scholar]
- 39.University of Chicago. Receiver Operating Characteristic program software downloads. http://xray.bsd.uchicago.edu/krl/KRL_ROC/software_index.htm.
- 40.Faul F, Erdfelder E, Lang AG, Buchner A. G*Power 3: a flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behav Res Methods. 2007;39:175–91. doi: 10.3758/bf03193146. [DOI] [PubMed] [Google Scholar]
- 41.Irisawa A, Mishra G, Hernandez LV, Bhutani MS. Quantitative analysis of endosonographic parenchymal echogenicity in patients with chronic pancreatitis. J Gastroenterol Hepatol. 2004;19:1199–205. doi: 10.1111/j.1440-1746.2004.03468.x. [DOI] [PubMed] [Google Scholar]
- 42.Irisawa A, Katakura K, Ohira H, Sato A, Bhutani MS, Hernandez LV, Koizumi M. Usefulness of endoscopic ultrasound to diagnose the severity of chronic pancreatitis. J Gastroenterol. 2007;42 17:90–4. doi: 10.1007/s00535-006-1916-9. [DOI] [PubMed] [Google Scholar]
- 43.Norton ID, Zheng Y, Wiersema MS, Greenleaf J, Clain JE, Dimagno EP. Neural network analysis of EUS images to differentiate between pancreatic malignancy and pancreatitis. Gastrointest Endosc. 2001;54:625–9. doi: 10.1067/mge.2001.118644. [DOI] [PubMed] [Google Scholar]
- 44.Das A, Nguyen CC, Li F, Li B. Digital image analysis of EUS images accurately differentiates pancreatic cancer from chronic pancreatitis and normal tissue. Gastrointest Endosc. 2008;67:861–7. doi: 10.1016/j.gie.2007.08.036. [DOI] [PubMed] [Google Scholar]
- 45.Loren DE, Seghal CM, Ginsberg GG, Kochman ML. Computer-assisted analysis of lymph nodes detected by EUS in patients with esophageal carcinoma. Gastrointest Endosc. 2002;56:742–6. doi: 10.1067/mge.2002.128920. [DOI] [PubMed] [Google Scholar]
- 46.Bhutani MS. Digital analysis of EUS images: “promising” method, but is it ready for “prime time”? Gastrointest Endosc. 2008;67:868–70. doi: 10.1016/j.gie.2007.12.061. [DOI] [PubMed] [Google Scholar]
- 47.Saftoiu A, Vilman P. Endoscopic ultrasound elastography-- a new imaging technique for the visualization of tissue elasticity distribution. J Gastrointestin Liver Dis. 2006;15:161–5. [PubMed] [Google Scholar]
- 48.Giovannini M, Hookey LC, Bories E, Pesenti C, Monges G, Delpero JR. Endoscopic ultrasound elastography: the first step towards virtual biopsy? Preliminary results in 49 patients. Endoscopy. 2006;38:344–8. doi: 10.1055/s-2006-925158. [DOI] [PubMed] [Google Scholar]
- 49.Saftoiu A, Vilmann P, Ciurea T, Popescu GL, Iordache A, Hassan H, Gorunescu F, Iordache S. Dynamic analysis of EUS used for the differentiation of benign and malignant lymph nodes. Gastrointest Endosc. 2007;66:291–300. doi: 10.1016/j.gie.2006.12.039. [DOI] [PubMed] [Google Scholar]
- 50.Janssen J, Dietrich CF, Will U, Greiner L. Endosonographic elastography in the diagnosis of mediastinal lymph nodes. Endoscopy. 2007;39:952–7. doi: 10.1055/s-2007-966946. [DOI] [PubMed] [Google Scholar]
- 51.Janssen J, Schlorer E, Greiner L. EUS elastography of the pancreas: feasibility and pattern description of the normal pancreas, chronic pancreatitis, and focal pancreatic lesions. Gastrointest Endosc. 2007;65:971–8. doi: 10.1016/j.gie.2006.12.057. [DOI] [PubMed] [Google Scholar]
- 52.Saftoiu A, Vilmann P, Gorunescu F, Gheonea DI, Gorunescu M, Ciurea T, Popescu GL, Iordache A, Hassan H, Iordache S. Neural network analysis of dynamic sequences of EUS elastography used for the differential diagnosis of chronic pancreatitis and pancreatic cancer. Gastrointest Endosc. 2008;68:1086–94. doi: 10.1016/j.gie.2008.04.031. [DOI] [PubMed] [Google Scholar]