Abstract
Green fluorescent protein (GFP) from a jellyfish, Aequorea victoria, and its mutants are widely used in biomedical studies as fluorescent markers. In spite of the enormous efforts of academia and industry toward generating its red fluorescent mutants, no GFP variants with emission maximum at more than 529 nm have been developed during the 15 years since its cloning. Here, we used a new strategy of molecular evolution aimed at generating a red-emitting mutant of GFP. As a result, we have succeeded in producing the first GFP mutant that substantially matures to the red-emitting state with excitation and emission maxima at 555 and 585 nm, respectively. A novel, nonoxidative mechanism for formation of the red chromophore in this mutant that includes a dehydration of the Ser65 side chain has been proposed. Model experiments showed that the novel dual-color GFP mutant with green and red emission is suitable for multicolor flow cytometry as an additional color since it is clearly separable from both green and red fluorescent tags.
Green fluorescent protein (GFP)1 and its mutants and homologues are widely known today due to their intensive use as in vivo fluorescent markers in biomedical sciences (1–3). Studies on the GFP family began 45 years ago when GFP from hydromedusa Aequorea aequorea (synonym Aequorea victoria) was discovered (4). This protein has been found to be a part of the bioluminescent system of the jellyfish A. victoria where GFP played a role of secondary emitter transforming blue light from the photoprotein aequorin into green light. Then, similar proteins were isolated from several bioluminescent coelenterates (5). All of these proteins fluoresced green (497–509 nm) and functioned as secondary emitters in bioluminescence.
In 1992, cDNA encoding A. victoria GFP was cloned (6). It transpired that this gene can be heterologically expressed in any organism, owing to the unique ability of GFP to form chromophore without the assistance of external enzymes or cofactors except for molecular oxygen (7). GFP amino acids Ser65-Tyr66-Gly67 undergo posttranslational modification (backbone cyclization and oxidation) resulting in a two-ring chromophore (8). One ring originates from Tyr66, and another five-membered heterocycle is formed after protein backbone cyclization. Wild-type GFP from A. victoria possesses a dual-peak absorption-excitation spectrum with a major peak at 396 nm and an approximately 3-fold smaller peak at 475 nm. It was found that the shorter wavelength peak corresponds to the chromophore with a neutral (protonated) Tyr66, while the longer wavelength peak corresponds to the anionic (deprotonated) state of the chromophore-forming Tyr66 (9, 10). Excitation at both peaks leads to green fluorescence at 503 or 509 nm.
In 1999, GFP homologues from nonbioluminescent Anthozoa species were cloned (11). Anthozoa-derived GFP-like proteins showed great spectral diversity including cyan, green, yellow, and red fluorescent proteins (FPs) and purple-blue nonfluorescent chromoproteins (CPs) (2). In addition, green-to-red photoconvertible fluorescent proteins were characterized (12, 13). Also, a broad spectral diversity of GFP-like proteins in Hydrozoa species was revealed (14). In particular, a yellow FP and a purple CP from hydroid jellyfishes were characterized.
Between 1994 and 1999, when only one fluorescent protein, GFP from A. victoria, was cloned, extensive efforts were applied to create GFP variants of different colors. As a result, blue and cyan mutants with Tyr66 substituted with Phe, His, or Trp were generated (9, 15). Also, it was demonstrated that certain point mutations can strongly influence a proportion of neutral/anionic chromophore. For example, substitution S65T leads to mainly anionic chromophore (absorption at ~490 nm) (16), while T203I stabilizes the neutral chromophore state (absorption at ~400 nm) (9). Finally, a yellow variant of GFP with excitation–emission maxima at 515–529 nm was obtained using the key mutation T203Y (17). In spite of enormous efforts of academia and industry toward generating a red fluorescent mutant, no further red-shifted variants have been developed on the basis of A. victoria GFP.
Here, basing our work on modern knowledge of mechanisms of red chromophore formation in GFP-like proteins, we used a new strategy of mutagenesis and succeeded in the creation of a GFP mutant that partially matures up to a red-emitting state.
MATERIALS AND METHODS
Mutagenesis and Protein Characterization
EGFP coding region with human codon usage (Clontech) was used as a template. Site-specific mutagenesis was performed using a QuickChange mutagensis kit (Stratagene). For simultaneous mutagenesis at several sites, including saturated mutagenesis at several sites, an overlap-extension approach was applied (18). Diversity PCR random mutagenesis kit (Clontech) was used for error-prone PCR in conditions optimal for seven mutations per 1000 bp. For bacterial expression, the full-length coding region was amplified using specific primers, subcloned into the pQE30 (Qiagen) using BamHI and HindIII sites, and was expressed with an N-terminal polyhistidine tag in Escherichia coli XL1 Blue strain (Invitrogen). Fluorescent stereomicroscope SZX-12 (Olympus) was used for visual screening of bacterial colonies expressing mutant proteins.
Recombinant proteins with a polyhistidine tag were purified with Ni-NTA agarose (Qiagen). To measure excitation and emission spectra, the FluoroMax-3 spectrofluorometer (Jobin Yvon) or Cary Eclipse spectrofluorometer (Varian) was applied. For absorbance measurements, a Hitachi U-2000 spectrophotometer was used. Oligomeric states of R10-3 and EGFP proteins were compared by size-exclusion gel filtration using an AKTAprime chromatograph (Amersham Pharmacia Biotech).
For expression in eukaryotic cells, the PCR-amplified fragment encoding R10-3 was inserted in lieu of the EGFP coding region in the pEGFP-C1 (Clontech) vector using AgeI and BglII sites. Human embryonic kidney 293 (HEK293) cell line was used. Cells were transfected using Lipo-fectamine reagent (Invitrogen) and were tested 16 h after transfection. A Leica confocal inverted microscope, DMIRE2 TCS SP2, equipped with an HCX PL APO lbd.BL 63X 1.4 NA oil objective and 125 mW Ar and 1 mW HeNe lasers was used for cell imaging.
Library Screening
For FACS screening, a coding region of GFP variants was PCR amplified as a BglII–EcoRI fragment and inserted into a pBAD/His-B vector (Invitrogen). Random mutagenesis was performed with a GeneMorph II random mutagenesis kit (Stratagene) under conditions resulting in a mutation frequency up to 16 mutations per 1000 base pairs. After ligation into pBAD/His-B vector (Invitrogen), a mixture of the random mutants was electroporated into TOP10 host cells (Invitrogen). Typical mutant libraries for each FACS screen consisted of approximately 107 independent clones. Protein expression in the libraries was induced overnight at 37 °C with 0.02% arabinose. The following morning the expressing bacteria were washed with phosphate-buffered saline (PBS) and then diluted with PBS for FACS sorting to optical densities of 0.015–0.02 at 600 nm. A MoFlo cell sorter (Dako) equipped with standard argon, krypton, and argon–krypton mixed-gas lasers was used. Typically, at least 10 sizes of each library were sorted on the FACS with 488 nm of argon, 407 nm of krypton, and 568 nm of mixed-gas excitation laser lines. The collected bacterial cells were rescued in a rich SOC medium at 37 °C for several hours and then plated on Petri dishes with 0.02% arabinose. The next day the dishes were studied with a Leica MZ16F fluorescence stereomicroscope using standard filter sets (Chroma).
RESULTS
Theoretical Considerations
Two main types of the red chromophores within the GFP-like proteins are known, specifically DsRed-like and Kaede-like. Compared to the GFP chromophore, the DsRed-like chromophore contains an additional acylimine group which is formed via dehydrogenation of the Cα—N bond of the first chromophore-forming residue with molecular oxygen (19, 20). A key role of some nearby residues including Ser68 (GFP numbering) in catalysis of this oxidation has been suggested (20, 21). In some proteins (e.g., asFP595, zFP538, mOrange, zFP574) the DsRed-like chromophore undergoes further chemical transformations (22–27).
The Kaede-like chromophore is characteristic for the green-to-red photoconvertible fluorescent proteins carrying His-Tyr-Gly chromophore-forming residues. In this case, after irradiation with UV or violet light (at approximately 350–420 nm) a GFP-like precursor undergoes a protein backbone break between N and Cα of His65 and converts into the red state where the GFP-like chromophore core is extended with a His65 heterocycle (28–30).
Importantly, in most cases both types of the red chromophores originate from a neutral state of the GFP-like chromophore, while anionic green chromophore cannot be converted into the red state (12, 31). The only known exceptions from this rule are the Kaede-like protein Dendra, which can undergo green-to-red photoconversion under blue light irradiation (32), and zFP574 protein in which an anionic green chromophore transforms into the red state via oxidation and decarboxylation of Asp65 (27, 33, 34). Taking into account these considerations, as well as previous unsuccessful attempts to achieve step-by-step red shift beyond YFP using mutagenesis of already red-shifted GFP variants with anionic chromophore, we concluded that GFP variants that contain chromophore mainly in the protonated state might be a promising template for construction of a red GFP mutant.
In the present study we suggest a new strategy for molecular evolution aimed at generating the red-emitting mutant of A. victoria GFP. As the first step, we introduced substitutions that were shown to be important for red chromophore formation in Anthozoa red FPs and CPs. Specifically, His65 and Asn68 or Ser68 were introduced for the following reasons. His65 is crucial for Kaede-like chromophore formation. Position 68 is usually occupied by Asn or Ser in red fluorescent proteins and chromoproteins. This step resulted in essentially nonfluorescent mutants. At the second step, random mutagenesis was used to restore green fluorescence in these mutants. Importantly, we selected mutants containing the GFP chromophore mainly in a neutral state (absorption maximum at about 400 nm). According to the mechanism previously described by us for chromophore formation in a majority of red fluorescent proteins, only such GFP chromophore could potentially form a red chromophore. Finally, we used random mutagenesis coupled with high-throughput screening to search for red fluorescent clones.
Mutagenesis
GFP mutants F64L/S65H/V68N and F64L/S65H/V68S obtained by site-directed mutagenesis were essentially nonfluorescent. These mutants together were used as templates for random mutagenesis. Screening of mutant libraries (about 30000 clones for each round) was performed visually using a fluorescent stereomicroscope; we selected clones that fluoresced under 405 nm excitation light. Also, fluorescence of clones in the red region and capacity for Kaede-type green-to-red photoconversion were evaluated. After three sequential rounds of random mutagenesis, the two best clones, named CR3 and CR4, were obtained. Both proteins contained target substitutions F64L/S66H/V68N (together with some randomly introduced mutations) and possessed excitation–emission spectra very similar to that of wild-type GFP; neither red emission nor green-to-red photoconversion was characteristic for CR3 and CR4. No clones carrying Ser68 were found.
A mix of plasmids CR3 and CR4 was used as a template for two further rounds of random mutagenesis. As a result, a clone named R5-1 showing not only green but also red fluorescence was found. Its red fluorescence was very weak and could be detected only after growth at 30 °C and incubation at 4 °C for 1 week. R5-1 contained the following substitutions, F64L, V68N, K162Q, I167V, L194P, and T230I, and additionally a stop codon after position 232 (six residues earlier than in GFP). Surprisingly, R5-1 carried Ser65; i.e., a reverse mutation of directly introduced His65 occurred via two nucleotide substitutions. Random mutagenesis of R5-1 resulted in mutant R6-31 with one additional substitution, A87G, which demonstrated the same spectral characteristics, but its red fluorescence could already be detected after overnight growth of E. coli colonies at 30 or 37 °C. Further rounds of random mutagenesis, even using high-throughput screening with a fluorescence-activated cell sorter (FACS), failed to enhance the red fluorescence of R6-31.
We then constructed a mutant carrying some substitutions that seem to be important for red fluorescence appearance in GFP, specifically F64L, V68N, K162Q, and I167V, and used this mutant as a template for sequential random mutagenesis followed by high-throughput screening using FACS. Three distinct phenotypes of the mutants were observed: green only, blue-green, and green-red. The first two random libraries resulted in a brighter mutant, R8-1, having an additional mutation, F46L. The next two rounds of random mutagenesis yielded an enhanced mutant, R10-3, with additional substitutions V163A and I171L (Figure 2A). The R10-3 demonstrated both green and red fluorescence (Figure 3) that was much brighter compared to R6-31. In comparison to GFP, the following substitutions were characteristic for R10-3: F46L, F64L, V68N, V163A, K162Q, I167V, and I171L (Figure 1).
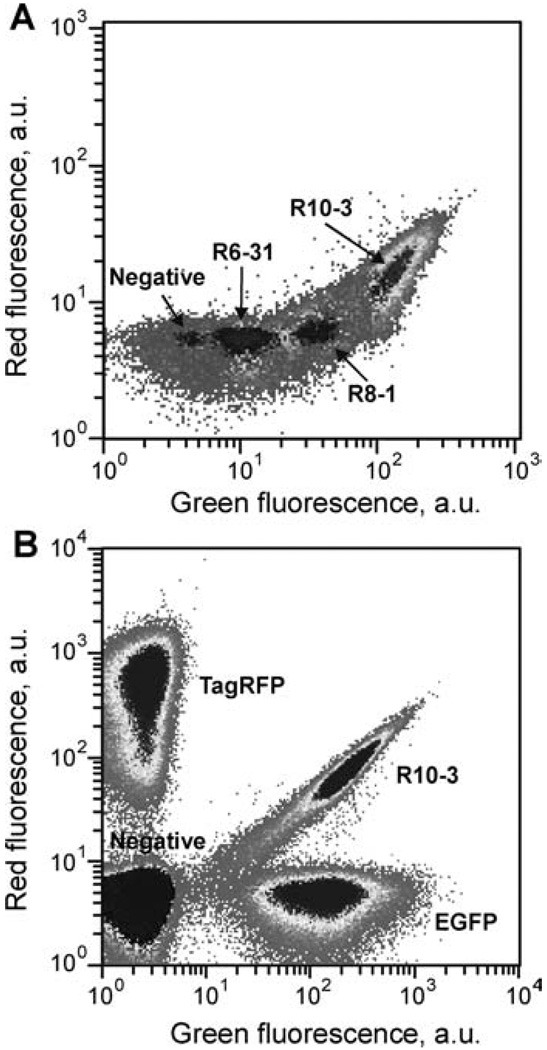
FIGURE 2.
Flow cytometry analysis. The 488 nm line of argon and 568 nm line of krypton lasers were used to excite green and red fluorescence, respectively. (A) Comparison of bacterial cells with empty plasmid or expressing R6-31, R8-1, R10-3 mutants obtained consequently during molecular evolution. (B) Sorting of a mixture of bacterial cells expressing TagRFP, EGFP, R10-3, or empty plasmid. Note the clear separation of R10-3 from both EGFP and TagRFP.
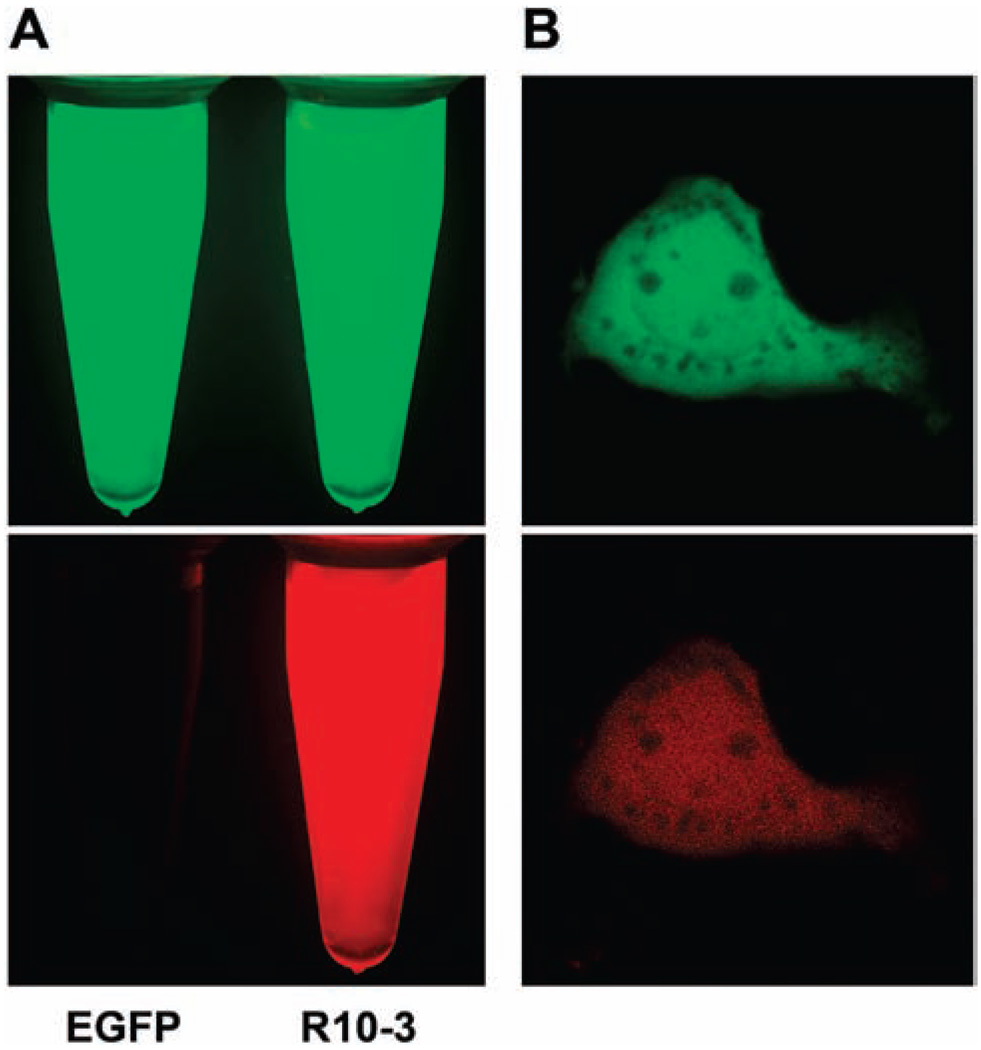
FIGURE 3.
Fluorescence microscopy of R10-3. (A) Comparison of EGFP (left tube) and R10-3 (right tube) fluorescence. Purified protein samples were photographed under a fluorescent stereomicroscope. Top: Excitation at 450–490 nm; detection above 510 nm (green channel). Bottom: Excitation at 540–580 nm; detection above 610 nm (red channel). (B) Confocal image of HEK293 cells expressing R10-3. Top: Excitation at 488 nm; detection at 500–530 nm (green channel). Bottom: Excitation at 543 nm; detection at 560–650 nm (red channel).
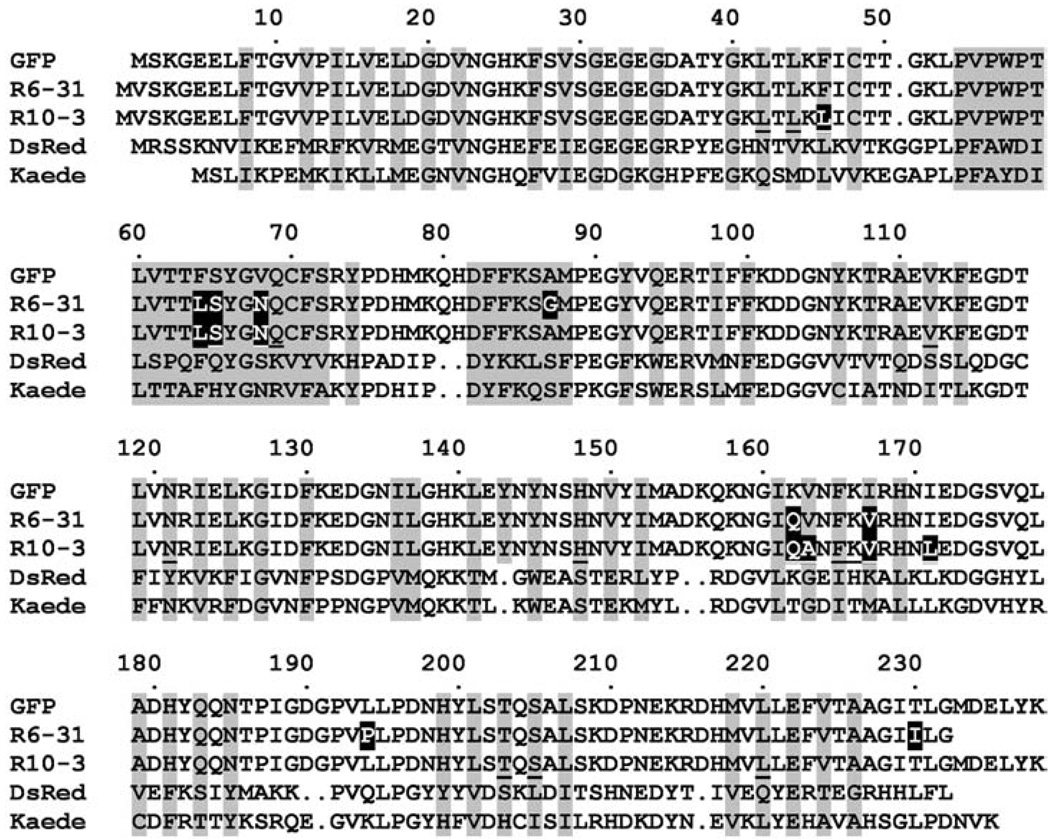
FIGURE 1.
Amino acid alignment of GFP, its mutants R6-31 and R10-3, Anthozoa red fluorescent protein DsRed, and green-to-red photoconvertible fluorescent protein Kaede. The residues whose side chains are in the interior of the β-can are shaded in gray. Introduced gaps are represented by dots. Substitutions in R6-31 and R10-3 are shaded in black. In R10-3, positions at which saturated mutagenesis was performed are underlined.
Further rounds of random mutagenesis followed by high-throughput screening did not provide improvement of R10-3. Also, we performed saturated mutagenesis at positions that are spatially close to the chromophore: 42, 44, 46, 65, 68, 69, 112, 121, 148, 163, 165, 167, 171, 203, 205, and 220. Several phenotypes have been observed at this step as a result of the single-point substitutions. For example, mutant L42Q possessed blue and green but no red fluorescence. A single mutation S65G gave the green-only variant. However, none of the mutants possessed red fluorescence brighter than that of the parental R10-3.
Properties of the Red Mutants
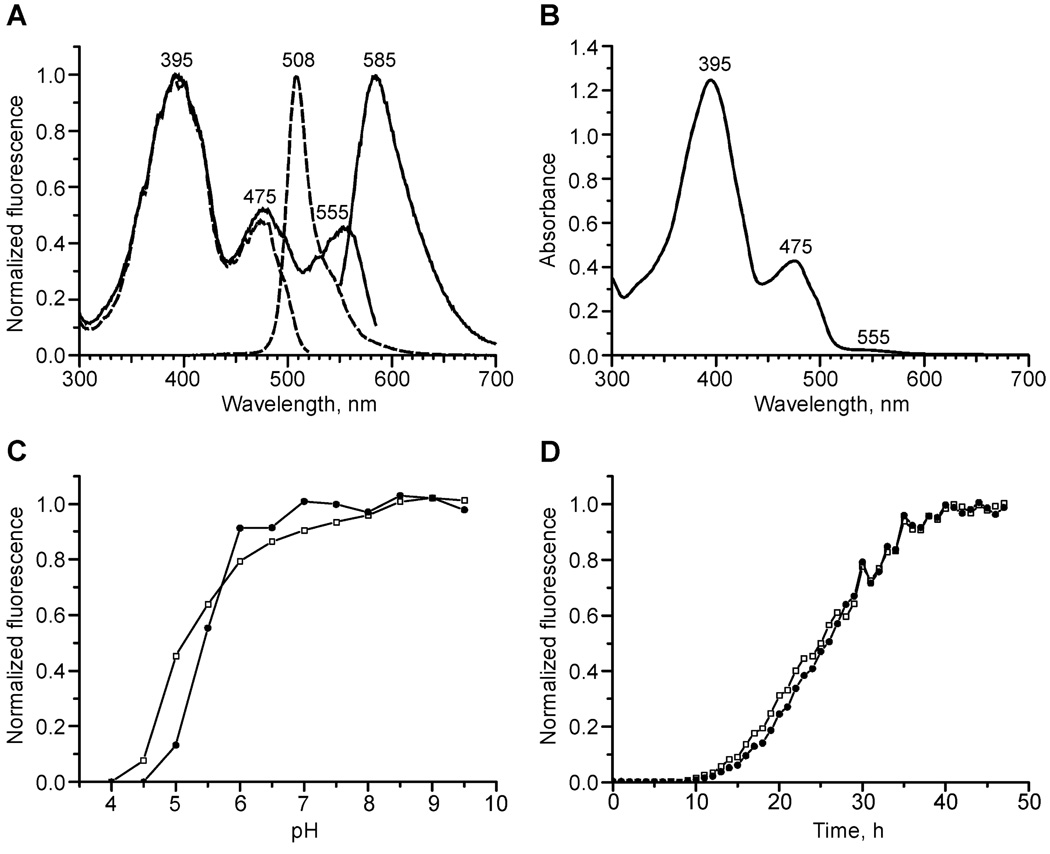
All mutants with red fluorescence (R5-1, R6-31, R8-1, and R10-3) possessed very similar shapes and maxima of excitation–emission in the red region. They differed considerably only in maturation rate and fluorescence brightness. Thus, we will focus here on the brightest variant, R10-3. Green fluorescence of R10-3 was characterized by a major excitation maximum at 395 nm (extinction coefficient 34000 M−1 cm−1), minor excitation maximum at 475 nm (extinction coefficient 12000 M−1 cm−1), and single emission maximum at 508 nm (fluorescence quantum yield 0.75), similarly to the wild-type GFP (Figure 4A). In addition, R10-3 demonstrated an excitation peak at 555 nm (extinction coefficient 800 M−1 cm−1) and a corresponding emission peak at 585 nm (fluorescence quantum yield 0.45). The absorption spectrum showed that the red form is present as a minor fraction (Figure 4B). Thus, only a small portion (1–3%) of R10-3 protein matures up to the red fluorescent state. Titration of R10-3 with buffers with different pH showed that pKa values of the green and red forms of R10-3 are very similar, 5.1 and 5.4, respectively (Figure 4C).
FIGURE 4.
Spectral properties of the R10-3 mutant. (A) Excitation and emission spectra for green (dashed lines) and red (solid lines) fluorescence. Within each pair of lines the emission spectrum is the one at longer wavelengths. (B) Absorbance spectrum. (C) pH dependence of green (open squares) and red (filled circles) fluorescence. (D) Development of green (open squares) and red (filled circles) fluorescence in growing E. coli streak expressing R10-3.
We have also tested the temperature dependence of R10-3 maturation. E. coli cells expressing R10-3 were grown at 22, 30, or 37 °C, and their fluorescence in green and red channels was compared. It was found that elevated temperature resulted in higher brightness of both green and red signals, while their ratio remained about the same at all three temperatures tested (not shown). Then we studied the kinetics of development of green and red fluorescence in E. coli streaks growing at 37 °C. Although several different processes, growth of the streak, protein synthesis, and R10-3 maturation, contributed to the fluorescence increase in this model, it was possible to compare maturation of the green and red forms since other processes were equal. It was found that the green signal developed slightly faster than the red (Figure 4D). So, the red to green ratio gradually increased approximately 1.5-fold during R10-3 maturation.
It was shown earlier that illumination with ~400 nm light greatly accelerates red fluorescence development in maturing DsRed (31). In contrast, under similar conditions we did not observe any increase in red fluorescence after 405 nm light irradiation of both immature and mature R10-3 samples.
We have successfully expressed R10-3 in mammalian cells. The next day following transient transfection, HEK293 cells demonstrated bright green fluorescence and weaker but clearly visible red fluorescence (Figure 3B).
Mutant R10-3 contains mainly inner substitutions. Two external mutations, specifically K162Q and I171L, should not change the oligomeric state of the protein because these positions do not participate in the GFP dimer interface. Indeed, size-exclusion chromatography (gel filtration) showed that both EGFP and R10-3 proteins at a concentration of 1 mg/mL migrate as a single peak corresponding to a monomer (not shown).
Possible Applications
Commonly, single-color tags with narrow spectra are considered to be ideal for multicolor labeling. However, dual-color fluorescent proteins can offer a clear advantage in some cases. For example, a tag with green plus red emission can be potentially discriminated from both green and red tags, using only two channels for detection. To demonstrate the usefulness of R10-3, we performed the following model experiment. E. coli cells expressing TagRFP (35) (single-color red tag), EGFP (single-color green tag), or R10-3 (dual-color green plus red tag) were mixed together. This mix was then analyzed by flow cytometry using two channels (green and red). Figure 2B shows that all three cell populations were clearly separated from each other (as well as from “empty” cells) due to unusual “diagonal” distribution of R10-3-expressing cells. Thus R10-3 appeared to be a useful extension to the available palette of fluorescent proteins for cell sorting applications.
DISCUSSION
Comparison of R10-3 Maturation with Green-to-Red GFP Photoconversion
Previously, a green-to-red photoconversion of GFP and some its variants has been described (36, 37). It was found that under anaerobic conditions irradiation by blue light results in the appearance of a GFP red fluorescent state with excitation and emission maxima at 525 and 600 nm, respectively. The red form is stable only in the absence of molecular oxygen, and it disappears after reoxygenation (38). No explanation of this phenomenon has been suggested to date.
Comparison of the light-induced red GFP state in anaerobic conditions with the R10-3 red form clearly shows that they have completely different properties. First, R10-3 develops red fluorescence spontaneously, while GFP requires light irradiation to become red fluorescent. Second, the R10-3 red state is formed and remains stable in the presence of oxygen, in contrast to the GFP photoconversion that requires anaerobic conditions. Finally, excitation and emission spectra for the red species in R10-3 and photoconverted red GFP are clearly different. Thus, the appearance of red fluorescence in R10-3 is unrelated to previously described GFP photo-conversion in a low oxygen environment.
Interpretation of Mutagenesis Results
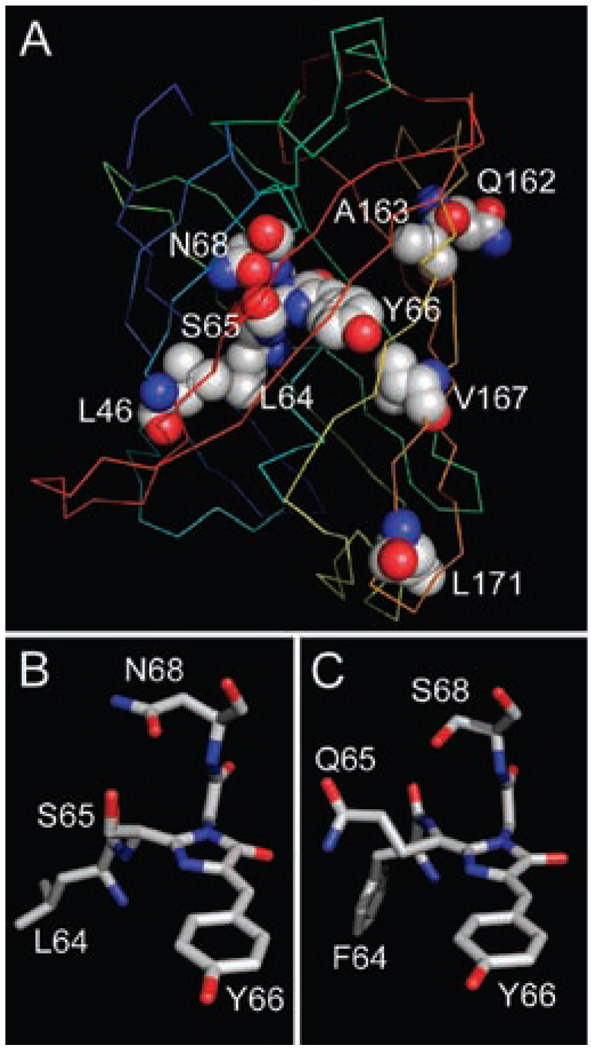
Figure 5A shows an overall GFP structure with modeled R10-3 substitutions. This model demonstrates that almost all mutated positions (except Leu171) are located in close proximity to the chromophore.
FIGURE 5.
Outline of the proposed three-dimensional structure of R10-3. (A) Ribbon scheme of the overall R10-3 structure modeled on the base of the GFP-F64 crystal structure (Protein Data Bank accession code 1EMM). The chromophore and residues, which are substituted in the R10-3 mutant, are labeled and shown in spacefilling representation. The protein backbone is colored from blue (N-end) to red (C-end). (B, C) Structural comparison of residues 64–68 in R10-3 (B) and DsRed (a monomer with presumably the catalytic conformation of Ser68 from Protein Data Bank accession code 1G7K) (C) shown in stick representation. Note the similar positioning of R10-3 Asn68 and DsRed Ser68 where oxygen atoms of the side chains are close to Cα at position 65. Amino acid substitutions followed by geometry optimization were performed using HyperChem software. Graphics are outputs of PyMOL software.
At the beginning of this work we supposed that generation of a Kaede-like GFP mutant might be easier, compared to a DsRed-like red fluorescent mutant, because UV irradiation provides an additional driving force for red chromophore formation in Kaede-like proteins. At the same time, His65 seemed to have no apparent negative effect on formation of a DsRed-type chromophore. Therefore, we directly introduced His65, which is invariant in Kaede-like fluorescent proteins. However, our assumption appeared to be incorrect, since no Kaede-type mutants with His65 were found. Instead, the reverse mutation to wild-type Ser65 was isolated (mutant R5-1). This roundabout approach added considerable work but also highlighted the importance of Ser65 in the formation of red chromophore within the GFP mutants obtained. However, Ser65 alone is undoubtedly insufficient for red phenotype appearance, since wild-type GFP never demonstrated red fluorescence. We believe that a combination of Ser65 and Asn68 plays a key role in red chromophore formation in the GFP mutants. In contrast to chemically inert valine, the Asn68 side chain may play a catalytic role (see below).
Mutations F46L and V163A were characterized earlier to greatly improve maturation of GFP variants (39, 40). Generally, substitutions F46L, V163A, and I167V are in line with the conclusion of Terskikh and co-workers that space around the chromophore accelerates red fluorescence development in DsRed (41). Substitution F46L is spatially very close to Ser65 (Figure 5A) and can provide additional flexibility required for chemical modification at the Ser65 backbone. Also, a smaller Leu residue may vacate space for an additional water molecule that can facilitate catalysis of red chromophore formation.
Possible Mechanisms for Red Chromophore Formation
Basing our views on the importance of Ser65 and Asn68 for development of red fluorescence in GFP mutants, we hypothesized possible structures and mechanisms of red chromophore formation in mutants R5-1, R6-31, and R10-3. Two distinct scenarios can be proposed. First, an oxidative pathway can be similar to that for DsRed. Yarbrough and co-workers suggested that Ser68 catalyzes proton abstraction from Cα of Gln65 and the formation of a carbanion intermediate (see Scheme 2 in ref 20). Then, molecular oxygen attacks the carbanion and forms a hydroperoxide adduct. Finally, a Cα=N double bond is formed at Gln65, and hydrogen peroxide is released. Analogously, in the case of red mutants of GFP, the same pathway can be proposed where Asn68 catalyzes proton abstraction instead of Ser68. Molecular modeling showed that Asn68 can potentially adopt a conformation similar to the presumably catalytic conformation of Ser68 in DsRed (Figure 5B,C).
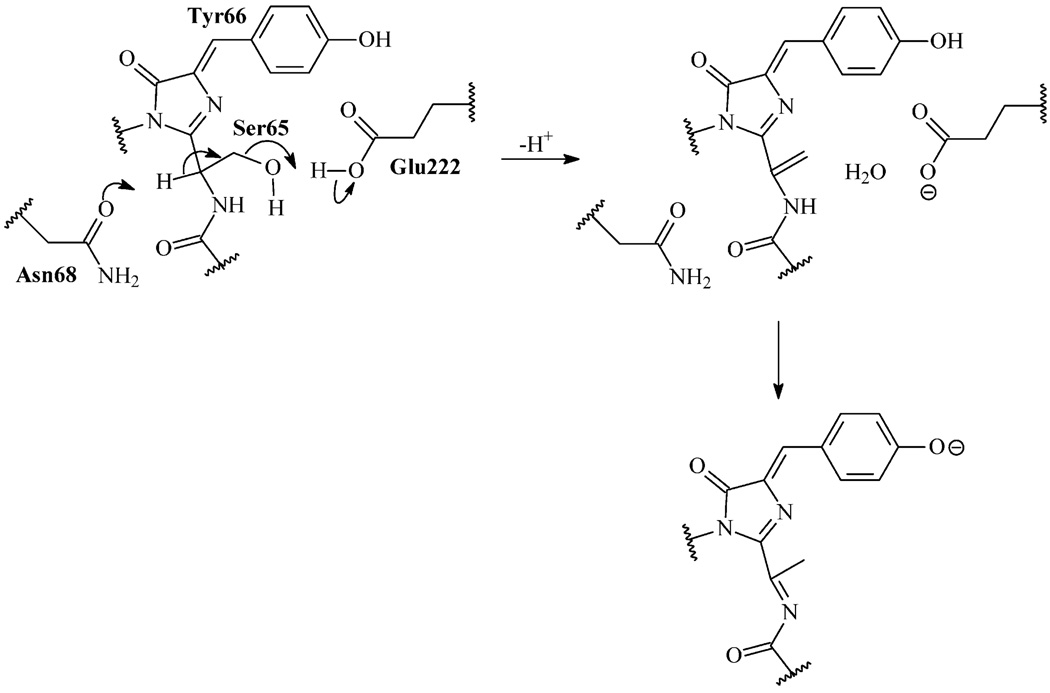
A second possible mechanism is nonoxidative and does not include reaction with molecular oxygen. Here, Asn68 catalyzes dehydration of Ser65 (Scheme 1). As a result, a Cα=Cβ double bond is formed at Ser65. Then, this double bond can isomerize to form acylimine. We prefer this mechanism because it clearly explains the irreplaceability of Ser65 in GFP red mutants that was observed experimentally. Notably, similar dehydration of the Ser residue is observed in the course of posttranslational maturation of histidine ammonia lyase (HAL) (42, 43), as well as in the S65A/Y66S mutant of GFP (44). Determination of the exact chromophore structure within R10-3 and the mechanism of its formation is outside the scope of the present work. Nonetheless, some possible ways to check our hypothesis can be suggested. First, Ser65 dehydration (loss of 18 Da) can be detected by mass spectroscopy. Also, the dependence or independence of red chromophore formation on molecular oxygen can be tested. Unfortunately, the R10-3 mutant contains only a minor fraction of red chromophore. Thus, experimental verification of the R10-3 chromophore structure appears to be a very difficult task.
Scheme 1.
Proposed Nonoxidative Mechanism of the Red Chromophore Formation in R10-3a
aDuring the first step, Asn68 and Glu222 promote dehydration of Ser65 resulting in a double bond between the Cα and Cβ atoms of Ser65. Then, an isomerization of this double bond and deprotonation of Tyr66 may lead to the DsRed-like chromophore where the GFP chromophore core is extended with the acylimine group.
The reason for incomplete maturation of the red chromophore in R10-3 is unclear. Red fluorescent proteins often contain a portion of immature molecules with GFP-like green chromophore. For example, DsRed samples contain roughly 50% of green monomers (19, 20, 45). However, in DsRed, this immature green chromophore is present exclusively in an anionic state. Earlier we have shown that the DsRed red chromophore is formed only from a neutral GFP-like intermediate that can be transiently detected in the course of maturation of DsRed and other red fluorescent proteins and chromoproteins (31). At the same time, R10-3 samples always show a major portion of neutral green chromophore, which potentially could be a source for red chromophore formation. Thus, instead of the absence of a GFP-like neutral chromophore precursor, some other reason should be suggested for incomplete maturation of R10-3. We speculate that Asn68 can adopt two distinct conformations. In one (minor) conformation, its side chain is close to Gln65 Cα (Figure 4B) and can play a catalytic role. In another (major) conformation, Asn68 is rotated away from Gln65 and is catalytically inert. Stochastic fixation of these conformations occurs immediately after the folding of the protein globule and does not change further. If so, the first conformation corresponds to the red species, while the second conformation corresponds to the green species in the R10-3 sample. Notably, such “catalytic” and “noncatalytic” conformations of Ser68 were experimentally detected in the crystal structure of DsRed (20).
Practical Applications
Apparently, R10-3 cannot be used simply as a red tag, due to its major green emission. However, it might be useful in some special cases. For example, we showed that R10-3 can serve as an additional color for flow cytometry due to a “diagonal” distribution between commonly used green and red channels that allows the discrimination of it from both green and red fluorescent proteins. As far as we know, a similar behavior has been described for only one fluorescent protein: Zs Yellow (excitation and emission maxima at 525 and 538 nm, respectively) from Clontech. Also, dual-color mutants of red fluorescent proteins such as DsRed-N42H have been described (46). However, similarly to many other coral fluorescent proteins, these proteins are tetramers and can aggregate at high concentration (and thus are potentially toxic); almost nothing is known about their behavior in different transgenics. In contrast, R10-3 is a close derivative of GFP, and thus it probably inherits GFP advantages: very low toxicity, excellent performance in protein fusions, very well documented behavior in different expression systems. So, we believe that R10-3 could find a niche among other fluorescent proteins used in practice. Also, we hope that R10-3 represents only a first step forward and that future work will generate a purely red GFP mutant.
In conclusion, the present work describes for the first time the mutants of A. victoria GFP that form the red-emitting spectral species with excitation and emission maxima at 555 and 585 nm, respectively. These mutants extend our knowledge about posttranslational chemistry of the widely used GFP protein. In particular, combination of Ser65 and Asn68 was found to play a crucial role in the formation of the red chromophore within this protein; a novel mechanism of the red chromophore formation based on the Asn68-induced dehydration of Ser65 has been proposed. Novel GFP mutants could find their use in multicolor flow cytometry.
ACKNOWLEDGMENT
We thank J. Zhang from the AECOM flow cytometry facility for excellent assistance with bacteria sorting.
Footnotes
This work was supported by National Institutes of Health Grants GM070358 and GM073913, Howard Hughes Medical Institute Grant HHMI 55005618, European Commission FP-6 Integrated Project LSHG-CT-2003-503259, and NATO Collaborative Linkage Grant CBP.NR.NRCLG 981752. K.A.L. is supported by the Russian Science Support Foundation.
Abbreviations: CP, chromoprotein; DsRed, red fluorescent protein from Discosoma; EGFP, enhanced green fluorescent proteins; FP, fluorescent protein; GFP, green fluorescent protein from Aequorea victoria.
REFERENCES
- 1.Lippincott-Schwartz J, Patterson GH. Development and use of fluorescent protein markers in living cells. Science. 2003;300:87–91. doi: 10.1126/science.1082520. [DOI] [PubMed] [Google Scholar]
- 2.Verkhusha VV, Lukyanov KA. The molecular properties and applications of Anthozoa fluorescent proteins and chromoproteins. Nat. Biotechnol. 2004;22:289–296. doi: 10.1038/nbt943. [DOI] [PubMed] [Google Scholar]
- 3.Chudakov DM, Lukyanov S, Lukyanov KA. Fluorescent proteins as a toolkit for in vivo imaging. Trends Biotechnol. 2005;23:605–613. doi: 10.1016/j.tibtech.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 4.Johnson FH, Shimomura O, Saiga Y, Gershman LC, Reynolds GT, Waters JR. Quantum efficiency of Cypridina luminescence, with a note on that of Aequorea. J. Cell. Comp. Physiol. 1962;60:85–104. [Google Scholar]
- 5.Chalfie M. Green fluorescent protein. Photochem. Photo-biol. 1995;62:651–656. doi: 10.1111/j.1751-1097.1995.tb08712.x. [DOI] [PubMed] [Google Scholar]
- 6.Prasher DC, Eckenrode VK, Ward WW, Prendergast FG, Cormier MJ. Primary structure of the Aequorea victoria green-fluorescent protein. Gene. 1992;111:229–233. doi: 10.1016/0378-1119(92)90691-h. [DOI] [PubMed] [Google Scholar]
- 7.Chalfie M, Tu Y, Euskirchen G, Ward WW, Prasher DC. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 8.Cody CW, Prasher DC, Westler WM, Prendergast FG, Ward WW. Chemical structure of the hexapeptide chromophore of the Aequorea green fluorescent protein. Biochemistry. 1993;32:1212–1218. doi: 10.1021/bi00056a003. [DOI] [PubMed] [Google Scholar]
- 9.Heim R, Prasher DC, Tsien RY. Wavelength mutations and posttranslational autoxidation of green fluorescent protein. Proc. Natl. Acad. Sci. U.S.A. 1994;91:12501–12504. doi: 10.1073/pnas.91.26.12501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brejc K, Sixma TK, Kitts PA, Kain SR, Tsien RY, Ormo M, Remington SJ. Structural basis for dual excitation and photoisomerization of the Aequorea victoria green fluorescent protein. Proc. Natl. Acad. Sci. U.S.A. 1997;94:2306–2311. doi: 10.1073/pnas.94.6.2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matz MV, Fradkov AF, Labas YA, Savitsky AP, Zaraisky AG, Markelov ML, Lukyanov SA. Fluorescent proteins from nonbioluminescent Anthozoa species. Nat. Biotechnol. 1999;17:969–973. doi: 10.1038/13657. [DOI] [PubMed] [Google Scholar]
- 12.Ando R, Hama H, Yamamoto-Hino M, Mizuno H, Miyawaki A. An optical marker based on the UV-induced green-to-red photoconversion of a fluorescent protein. Proc. Natl. Acad. Sci. U.S.A. 2002;99:12651–12656. doi: 10.1073/pnas.202320599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wiedenmann J, Ivanchenko S, Oswald F, Schmitt F, Rocker C, Salih A, Spindler KD, Nienhaus GU. EosFP, a fluorescent marker protein with UV-inducible green-to-red fluorescence conversion. Proc. Natl. Acad. Sci. U.S.A. 2004;101:15905–15910. doi: 10.1073/pnas.0403668101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shagin DA, Barsova EV, Yanushevich YG, Fradkov AF, Lukyanov KA, Labas YA, Semenova TN, Ugalde JA, Meyers A, Nunez JM, Widder EA, Lukyanov SA, Matz MV. GFP-like proteins as ubiquitous metazoan superfamily: evolution of functional features and structural complexity. Mol. Biol. Evol. 2004;21:841–850. doi: 10.1093/molbev/msh079. [DOI] [PubMed] [Google Scholar]
- 15.Heim R, Tsien RY. Engineering green fluorescent protein for improved brightness, longer wavelengths and fluorescence resonance energy transfer. Curr. Biol. 1996;6:178–182. doi: 10.1016/s0960-9822(02)00450-5. [DOI] [PubMed] [Google Scholar]
- 16.Heim R, Cubitt AB, Tsien RY. Improved green fluorescence. Nature. 1995;373:663–664. doi: 10.1038/373663b0. [DOI] [PubMed] [Google Scholar]
- 17.Ormo M, Cubitt AB, Kallio K, Gross LA, Tsien RY, Remington SJ. Crystal structure of the Aequorea victoria green fluorescent protein. Science. 1996;273:1392–1395. doi: 10.1126/science.273.5280.1392. [DOI] [PubMed] [Google Scholar]
- 18.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 19.Gross LA, Baird GS, Hoffman RC, Baldridge KK, Tsien RY. The structure of the chromophore within DsRed, a red fluorescent protein from coral. Proc. Natl. Acad. Sci. U.S.A. 2000;97:11990–11995. doi: 10.1073/pnas.97.22.11990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yarbrough D, Wachter RM, Kallio K, Matz MV, Remington SJ. Refined crystal structure of DsRed, a red fluorescent protein from coral, at 2.0 Å resolution. Proc. Natl. Acad Sci. U.S.A. 2001;98:462–467. doi: 10.1073/pnas.98.2.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gurskaya NG, Savitsky AP, Yanushevich YG, Lukyanov SA, Lukyanov KA. Color transitions in coral’s fluorescent proteins by site-directed mutagenesis. BMC Biochem. 2001;2(6):1–10. doi: 10.1186/1471-2091-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Quillin ML, Anstrom DM, Shu X, O’Leary S, Kallio K, Chudakov DM, Remington SJ. Kindling fluorescent protein from Anemonia sulcata: dark-state structure at 1.38 Å resolution. Biochemistry. 2005;44:5774–5787. doi: 10.1021/bi047644u. [DOI] [PubMed] [Google Scholar]
- 23.Tretyakova YA, Pakhomov AA, Martynov VI. Chromophore structure of the kindling fluorescent protein asFP595 from Anemonia sulcata. J. Am. Chem. Soc. 2007;129:7748–7749. doi: 10.1021/ja071992c. [DOI] [PubMed] [Google Scholar]
- 24.Remington SJ, Wachter RM, Yarbrough DK, Branchaud B, Anderson DC, Kallio K, Lukyanov KA. zFP538, a yellow-fluorescent protein from Zoanthus, contains a novel three-ring chromophore. Biochemistry. 2005;44:202–212. doi: 10.1021/bi048383r. [DOI] [PubMed] [Google Scholar]
- 25.Shu X, Shaner NC, Yarbrough CA, Tsien RY, Remington SJ. Novel chromophores and buried charges control color in mFruits. Biochemistry. 2006;45:9639–9647. doi: 10.1021/bi060773l. [DOI] [PubMed] [Google Scholar]
- 26.Remington SJ. Fluorescent proteins: maturation, photochemistry and photophysics. Curr. Opin. Struct. Biol. 2006;16:714–721. doi: 10.1016/j.sbi.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 27.Pletneva N, Pletnev S, Tikhonova T, Popov V, Martynov V, Pletnev V. Structure of a red fluorescent protein from Zoanthus, zRFP574, reveals a novel chromophore. Acta Crystallogr. 2006;D62:527–532. doi: 10.1107/S0907444906007852. [DOI] [PubMed] [Google Scholar]
- 28.Mizuno H, Mal TK, Tong KI, Ando R, Furuta T, Ikura M, Miyawaki A. Photo-induced peptide cleavage in the green-to-red conversion of a fluorescent protein. Mol. Cell. 2003;12:1051–1058. doi: 10.1016/s1097-2765(03)00393-9. [DOI] [PubMed] [Google Scholar]
- 29.Nienhaus K, Nienhaus GU, Wiedenmann J, Nar H. Structural basis for photo-induced protein cleavage and green-to-red conversion of fluorescent protein EosFP. Proc. Natl. Acad. Sci. U.S.A. 2005;102:9156–9159. doi: 10.1073/pnas.0501874102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hayashi I, Mizuno H, Tong KI, Furuta T, Tanaka F, Yoshimura M, Miyawaki A, Ikura M. Crystallographic evidence for water-assisted photo-induced peptide cleavage in the stony coral fluorescent protein Kaede. J. Mol. Biol. 2007;372:918–926. doi: 10.1016/j.jmb.2007.06.037. [DOI] [PubMed] [Google Scholar]
- 31.Verkhusha VV, Chudakov DM, Gurskaya NG, Lukyanov S, Lukyanov KA. Common pathway for the red chromophore formation in fluorescent proteins and chromoproteins. Chem. Biol. 2004;11:845–854. doi: 10.1016/j.chembiol.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 32.Gurskaya NG, Verkhusha VV, Shcheglov AS, Staroverov DB, Chepurnykh TV, Fradkov AF, Lukyanov S, Lukyanov KA. Engineering of a monomeric green-to-red photoactivatable fluorescent protein induced by blue light. Nat. Biotechnol. 2006;24:461–465. doi: 10.1038/nbt1191. [DOI] [PubMed] [Google Scholar]
- 33.Pakhomov AA, Martynov VI. Chromophore aspartate oxidation-decarboxylation in the green-to-red conversion of a fluorescent protein from Zoanthus sp. 2. Biochemistry. 2007;46:11528–11535. doi: 10.1021/bi700721x. [DOI] [PubMed] [Google Scholar]
- 34.Pletneva N, Pletnev V, Tikhonova T, Pakhomov AA, Popov V, Martynov VI, Wlodawer A, Dauter Z, Pletnev S. Refined crystal structures of red and green fluorescent proteins from the button polyp Zoanthus. Acta Crystallogr. 2007;D63:1082–1093. doi: 10.1107/S0907444907042461. [DOI] [PubMed] [Google Scholar]
- 35.Merzlyak EM, Goedhart J, Shcherbo D, Bulina ME, Shcheglov AS, Fradkov AF, Gaintzeva A, Lukyanov KA, Lukyanov S, Gadella TWJ, Chudakov DM. Bright monomeric red fluorescent protein with an extended fluorescence lifetime. Nat. Methods. 2007;4:555–557. doi: 10.1038/nmeth1062. [DOI] [PubMed] [Google Scholar]
- 36.Elowitz MB, Surette MG, Wolf PE, Stock J, Leibler S. Photoactivation turns green fluorescent protein red. Curr. Biol. 1997;7:809–812. doi: 10.1016/s0960-9822(06)00342-3. [DOI] [PubMed] [Google Scholar]
- 37.Sawin KE, Nurse P. Photoactivation of green fluorescent protein. Curr. Biol. 1997;7:R606–R607. doi: 10.1016/s0960-9822(06)00313-7. [DOI] [PubMed] [Google Scholar]
- 38.Takahashi E, Takano T, Nomura Y, Okano S, Nakajima O, Sato M. In vivo oxygen imaging using green fluorescent protein. Am. J. Physiol. Cell Physiol. 2006;291:C781–C787. doi: 10.1152/ajpcell.00067.2006. [DOI] [PubMed] [Google Scholar]
- 39.Crameri A, Whitehorn EA, Tate E, Stemmer WPC. Improved green fluorescent protein by molecular evolution using DNA shuffling. Nat. Biotechnol. 1996;14:315–319. doi: 10.1038/nbt0396-315. [DOI] [PubMed] [Google Scholar]
- 40.Nagai T, Ibata K, Park ES, Kubota M, Mikoshiba K, Miyawaki A. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 2002;20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- 41.Terskikh AV, Fradkov AF, Zaraisky AG, Kajava AV, Angres B. Analysis of DsRed mutants. Space around the fluorophore accelerates fluorescence development. J. Biol. Chem. 2002;277:7633–7636. doi: 10.1074/jbc.C100694200. [DOI] [PubMed] [Google Scholar]
- 42.Schwede TF, Retey J, Schulz GE. Crystal structure of histidine ammonia-lyase revealing a novel polypeptide modification as the catalytic electrophile. Biochemistry. 1999;38:5355–5361. doi: 10.1021/bi982929q. [DOI] [PubMed] [Google Scholar]
- 43.Baedeker M, Schulz GE. Autocatalytic peptide cyclization during chain folding of histidine ammonia-lyase. Structure. 2002;10:61–67. doi: 10.1016/s0969-2126(01)00692-x. [DOI] [PubMed] [Google Scholar]
- 44.Barondeau DP, Kassmann CJ, Tainer JA, Getzoff ED. Understanding GFP chromophore biosynthesis: controlling backbone cyclization and modifying post-translational chemistry. Biochemistry. 2005;44:1960–1970. doi: 10.1021/bi0479205. [DOI] [PubMed] [Google Scholar]
- 45.Garcia-Parajo MF, Koopman M, van Dijk EM, Subramaniam V, van Hulst NF. The nature of fluorescence emission in the red fluorescent protein DsRed, revealed by single-molecule detection. Proc. Natl. Acad. Sci. U.S.A. 2001;98:14392–14397. doi: 10.1073/pnas.251525598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bevis BJ, Glick BS. Rapidly maturing variants of the Discosoma red fluorescent protein (DsRed) Nat. Biotechnol. 2002;20:83–87. doi: 10.1038/nbt0102-83. [DOI] [PubMed] [Google Scholar]