Abstract
HPV infection of the genital tract is very common and normally follows a benign clinical course, however in an unfortunate minority of infected individuals it can cause disease which sometimes leads to cancer. It is accepted that HPV DNA testing has a role in the management of cervical disease both in a pre and post vaccination era, however to improve the specificity of this approach, there is a requirement to develop and validate tools/assays that can identify women at risk for progressive disease. There is evidence to suggest that detection of viral gene expression both directly and indirectly may constitute a more specific approach for delineating clinically significant infection compared with HPV DNA based assays. HPV oncogene expression and evidence of its deregulation can be monitored through direct detection of viral mRNA transcripts or through detection of the cellular protein p16. For both approaches, commercial assays have been introduced and numerous studies have been conducted. The present article describes the scientific theory underpinning these approaches, their amenability to routine-diagnostic specimens/settings and the clinical data that has been garnered through their application thus far. Currently, there is promising data indicating that HPV mRNA and p16 might play an important role in future cervical cancer screening scenarios. Still, large randomized studies are necessary to confirm the preliminary data
Keywords: cervical cancer, HPV, E6, E7, mRNA, p16, biomarker, screening
Methods
PubMed and OVID were interrogated with search terms: “HPV RNA”, “HPV mRNA”, “HPV transcript; - detection, testing and methods”, “p16” AND “cervical cancer”; “p16” AND “CIN”; “p16” AND “histology”; “p16” AND “cytology”; “p16” and “screening”.
Background
Human Papillomavirus (HPV) DNA testing is already being performed in the US for the management of cervical disease. Moreover, the data generated from the large randomised controlled trials of HPV testing in screening settings –either recently completed or ongoing in Europe will inform its use in the appropriate countries therein (1–3). While it is accepted that HPV DNA testing is more sensitive for the detection of cervical disease compared with cytology, its specificity is less so, especially in women under 30 years of age; largely due to the prevalence of transient, clinically benign infection (4). As a consequence, appropriate contextualisation of HPV DNA testing is paramount e.g. age restricting testing and interpreting the result alongside concurrent pathological diagnosis is required to avoid unnecessary testing for what is a prevalent viral infection (5).
HPV DNA based tests
The majority of HPV detection tests and certainly the clinically validated ones have incorporated (largely), aggregate detection of a group of HPV types and can be termed presence/absence tests. These have relied on DNA based detection of a portion of the gene that encodes the major structural protein of HPV: L1. Presence/absence tests have the benefit of being amenable to high-throughput and being easy to interpret and indeed have generated the majority of clinical data which have informed the use of HPV testing in clinical contexts (6).
However, such tests cannot delineate between transient and potentially transforming infection. To improve the specificity of an HPV test would clearly be of clinical value and suggested candidates, to this end, have included measurement of viral load and the use of HPV type-specific detection via genotyping (7;8). With respect to the latter there is good evidence to suggest that HPV 16 confers a significantly higher absolute risk for the development of significant lesions compared to other high risk types (9). One of the issues of type specific testing however is the complexity of the data/result it may generate. Multiple infections are common and also the significance of one non HPV-16 high-risk type over another is poorly understood (10).
Viral load has been shown in some cross-sectional studies to be a useful indicator of clinically significant infection and lesion severity although there is evidence to suggest that 1) its usefulness may alter according to infecting HPV type and that 2) the wide dynamic range of what constitutes a “significant” viral load currently precludes its use, practically, as a prognostic indicator, especially for types other than HPV 16 (11;12). Also, accurate measurement requires standardisation of sample input to the assay – a significant technical challenge. Testing algorithms are being designed to ameliorate the complexity of testing and result dissemination using typing and load. However the onus is still on the HPV community to strive for more specific, (bio) markers of significant infection, the detection and interpretation of which can be performed in as straight-forward a manner as possible.
Alternatives to HPV DNA testing
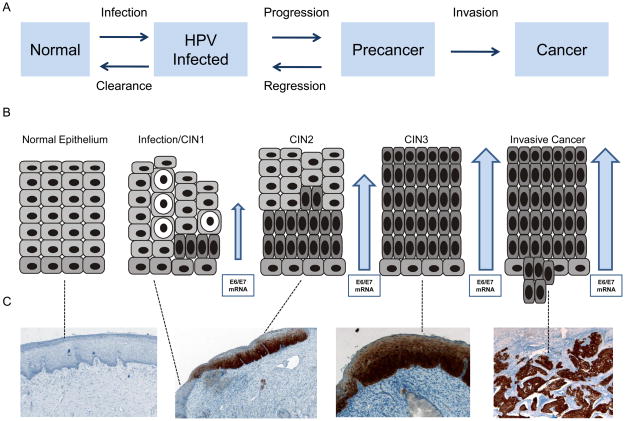
At the 2006 meeting of the European Research Organization on Genital Infection and Neoplasia (EUROGIN) a working group was convened with the remit to compile a consensus report on how to apply (HPV associated) molecular (bio)markers in practice. The authors concluded that four main lines of research were being followed to identify appropriate markers, namely: 1) detection/measurement of E6 and E7 HPV mRNA transcripts, 2) alterations of the methylation pattern of several genes, 3) alterations of viral (e.g. viral integration) and host (e.g. chromosomal gains and losses) genomes and 4) detection of cellular proteins that are over-expressed by HPV infected cells, eg the p16ink4A gene product (13). While biomarkers within (above) categories 2 and 3 show demonstrable proof of concepts, experimental work has occurred in academic laboratories on small numbers of samples. Few studies have been reported where they have been applied to sizeable clinical sample sets and fewer still where their performance has been compared with validated HPV DNA tests. The focus of this article is to describe the state of the art of two biomarkers, for which standardized, commercial assays are available and which have been the most interrogated in clinical studies, thus far namely, detection of oncogenic mRNA transcripts and the detection of a protein - p16 - that is produced in response to their deregulated expression (Figure 1). Other biomarkers are currently emerging, among them a standardized commercial assay for histology and cytology applications, ProExC, which detects two replication associated proteins, MCM2 and Top2A (14;15). However, limited data precludes thorough assessment of these markers.
Figure 1.
HPV oncogene expression
It is widely accepted that for HPV to cause cancer, persistent infection and a cellular environment which allows for high level expression of viral oncogenes E6 and E7 (initially in the basal cell layer and then throughout the epithelium) is necessary. The E6 and E7 proteins are necessary for the virus to replicate itself and are expressed during the productive “normal” life cycle, where their regulation is tightly controlled. When this regulation is disrupted and E6 and E7 are over expressed they can abrogate normal tumour suppressive function and cell cycling and this process is considered necessary for incurring a malignant phenotype. This de-regulation is evidenced particularly by the presence of E7 throughout the epithelium and in the surface layers (16). It is therefore logical to hypothesise that detection of E6/E7 mRNA may identify more clinically significant infection than a DNA approach where a structural gene is targeted, particularly if the surface layers of the epithelium are sampled for analysis as is the case when a cervical cytology specimen is taken
Technical issues relating to RNA detection
RNA, as a single stranded moiety, is less stable than DNA Thus, although conceptually E6 and E7 mRNA may be suitable targets, is their detection practically feasible in routinely collected clinical samples? This is an especially important point considering that certain types of liquid based cytology (LBC) medium appear to preserve HPV DNA in exfoliated cervical cells very well (17).
Accumulated, indirect evidence suggests that E6 and E7 mRNA are detectable in stored clinical samples. In a key paper by Stoler et al 3H-labeled riboprobes specific to individual mRNA families were generated, in vitro, to examine serial sections of archived biopsy specimens (18). In the early 90s Rose et al successfully amplified HPV 16 E6/E7 transcripts from paraffin embedded tissue (19).
More direct evidence on the stability of HPV mRNA came from the work of Habis et al who showed that a widely used cytological collection fluid - PreservCyt, (Cytyc Corp., Boston, MA) produced a high yield of RNA and fared better when compared to a high-salt solution designed for RNA preservation (20). Tarkowski et al performed a prospective study by seeding PreservCyt media with an HPV-16 containing cell line and found that HPV E6/E7 transcripts were still detectable after one year of storage, with 1HPV +ve cell in 30,000 being consistently detected (21). A later, prospective study which involved the longitudinal detection of E6/E7 transcripts in cytology samples (PreservCyt) collected from women attending colposcopy showed no loss of HPV transcript detection over a 2-week time course (22).
These data are encouraging, however we cannot conclude that all alcohol based cytology media will constitute suitable buffers for HPV RNA preservation. In a recent article by Powell et al, yields of RNA from an HPV cell line spiked into SurePath medium diminished substantially over a 600 h time course (23). Poor recovery of RNA from SurePath has been documented elsewhere also (24). Clearly technical validation of any viral assay is necessary with respect to different specimen types; nevertheless the evidence would indicate that HPV mRNA appears to be detectable and stable in certain routinely taken cervical specimens.
HPV E6 and E7 mRNA detection methods - in house
The in-house RNA detection methods described to date have tended to be based on reverse transcription PCR and the minority have incorporated a quantitative element. Table 1 depicts the remit of a selection of these assays (18;25–31) and real-time PCR approaches continue to be employed for detection of HPV RNA in ano-genital region and in other sites (32;33).
Table 1.
Detection rate of E6/E7 transcipts in cervical cancers and CIN2/3
| Author | Cancer Cases assessed (n) | E6/E7 transcript positive cancers (%) | CIN2/3 cases assessed (n) | E6/E7 transcript positive CIN2/3 (%) |
|---|---|---|---|---|
| Rose 1995 {Rose, 1995 1673/id} | 28 | 28 (100) | N/A | N/A |
| Nakagawa 1995 {Nakagawa, 2000 1674/id} | 31 | 31 (100) | 19 | 19 (100) |
| Kraus 2006 {Kraus, 2006 1675/id} | 204 | 199 (92) | N/A | N/A |
| Lie 2005 {Lie, 2005 1676/id} | 20 | 20 (100) | 291 | 225 (77) |
| Molden 2005 {Molden, 2005 1680/id} | N/A | N/A | 14 | 13 (93) |
| Sotlar 2004 {Sotlar, 2004 1677/id} | N/A | N/A | 109 | 95 (87) |
| Total | 283 | 98% | 433 | 81% |
Published in-house assays have differed from each other with respect to the nature of the transcripts which have been targeted. E6 and E7 proteins arise from polycistronic transcripts. The full length E6/E7 transcript is required to encode the complete E6 protein while, through splicing in the E6 ORF, a shorter E6/E7 transcript, often referred to as E6* encodes the E7 protein. HPV 16 has two splice acceptor sites in the E6 ORF and can generate two E7 transcripts E6I* and E6II*. Most of the other HR-HPV types have just the one E7 transcript (34).
The point in relation to diagnostic HPV mRNA assays is that their performance may be related the nature of the transcript they detect e.g. in a study by Sotlar et al, the authors found that spliced transcripts were detected less frequently than full-length transcripts in pathological samples ranging from no CIN to CIN3(35).
In addition, few of the published in-house HPV mRNA assays described have been internally controlled. The need for internal control is justified so inhibitory and/or inadequate specimens are identified. Moreover, quantitative measurement of expression requires normalisation through measurement of a reference gene with a predictable and stable expression in order to control for the input into the assay, again it is expedient to evaluate these with respect to specimen type.
HPV E6 and E7 mRNA detection methods - commercial
Two commercial HPV mRNA assays are available – PreTect HPV Proofer (Norchip) and the APTIMA ® HPV Assay (GenProbe). The former is a qualitative assay based on a NASBA RNA amplification (of full length E6/E7 transcript) prior to detection of amplified RNA with molecular beacons corresponding to HPV types 16,18,31,33 and 45. The HPV Proofer is CE marked, resolves type, is designed primarily to be compatible with LBC samples and is internally controlled. The latter assay was launched in Europe in May 2008 as a CE marked test, is also qualitative and relies on aggregate detection of 14 HR types. . It too is compatible with LBC samples and is internally controlled. The chemistry is based on transcription mediated amplification of full length E6/E7 transcript pre-empted by target capture.
Overview of E6 and E7 mRNA detection in clinical studies
Few clinical studies in which HPV RNA testing has been performed have been published. Undoubtedly, this will change as more commercial assays become available, recently developed assays (ie the APTIMA test) are evaluated, and detection of HPV mRNA expression is not seen exclusively as a means to facilitate basic research. Currently, however, several factors hamper establishing consensus findings across published clinical studies - they include: different types of clinical specimens tested, lack of demographic data, different type detection remits of mRNA assays and comparator DNA assays (if used) and the different species of mRNA transcript targeted (i.e. full length vs spliced etc). Moreover, the few studies where DNA and RNA testing has been performed on the same sample set, have (largely) not constituted direct comparisons of HPV DNA and RNA for the detection of clinical endpoints - rather, they have been correlative - i.e. samples/cases are already selected due to their HPV DNA positivity so that the corresponding transcript can be looked for.
Cross Sectional Clinical Studies
Few would doubt the sensitivity of HPV DNA testing for the detection of infection associated with significant lesions and cervical cancer, thus an important initial issue to address is how does RNA testing compare? Evidence collated thus far would indicate that e6/e7 detection in cancers is high (%) as depicted in Table 2.
Table 2.
p16 staining in immunohistochemistry
| Author | Non-dysplastic* | CIN1 | CIN2 | CIN3 | Invasive CA |
|---|---|---|---|---|---|
| Sano 1998 | 0/15 (0%) | 8/15 (53%) | 16/17 (94%) | 27/27 (100%) | 38/39 (97%) |
| Klaes 2001 | 1/111 (1%) | 29/47 (61%) | 32/32 (100%) | 60/60 (100%) | 58/60 (97%) |
| Klaes 2002 | 7/58 (12%) | 15/17 (88%) | 10/10 (100%) | 43/43 (100%) | 46/46 (100%) |
| Agoff 2003 | 30/247 (12%) | 43/76 (57%) | 60/80 (75%) | 103/113 (91%) | 47/53 (89%) |
| Wang 2004§ | 12/179 (7%) | 27/75 (36%) | 12/19 (63%) | 19/19 (100%) | na |
| Hu 2005 #& | na | 20/45 (44%) | 43/46 (93%) | 51/51 (100%) | na |
| Benevolo 2006 # | 0/17 (0%) | 17/54 (31%) | 9/10 (90%) | 11/11 (100%) | 8/8 (100%) |
| Ishikawa 2006 # | 0/7 (0%) | 13/53 (25%) | 32/40 (80%) | 45/48 (94%) | 16/16 (100%) |
| Focchi 2007 i# | 0/114 (0%) | 80/88 (91%) | 33/33 (100%) | 32/32 (100%) | 44/47 (94%) |
| Total | 50/748 (7%) | 252/470 (54%) | 247/287 (86%) | 391/404 (96%) | 257/269 (96%) |
includes normal, reactive, inflammation, hyperplasia, atypical, equivocal
population based study
criteria modified from Klaes et al.
adolescents
With respect to high-grade lesions, frequently CIN 3 and CIN 2 or worse are used as a surrogate marker for cancer and as an end point for invasive clinical intervention respectively. Consequently, sensitivity and specificity for the detection of such are used to assess the performance of HPV detection tools, data relating to E6 E7 detection in CIN2+ can be seen in Table 2. More specifically, Lie et al performed an evaluation of samples collected from 383 women attending colposcopy. HPV DNA and RNA testing was performed by the hybrid capture 2 test (13 type range) and the Proofer respectively. Histology ranged from no disease (61 cases) to invasive carcinoma (20 cases) (36). Overall, agreement between DNA and mRNA positivity was reached in 72%. HPV DNA was detected in a significantly higher number of benign and low-grade lesions lending a higher specificity to the mRNA test. However 95% and 77% of CIN2 + lesions were HPV DNA and RNA positive respectively - a significant difference. It is difficult to attribute whether this difference is solely as a result of mRNA expression being absent/undetectable or, whether it was due to the larger type detection range of the hc2. The study of Nakagawa et al allies with the latter explanation as they found that E6/E7 expression in 19/19 high grade lesions when a broad spectrum, consensus approach for E6/E7 detection was employed (26). Lie et al also suggested that the RNA negative, DNA positive high-grade cases would be those more likely to regress without intervention but prospective data would need to be captured to show this conclusively.
Sotlar et al tested 779 HPV DNA positive cervical cytological samples and looked for E6/E7 oncogene (spliced and unspliced) transcripts via a nested type specific RT-PCR for 14 high-risk HPV types. Transcript detection was positively correlated with severity of the abnormality, with 18% and 84% of cases of no CIN and CIN 3 testing mRNA positive respectively. It could be argued that these figures may not represent prevalence of mRNA in cervical samples wholly accurately as HPV DNA positive samples had been specifically selected pre-emptively. However, the design of this study does allow for the correlation of DNA and RNA testing approaches that have the same detection range - and the two correlated closely at the higher disease grades, with 103 and 95 cases testing positive for HR-HPV DNA (via multiplex PCR) and mRNA (spliced and/or unspliced transcript) respectively out of a total of 109 cases with CIN2 or worse. The correlation between RNA and DNA was less so in normal and CIN I lesions with 27.7 % and 82 % of such testing DNA positive respectively compared with 9.1% and 62.5 % testing positive for mRNA. Again these data show that the specificity of an mRNA approach for the detection of significant disease may be higher than a DNA approach (30).
Earlier, we discussed the relevance of a quantitative element to an E6/E7 test, in terms of clinical studies that have involved their application. Wang Johanning et al found that HPV 16 E6 and E7 mRNA levels positively correlated with increasing severity of cytological diagnosis using a type specific RT-PCR (31). However Scheurer et al, by employing a RT-PCR for HPV 16 and HPV 18 E7 transcripts, found such a correlation existed with HPV 18 only (28). The difference in the findings of these two studies could be attributable to the length of storage of the RNA extract and sampling method. In the former study, the residual volume of cytological samples was used directly for testing whereas the latter took separate samples for cytological analysis, HPV DNA testing and HPV RNA testing (with the RNA sample being taken last). Molden et al performed the largest, clinical, cross sectional study where HPV mRNA and DNA detection was performed, evaluated and compared (37). A total of 4,136 women over 30 years of age attending a gynaecology clinic were tested using the Proofer for HPV mRNA and type specific and consensus (GP5+6+) PCR for HPV DNA. When HPV positivity was related to cytological status, 2.4% and 9.3% of cytologically normal women were RNA and DNA positive respectively. When the authors confined the RNA vs. DNA comparison to types within the range of the Proofer by using type specific DNA PCR, overall, 3.1% and 4.4% of samples tested positive for HPV RNA and DNA respectively compared to the 10.4% that tested positive by consensus PCR. The authors found that HPV DNA (via GP5+) was significantly higher than detection of RNA in all grades of cytology except for HSIL and of the HSIL cases that were confirmed to be CIN 2 + through histology, 13/14 tested mRNA positive compared to 14/14 that were DNA positive. These results could imply that the mRNA approach was specific for the detection “significant” disease when compared to a DNA approach.
In a recent study by Castle et al the authors evaluated the prototype Aptima assay with a DNA line blot assay using 531 cytology samples (with histology confirmation) (38). Two cut-off values for positivity were assessed and related to clinical findings. Significantly fewer women tested positive using the RNA compared with the DNA approach (at both cut offs) for <CIN1 and at the higher cut off for CIN 1. At the lower cut-off the same number of CIN2+ cases (95/103) were detected by the RNA approach as the DNA approach and at the higher cutoff, 95/103 and 93/103 cases were detected by DNA and RNA detection respectively. These data led to the authors preliminary optimism about the improved specificity of the RNA approach with the caveat that more longitudinal clinical data were needed.
Prospective Clinical Studies
Few studies have investigated the prospective sensitivity and specificity of diagnostic mRNA detection. One published study examined a prospective cohort of HPV DNA positive women with normal cytology. HPV mRNA detection was performed on the same specimen that tested DNA positive and follow-up HPV DNA and RNA testing was performed 2–3 years post the normal diagnosis (39). The main finding of this study was that women who were HPV mRNA positive at baseline were significantly more likely to have a persistent infection than women who were HPV DNA positive only. Another study looked to assess the predictive value of HPV mRNA and DNA detection for the detection of CIN 2 or worse in 77 women with cytological low-grade disease, at ~ 2 years. RNA and DNA detection was performed by the Proofer and consensus (GP5+6+) PCR respectively. The main finding was that HPV mRNA detection was more specific for the detection of CIN 2 at 2 years than the DNA approach (84.9% compared to 50%) and that the two approaches were equally sensitive in detecting the 7/77 women who did develop a minimum of CIN2 (40). Lastly, an RNA assay has been used to test samples collected as part of the Dutch randomised controlled trial POBASCAM - these data are awaited and will be welcome.
p16 expression in HPV-related disease
As discussed, alterations in the viral gene expression pattern mark the progression of a productive to a transforming infection, the latter conferring a higher risk for cervical precancer and cancer. p16 is a cellular correlate of the increased expression of oncogenic E6/E7 mRNA: The main actions of the HPV oncogenes are the degradation of p53 by E6 and thereby the abrogation of apoptosis as well as the release of E2F from pRb that leads to continuous activation of the cell cycle (41;42). Physiologically, E2F activation is mediated by phosphorylation of the Rb protein. This pathway is strictly regulated by a set of cyclin dependent kinase inhibitors, among them p16, that block enzymes phosphorylating pRB (cyclin dependent kinases). In cells with transforming HPV infections, the regulation of the Rb-E2F pathway is disturbed by E7 and the activation of p16 has no downstream effect (43). As a result, p16 is strongly overexpressed and accumulates in the cells (44). p16 overexpression has been demonstrated in the vast majority of cervical precancers and cancers while in normal tissue, p16 expression is found only rarely (45).
Several properties of p16 make this protein a promising biomarker for HPV-related cancers: The expression is directly linked to the HPV oncogene action, since continuous expression of E7 is necessary to maintain a malignant phenotype in HPV-associated cancer (46). The expression of p16 seems to be independent of the HPV type causing the oncogenic infection, obviating the need to detect different HPV types in DNA and RNA assays. Also, in contrast to many classic tumor markers such as ki67 or MYC, p16 is not associated with proliferation, but rather with senescence and cell cycle arrest (47) and is not found expressed in normal basal cells or in other cells with proliferative capacity.
Role of p16 in the histological diagnosis of neoplasia
Methodology
The majority of the p16 histology and cytology studies discussed here are based on CE-labeled kits for p16 immunostaining (CINtec, mtm Laboratories, Heidelberg, Germany), although studies where in-house protocols were used have also been described. In normal cervical histology specimens, barely any p16 expression is observed. Occasionally, metaplastic and endocervical cells may display p16 staining. This is contrasted by strong expression of the protein in cervical precancer and cancer (45). The typical staining pattern of HPV-related transformation originates in the basal epithelial cells and extends to the upper layers. Three categories have been used to describe p16 staining: no staining, focal staining of single metaplastic or endocervical cells (independent of HPV), and diffuse staining indicative of a HPV associated lesion. Specifically, focal staining is defined as non-continuous staining of isolated cells or small cell clusters, usually not located in the basal and parabasal layers.
Diffuse staining is defined as a continuous staining of cells in the basal and parabasal layers (with or without staining of superficial squamous cell layers). The focal staining pattern is considered negative for HPV associated disease, resulting in a dichotomous evaluation system of positive (diffuse) and negative (negative and focal) staining (48).
Improvement of interobserver variation in cervical histology
An initial study on interobserver variation of cervical histology showed a higher consistency in the assessment of p16 stained specimens compared to conventional H&E slides (48). In a recent study, 247 punch biopsies and 249 cone biopsies were analyzed by 6 pathologists whose initial assessment was based on H&E slides only. Inclusion of a p16 stained slide in the evaluation after a washout phase led to a significant increase in inter-observer agreement for both punch and cone biopsies (from 0.49 to 0.63 and 0.63 to 0.70, respectively) (49).
Zhang et al. used p16 to refine the histological gold standard of a large cervical cancer screening study comparing cytology, HPV testing and VIA (50). Only CIN2/3 histology results that exhibited diffuse p16 staining were considered “real” cases. Using this refined case definition, the sensitivity of HPV DNA testing and cytology was found to be increased, while the sensitivity of VIA was decreased. The p16 negative CIN2/3 cases showed characteristics of immature squamous metaplasia accompanied by unusual cellular atypia, most probably leading to false positive histology results.
Cross sectional studies of p16 histology
Numerous studies have been conducted analyzing p16 as a biomarker for cervical precancer and cancer in immunohistochemistry, most of them based on convenience samples. A limitation in comparing the studies is related to the different evaluation/scoring strategies applied. The studies using the criteria described earlier (48) with a minimum sample size of 100 are summarized in Table 3, but also within this set, there is considerable heterogeneity in the populations analyzed and the slide evaluation process (44;45;48;51–56).
Table 3.
p16 staining in immunocytochemistry
| Author | Platform | Cutoff | NILM | ASCUS | LSIL | HSIL | SCC |
|---|---|---|---|---|---|---|---|
| Bibbo 2002 | Thinprep LBC | >10/slide | 14/19 (74%) | 25/26 (97%) | |||
| Saqi 2002 | Surepath LBC | >10/slide | 5/28 (18%) | 20/24 (74%) | 9/10 (90%) | 1/1 (100%) | |
| Pientong 2003 | Smear dissolved in PBS | >3/slide | 17/30 (57%) | 10/30 (33%) | 28/30 (98%) | 29/30 (97%) | |
| Yoshida 2004 | Destained Pap slides | atypical | 5/38 (13%) | 7/12 (58%) | 33/33 (100%) | 12/12 (100%) | |
| Pientong 2004 | Smear dissolved in PBS | >3/slide | 0/30 (0%) | 21/40 (53%) | 19/35 (54%) | 29/30 (97%) | 30/30 (100%) |
| Bose 2005 | Conventional smear | not specified | 3/23 (13%) | 10/34 (29%) | 7/34 (21%) | 13/16 (81%) | |
| Wentzensen 2005 | Cytoscreen LBC | Morphology based score | 1/108 (1%) | 5/52 (10%) | 49/50 (98%) | ||
| Ekalaksananan 2006 | Conventional smear | >10/slide | 11/148 (7%) | 12/19 (63%) | 3/5 (60%) | 12/12 (100%) | 2/2 (100%) |
| Wentzensen 2007 | Cytoscreen LBC | Morphology based score | 37/137 (27%) | 21/88 (24%) | |||
| Meyer 2007 | Thinprep LBC | atypical | 9/235 (4%) | 9/49 (18%) | 24/57 (42%) | 43/53 (81%) | |
| Total | 29/572 (5%) | 111/347 (32%) | 130/356 (37%) | 241/260 (93%) | 74/75 (99%) |
LBC= liquid based cytology, NILM=Negative for intraepithelial lesion or malignancy, ASCUS=Atypical squamous cells of undetermined significance, LSIL=Low grade squamous intraepithelial lesion, HSIL=High grade squamous intraepithelial lesion, SCC=Squamous cell carcinoma
Murphy and colleagues used a system to analyze p16 histology, assigning a score of 1 for <10% positive cells, a score of 2 for 10–50% positive cells and a score of 3 for > 50% positive cells (57–59). In a study of 149 cases with varying histological diagnosis, the authors found that p16 histology delineated dysplastic squamous and glandular cells with good sensitivity and specificity and that p16 out-performed the concomitantly analyzed proliferation markers MCM5 and CDC6 (59). However, since no qualitative distinction between focal and diffuse staining patterns was made, the authors found a higher percentage of positive CIN1 and non dysplastic cases..
Based on a summary of 9 studies including 2178 cases and applying the criteria from (48)(or slight variations thereof), 7% of the non-dysplastic cases, 54% of the CIN1 cases, 86% of the CIN2 cases and each 96% of the CIN3 and cancer cases showed diffuse p16 expression (Table 3).
Prospective studies of p16 histology
In an extension of the cross-sectional study by Wang et al., the prospective value of p16 IHC was determined for a subset of women for whom follow up data was available (56). In these women, p16 staining showed a positive predictive value of 44% and a negative predictive value of 85% for CIN progression or HPV persistence. Negri et al. performed a study analyzing CIN1 lesions categorised into three groups according to follow-up data (60), (1) cases with spontaneous regression, (2) cases with progression to CIN3, (3) cases selected irrespective of progression status. All biopsies were stained with p16 and evaluated according to Klaes et al. (48). In the regressor group, 44% were diffusely p16 positive, while 74% of the progressing cases showed diffuse p16 expression. In the unselected group, 58% showed diffuse p16 staining, a percentage in keeping with most studies where p16 expression has been assessed in CIN1.
Hariri et al. collected 100 CIN1, 50 HGCIN and 50 non-dysplastic lesions and performed a 5–7 year follow up (61). All p16 negative CIN1 cases showed regression during follow-up, while 45% of the p16 positive CIN1 cases progressed or had persistent CIN1.
In conclusion, the studies with prospective endpoints suggest that CIN1 lesions exhibiting diffuse p16 expression may have a higher risk of progression in comparison to p16 negative lesions, but more prospective data is necessary to support this notion.
Role of p16 in the cytological detection of neoplasia
Methodology
p16 immunocytochemistry has been used with several liquid based cytology systems, including Thinprep, SurePath, and SEROA. In cytology, the spatial context of single cells that is preserved in histology is lost. Therefore, the distinction between focal and diffuse p16 staining cannot be transferred to cytology. Different approaches have been used to identify dysplastic lesions by p16 cytology. In several studies, p16 positive cells were counted and a cutoff level was defined to ascribe a specimen as p16 positive. Bibbo et al. used 10 p16 positive cells per total slide as a cutoff for detecting high grade dysplasias (62), while Sahebali et al. have used 1.87 p16 positive cells per 1000 cells (63). A different approach is the qualitative assessment of p16 positive cells. Trunk et al. described characteristics of different p16 positive cell types (64) and demonstrated that p16 stained specimens can be analyzed based on morphological criteria.
In contrast to conventional Pap staining, in p16 cytology, only a fraction of the cells on a slide (those that are stained by p16) need to be analyzed. In order to facilitate this approach, a scoring system was developed on which to base the assessment of p16 positive cells (65). Using the score, the specificity of p16 aided detection of HGCIN without sacrificing sensitivity. In direct comparison with counting p16 positive cells, the qualitative assessment had a better overall performance (66). Recently, modified p16 cytology staining protocols have been presented. Negri et al. used p16 immunocytology combined with a Pap counterstain (67). Baak et al. have proposed a concomitant staining approach using antibodies directed against p16 and ki-67 in order to specifically highlight HPV-transformed proliferating cells (68).
Cross sectional studies of p16 cytology
Many studies on p16 cytology have been published, most of them using convenience samples or consecutive specimens of a particular cytological diagnosis. Table 4 summarizes 10 large studies incorporating 1610 specimens (62;65;66;69–75). About 5% of the normal cytology specimens, 32% of the ASCUS (including also ASC-US and ASC-H), 37% of the LSIL, 93% of the HSIL, and 99% of the cancer specimens were found to be positive in p16 cytology (Table 4). These data are summarised with the caveat that different cytology platforms and systems for p16 cytology assessment were used. Some p16 cytology studies could not be summarized in table 4: Sahebali et al. determined p16 positive cell counts in consecutive slides from a routine cytology laboratory(63, 76). ASC-H and HSIL cases had significantly higher p16 counts as compared to the LSIL, ASC-US and negative cytology cases. Akpolat et al prepared and analysed paraffin embedded blocks from residual Thinprep specimens (77) and found a good correlation between p16 positivity and Pap cytology. In addition to the identification of squamous cervical lesions, p16 cytology has also been used to identify glandular lesions with promising results (78–80).
The data described above indicate that the performance of p16 cytology is dependent on the evaluation strategy applied. Currently, a qualitative analysis of p16 positive cells either by standard cytological criteria or by using a simplified score based on cytological abnormalities associated with p16 positive cells can substantially increase the specificity of p16 cytology without sacrificing sensitivity (66). In time, the combination of p16 with other biomarkers might further eliminate subjectivity in cytological assessment.
Prospective studies of p16 cytology, triage studies
Bibbo et al. analysed a series of cytology specimens with a HSIL result, for which follow-up biopsy information was available (81). In most of the p16 cytology positive cases, high-grade lesions were detected in the biopsy, while the p16 negative-cytology cases were confirmed as cervicitis, squamous metaplasia, and CIN1. Nieh et al. performed p16 cytology on a series of conventional smears with related follow-up biopsies (82;83). Using p16 cytology, 95% sensitivity and 96% specificity was achieved for the detection of HGCIN and greater, compared to 86% and 31% for HPV DNA testing.
In a study by Ziemke, p16 cytology was used to triage mild-moderate Pap results. All women with persistent abnormalities (PapIIID in the Munich classification) were positive for p16, while only 23% of those that had subsequent regression were positive for p16 cytology (84).
Carrozzi et al. used p16 cytology on liquid based cytology samples obtained from 283 women with abnormal Pap results (85). The authors analyzed p16 cytology as an adjunct to HPV testing to prompt colposcopy and found a better PPV for the combined marker set albeit at a decreased detection rate of CIN2+.
In a recent study, p16 cytology was used to triage ASC-US and LSIL cytology specimens in a reflex triage setting (66). Overall, a sensitivity of over 90% and a specificity of over 80% for the detection of HGCIN were achieved. The triage worked equally well in both the ASCUS and LSIL cytology groups, making p16 an interesting marker for the triage of LSIL cytology, particularly when compared with HPV DNA testing.
Role of p16 protein quantification for the detection of cervical precancer
Recently, a biochemical assay to detect p16 levels in solubilized cervical cells has been developed. In contrast to morphological evaluation, the measurement of protein levels is independent of the observer’s education and training. This assay was performed in high-risk populations and was shown to have a good sensitivity for the detection of HGCIN in two independent studies(86,87). These preliminary data are promising, but further population-based studies are necessary to evaluate the value of p16 quantification in cervical cancer screening.
Conclusions
Many biomarkers have been proposed for cervical cancer screening, a recent overview is given in (88). Most of the markers have been analyzed in a few small studies only and their value cannot be assessed properly at the moment. Here, we have described two related biomarkers of transforming HPV infections, E6/E7 mRNA expression and detection of the p16 protein. The candidacy of these as relevant biomarkers has been justified through our knowledge of basic HPV virology and in-vitro studies. While both HPV mRNA and p16 have been analyzed in a number of studies, there is clearly some way to go before we can say how they will “best fit” to improve the diagnosis of cervical neoplasia, as either stand alone or as adjunctive tests, in triage or in primary screening contexts. For this we need more clinical data, particularly sufficiently powered, longitudinal studies where the candidates are assessed alongside concurrent pathology. The preliminary clinical data look promising although more work is needed to demonstrate sufficient sensitivity and specificity for the detection of high-grade lesions in real-life clinical situations. An appealing feature of both approaches is that potentially, they would not require highly specialised platforms or a technical sea-change for future operators. For example diagnostic platforms already exist for high-throughput testing of viral RNA that could be adapted to HPV as necessary. p16, if used to inform/refine traditional cytology and histology builds on existing skills and if the p16 ELISA approach proves robust, ELISA technology and result interpretation is a mainstay of many diagnostic laboratories. There is currently only one FDA approved test for HPV infection, this is an unusual situation for what is a stable and moreover clinically significant virus. There is therefore a requirement on the research community to seek out and validate new HPV tests or markers of significant infection that are robust and informative but at the same time feasible in routine diagnostic contexts – in this light those we have discussed in this article show promise.
Contributor Information
Kate Cuschieri, Email: Kate.Cuschieri@luht.scot.nhs.uk, Specialist Virology Centre, Royal Infirmary of Edinburgh, UK EH16 4SA, Tel: 00 44 131 242 6039.
Nicolas Wentzensen, Email: wentzenn@mail.nih.gov, Division of Cancer Epidemiology and Genetics, National Cancer Institute, 6120 Executive Blvd, Room 5012, Rockville, MD 20854-7234.
Reference List
- 1.Bulkmans NW, Berkhof J, Rozendaal L, et al. Human papillomavirus DNA testing for the detection of cervical intraepithelial neoplasia grade 3 and cancer: 5-year follow-up of a randomised controlled implementation trial. Lancet. 2007 Nov 24;370(9601):1764–72. doi: 10.1016/S0140-6736(07)61450-0. [DOI] [PubMed] [Google Scholar]
- 2.Davies P, Arbyn M, Dillner J, et al. A report on the current status of European research on the use of human papillomavirus testing for primary cervical cancer screening. Int J Cancer. 2006 Feb 15;118(4):791–6. doi: 10.1002/ijc.21611. [DOI] [PubMed] [Google Scholar]
- 3.Mayrand MH, Duarte-Franco E, Rodrigues I, et al. Human papillomavirus DNA versus Papanicolaou screening tests for cervical cancer. N Engl J Med. 2007 Oct 18;357(16):1579–88. doi: 10.1056/NEJMoa071430. [DOI] [PubMed] [Google Scholar]
- 4.Bosch FX, de SS. The epidemiology of human papillomavirus infection and cervical cancer. Dis Markers. 2007;23(4):213–27. doi: 10.1155/2007/914823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schiffman M, Castle PE, Jeronimo J, Rodriguez AC, Wacholder S. Human papillomavirus and cervical cancer. Lancet. 2007 Sep 8;370(9590):890–907. doi: 10.1016/S0140-6736(07)61416-0. [DOI] [PubMed] [Google Scholar]
- 6.Brink AA, Snijders PJ, Meijer CJ. HPV detection methods. Dis Markers. 2007;23(4):273–81. doi: 10.1155/2007/147429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Castle PE. The potential utility of HPV genotyping in screening and clinical management. J Natl Compr Canc Netw. 2008 Jan;6(1):83–95. doi: 10.6004/jnccn.2008.0008. [DOI] [PubMed] [Google Scholar]
- 8.Snijders PJ, Hogewoning CJ, Hesselink AT, et al. Determination of viral load thresholds in cervical scrapings to rule out CIN 3 in HPV 16, 18, 31 and 33-positive women with normal cytology. Int J Cancer. 2006 Sep 1;119(5):1102–7. doi: 10.1002/ijc.21956. [DOI] [PubMed] [Google Scholar]
- 9.Khan MJ, Castle PE, Lorincz AT, et al. The elevated 10-year risk of cervical precancer and cancer in women with human papillomavirus (HPV) type 16 or 18 and the possible utility of type-specific HPV testing in clinical practice. J Natl Cancer Inst. 2005 Jul 20;97(14):1072–9. doi: 10.1093/jnci/dji187. [DOI] [PubMed] [Google Scholar]
- 10.Bulkmans NW, Berkhof J, Bulk S, et al. High-risk HPV type-specific clearance rates in cervical screening. Br J Cancer. 2007 May 7;96(9):1419–24. doi: 10.1038/sj.bjc.6603653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gravitt PE, Kovacic MB, Herrero R, et al. High load for most high risk human papillomavirus genotypes is associated with prevalent cervical cancer precursors but only HPV16 load predicts the development of incident disease. Int J Cancer. 2007 Dec 15;121(12):2787–93. doi: 10.1002/ijc.23012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hesselink AT, Berkhof J, Bulkmans NW. The value of viral load assessment in the triage of HRHPV positive women to detect CIN2+: A cohort study. 2007 [Google Scholar]
- 13.von Knebel Doeberitz M, Syrjanen KJ. Molecular markers: how to apply in practice. Gynecol Oncol. 2006 Oct;103(1):18–20. doi: 10.1016/j.ygyno.2006.07.032. [DOI] [PubMed] [Google Scholar]
- 14.Dehn D, Torkko KC, Shroyer KR. Human papillomavirus testing and molecular markers of cervical dysplasia and carcinoma. Cancer. 2007 Feb 25;111(1):1–14. doi: 10.1002/cncr.22425. [DOI] [PubMed] [Google Scholar]
- 15.Kelly D, Kincaid E, Fansler Z, Rosenthal DL, Clark DP. Detection of cervical high-grade squamous intraepithelial lesions from cytologic samples using a novel immunocytochemical assay (ProEx C) Cancer. 2006 Dec 25;108(6):494–500. doi: 10.1002/cncr.22288. [DOI] [PubMed] [Google Scholar]
- 16.Doorbar J. Molecular biology of human papillomavirus infection and cervical cancer. Clin Sci (Lond) 2006 May;110(5):525–41. doi: 10.1042/CS20050369. [DOI] [PubMed] [Google Scholar]
- 17.Castle PE, Solomon D, Hildesheim A, et al. Stability of archived liquid-based cervical cytologic specimens. Cancer. 2003 Apr 25;99(2):89–96. doi: 10.1002/cncr.11058. [DOI] [PubMed] [Google Scholar]
- 18.Stoler MH, Rhodes CR, Whitbeck A, Wolinsky SM, Chow LT, Broker TR. Human papillomavirus type 16 and 18 gene expression in cervical neoplasias. Hum Pathol. 1992 Feb;23(2):117–28. doi: 10.1016/0046-8177(92)90232-r. [DOI] [PubMed] [Google Scholar]
- 19.Rose BR, Jiang XM, Thompson CH, Tattersall MH, Cossart YE. Detection of human papillomavirus type 16 E6/E7 transcripts in fixed paraffin-embedded cervical cancers by the polymerase chain reaction. J Virol Methods. 1991 Dec;35(3):305–13. doi: 10.1016/0166-0934(91)90072-8. [DOI] [PubMed] [Google Scholar]
- 20.Habis AH, Vernon SD, Lee DR, Verma M, Unger ER. Molecular quality of exfoliated cervical cells: implications for molecular epidemiology and biomarker discovery. Cancer Epidemiol Biomarkers Prev. 2004 Mar;13(3):492–6. [PubMed] [Google Scholar]
- 21.Tarkowski TA, Rajeevan MS, Lee DR, Unger ER. Improved detection of viral RNA isolated from liquid-based cytology samples. Mol Diagn. 2001 Jun;6(2):125–30. doi: 10.1054/modi.2001.25320. [DOI] [PubMed] [Google Scholar]
- 22.Cuschieri KS, Beattie G, Hassan S, Robertson K, Cubie H. Assessment of human papillomavirus mRNA detection over time in cervical specimens collected in liquid based cytology medium. J Virol Methods. 2005 Mar;124(1–2):211–5. doi: 10.1016/j.jviromet.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 23.Powell N, Smith K, Fiander A. Recovery of human papillomavirus nucleic acids from liquid-based cytology media. J Virol Methods. 2006 Oct;137(1):58–62. doi: 10.1016/j.jviromet.2006.05.033. [DOI] [PubMed] [Google Scholar]
- 24.Horvath CA, Boulet G, Sahebali S, et al. Effects of fixation on RNA integrity in a liquid-based cervical cytology setting. J Clin Pathol. 2008 Jan;61(1):132–7. doi: 10.1136/jcp.2007.047266. [DOI] [PubMed] [Google Scholar]
- 25.Lamarcq L, Deeds J, Ginzinger D, Perry J, Padmanabha S, Smith-McCune K. Measurements of human papillomavirus transcripts by real time quantitative reverse transcription-polymerase chain reaction in samples collected for cervical cancer screening. J Mol Diagn. 2002 May;4(2):97–102. doi: 10.1016/S1525-1578(10)60687-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakagawa S, Yoshikawa H, Yasugi T, et al. Ubiquitous presence of E6 and E7 transcripts in human papillomavirus-positive cervical carcinomas regardless of its type. J Med Virol. 2000 Oct;62(2):251–8. doi: 10.1002/1096-9071(200010)62:2<251::aid-jmv18>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 27.Rose BR, Thompson CH, Tattersall MH, Elliott PM, Dalrymple C, Cossart YE. Identification of E6/E7 transcription patterns in HPV 16-positive cervical cancers using the reverse transcription/polymerase chain reaction. Gynecol Oncol. 1995 Feb;56(2):239–44. doi: 10.1006/gyno.1995.1039. [DOI] [PubMed] [Google Scholar]
- 28.Scheurer ME, Tortolero-Luna G, Guillaud M, et al. Correlation of human papillomavirus type 16 and human papillomavirus type 18 e7 messenger RNA levels with degree of cervical dysplasia. Cancer Epidemiol Biomarkers Prev. 2005 Aug;14(8):1948–52. doi: 10.1158/1055-9965.EPI-05-0073. [DOI] [PubMed] [Google Scholar]
- 29.Smits HL, van GB, Schukkink R, et al. Application of the NASBA nucleic acid amplification method for the detection of human papillomavirus type 16 E6-E7 transcripts. J Virol Methods. 1995 Jul;54(1):75–81. doi: 10.1016/0166-0934(95)00032-p. [DOI] [PubMed] [Google Scholar]
- 30.Sotlar K, Stubner A, Diemer D, et al. Detection of high-risk human papillomavirus E6 and E7 oncogene transcripts in cervical scrapes by nested RT-polymerase chain reaction. J Med Virol. 2004 Sep;74(1):107–16. doi: 10.1002/jmv.20153. [DOI] [PubMed] [Google Scholar]
- 31.Wang-Johanning F, Lu DW, Wang Y, Johnson MR, Johanning GL. Quantitation of human papillomavirus 16 E6 and E7 DNA and RNA in residual material from ThinPrep Papanicolaou tests using real-time polymerase chain reaction analysis. Cancer. 2002 Apr 15;94(8):2199–210. doi: 10.1002/cncr.10439. [DOI] [PubMed] [Google Scholar]
- 32.Dang C, Koehler A, Forschner T, et al. E6/E7 expression of human papillomavirus types in cutaneous squamous cell dysplasia and carcinoma in immunosuppressed organ transplant recipients. Br J Dermatol. 2006 Jul;155(1):129–36. doi: 10.1111/j.1365-2133.2006.07378.x. [DOI] [PubMed] [Google Scholar]
- 33.Scott IU, Karp CL, Nuovo GJ. Human papillomavirus 16 and 18 expression in conjunctival intraepithelial neoplasia. Ophthalmology. 2002 Mar;109(3):542–7. doi: 10.1016/s0161-6420(01)00991-5. [DOI] [PubMed] [Google Scholar]
- 34.Thomas M, Pim D, Banks L. In: The role of the HPV E6 Oncoprotiein in Malignant Progression. Campo M, editor. 2008. [Google Scholar]
- 35.Sotlar K, Diemer D, Dethleffs A, et al. Detection and typing of human papillomavirus by e6 nested multiplex PCR. J Clin Microbiol. 2004 Jul;42(7):3176–84. doi: 10.1128/JCM.42.7.3176-3184.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lie AK, Risberg B, Borge B, et al. DNA- versus RNA-based methods for human papillomavirus detection in cervical neoplasia. Gynecol Oncol. 2005 Jun;97(3):908–15. doi: 10.1016/j.ygyno.2005.02.026. [DOI] [PubMed] [Google Scholar]
- 37.Molden T, Kraus I, Karlsen F, Skomedal H, Nygard JF, Hagmar B. Comparison of human papillomavirus messenger RNA and DNA detection: a cross-sectional study of 4,136 women >30 years of age with a 2-year follow-up of high-grade squamous intraepithelial lesion. Cancer Epidemiol Biomarkers Prev. 2005 Feb;14(2):367–72. doi: 10.1158/1055-9965.EPI-04-0410. [DOI] [PubMed] [Google Scholar]
- 38.Castle PE, Dockter J, Giachetti C, et al. A cross-sectional study of a prototype carcinogenic human papillomavirus E6/E7 messenger RNA assay for detection of cervical precancer and cancer. Clin Cancer Res. 2007 May 1;13(9):2599–605. doi: 10.1158/1078-0432.CCR-06-2881. [DOI] [PubMed] [Google Scholar]
- 39.Cuschieri KS, Whitley MJ, Cubie HA. Human papillomavirus type specific DNA and RNA persistence--implications for cervical disease progression and monitoring. J Med Virol. 2004 May;73(1):65–70. doi: 10.1002/jmv.20062. [DOI] [PubMed] [Google Scholar]
- 40.Molden T, Nygard JF, Kraus I, et al. Predicting CIN2+ when detecting HPV mRNA and DNA by PreTect HPV-proofer and consensus PCR: A 2-year follow-up of women with ASCUS or LSIL Pap smear. Int J Cancer. 2005 May 10;114(6):973–6. doi: 10.1002/ijc.20839. [DOI] [PubMed] [Google Scholar]
- 41.Mantovani F, Banks L. The human papillomavirus E6 protein and its contribution to malignant progression. Oncogene. 2001 Nov 26;20(54):7874–87. doi: 10.1038/sj.onc.1204869. [DOI] [PubMed] [Google Scholar]
- 42.Munger K, Basile JR, Duensing S, et al. Biological activities and molecular targets of the human papillomavirus E7 oncoprotein. Oncogene. 2001 Nov 26;20(54):7888–98. doi: 10.1038/sj.onc.1204860. [DOI] [PubMed] [Google Scholar]
- 43.Khleif SN, DeGregori J, Yee CL, et al. Inhibition of cyclin D-CDK4/CDK6 activity is associated with an E2F-mediated induction of cyclin kinase inhibitor activity. Proc Natl Acad Sci U S A. 1996 Apr 30;93(9):4350–4. doi: 10.1073/pnas.93.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sano T, Oyama T, Kashiwabara K, Fukuda T, Nakajima T. Expression status of p16 protein is associated with human papillomavirus oncogenic potential in cervical and genital lesions. Am J Pathol. 1998;153(6):1741–8. doi: 10.1016/S0002-9440(10)65689-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Klaes R, Friedrich T, Spitkovsky D, et al. Overexpression of p16(INK4A) as a specific marker for dysplastic and neoplastic epithelial cells of the cervix uteri. Int J Cancer. 2001 Apr 15;92(2):276–84. doi: 10.1002/ijc.1174. [DOI] [PubMed] [Google Scholar]
- 46.von Knebel DM, Rittmuller C, Zur HH, Durst M. Inhibition of tumorigenicity of cervical cancer cells in nude mice by HPV E6-E7 anti-sense RNA. Int J Cancer. 1992 Jul 9;51(5):831–4. doi: 10.1002/ijc.2910510527. [DOI] [PubMed] [Google Scholar]
- 47.Beausejour CM, Krtolica A, Galimi F, et al. Reversal of human cellular senescence: roles of the p53 and p16 pathways. EMBO J. 2003 Aug 15;22(16):4212–22. doi: 10.1093/emboj/cdg417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Klaes R, Benner A, Friedrich T, et al. p16INK4a immunohistochemistry improves interobserver agreement in the diagnosis of cervical intraepithelial neoplasia. Am J Surg Pathol. 2002 Nov;26(11):1389–99. doi: 10.1097/00000478-200211000-00001. [DOI] [PubMed] [Google Scholar]
- 49.Horn LC, Reichert A, Oster A, et al. Immunostaining for p16INK4a Used as a Conjunctive Tool Improves Interobserver Agreement of the Histologic Diagnosis of Cervical Intraepithelial Neoplasia. Am J Surg Pathol. 2008 Jan 24; doi: 10.1097/PAS.0b013e31815ac420. [DOI] [PubMed] [Google Scholar]
- 50.Zhang Q, Kuhn L, Denny LA, De SM, Taylor S, Wright TC., Jr Impact of utilizing p16INK4A immunohistochemistry on estimated performance of three cervical cancer screening tests. Int J Cancer. 2007 Jan 15;120(2):351–6. doi: 10.1002/ijc.22172. [DOI] [PubMed] [Google Scholar]
- 51.Agoff SN, Lin P, Morihara J, Mao C, Kiviat NB, Koutsky LA. p16(INK4a) expression correlates with degree of cervical neoplasia: a comparison with Ki-67 expression and detection of high-risk HPV types. Mod Pathol. 2003 Jul;16(7):665–73. doi: 10.1097/01.MP.0000077518.78046.0C. [DOI] [PubMed] [Google Scholar]
- 52.Benevolo M, Mottolese M, Marandino F, et al. Immunohistochemical expression of p16(INK4a) is predictive of HR-HPV infection in cervical low-grade lesions. Mod Pathol. 2006 Mar;19(3):384–91. doi: 10.1038/modpathol.3800551. [DOI] [PubMed] [Google Scholar]
- 53.Focchi GR, Silva ID, Nogueira-de-Souza NC, Dobo C, Oshima CT, Stavale JN. Immunohistochemical expression of p16(INK4A) in normal uterine cervix, nonneoplastic epithelial lesions, and low-grade squamous intraepithelial lesions. J Low Genit Tract Dis. 2007 Apr;11(2):98–104. doi: 10.1097/01.lgt.0000245042.29847.dd. [DOI] [PubMed] [Google Scholar]
- 54.Hu L, Guo M, He Z, Thornton J, McDaniel LS, Hughson MD. Human papillomavirus genotyping and p16INK4a expression in cervical intraepithelial neoplasia of adolescents. Mod Pathol. 2005 Feb;18(2):267–73. doi: 10.1038/modpathol.3800290. [DOI] [PubMed] [Google Scholar]
- 55.Ishikawa M, Fujii T, Saito M, et al. Overexpression of p16 INK4a as an indicator for human papillomavirus oncogenic activity in cervical squamous neoplasia. Int J Gynecol Cancer. 2006 Jan;16(1):347–53. doi: 10.1111/j.1525-1438.2006.00355.x. [DOI] [PubMed] [Google Scholar]
- 56.Wang SS, Trunk M, Schiffman M, et al. Validation of p16INK4a as a marker of oncogenic human papillomavirus infection in cervical biopsies from a population-based cohort in Costa Rica. Cancer Epidemiol Biomarkers Prev. 2004 Aug;13(8):1355–60. [PubMed] [Google Scholar]
- 57.Murphy N, Ring M, Killalea AG, et al. p16INK4A as a marker for cervical dyskaryosis: CIN and cGIN in cervical biopsies and ThinPrep smears. J Clin Pathol. 2003 Jan;56(1):56–63. doi: 10.1136/jcp.56.1.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Murphy N, Heffron CC, King B, et al. p16INK4A positivity in benign, premalignant and malignant cervical glandular lesions: a potential diagnostic problem. Virchows Arch. 2004 Dec;445(6):610–5. doi: 10.1007/s00428-004-1111-4. [DOI] [PubMed] [Google Scholar]
- 59.Murphy N, Ring M, Heffron CC, et al. p16INK4A, CDC6, and MCM5: predictive biomarkers in cervical preinvasive neoplasia and cervical cancer. J Clin Pathol. 2005 May;58(5):525–34. doi: 10.1136/jcp.2004.018895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Negri G, Vittadello F, Romano F, et al. P16(INK4a) expression and progression risk of low-grade intraepithelial neoplasia of the cervix uteri. Virchows Arch. 2004 Oct 9; doi: 10.1007/s00428-004-1127-9. [DOI] [PubMed] [Google Scholar]
- 61.Hariri J, Oster A. The negative predictive value of p16INK4a to assess the outcome of cervical intraepithelial neoplasia 1 in the uterine cervix. Int J Gynecol Pathol. 2007 Jul;26(3):223–8. doi: 10.1097/01.pgp.0000236942.51840.56. [DOI] [PubMed] [Google Scholar]
- 62.Bibbo M, Klump WJ, DeCecco J, Kovatich AJ. Procedure for immunocytochemical detection of P16INK4A antigen in thin-layer, liquid-based specimens. Acta Cytol. 2002 Jan;46(1):25–9. doi: 10.1159/000326711. [DOI] [PubMed] [Google Scholar]
- 63.Sahebali S, Depuydt CE, Segers K, et al. P16INK4a as an adjunct marker in liquid-based cervical cytology. Int J Cancer. 2004 Mar 1;108(6):871–6. doi: 10.1002/ijc.11589. [DOI] [PubMed] [Google Scholar]
- 64.Trunk MJ, Dallenbach-Hellweg G, Ridder R, et al. Morphologic Characteristics of p16INK4a-Positive Cells in Cervical Cytology Samples. Acta Cytol. 2004 Nov 1;48(6):771–82. doi: 10.1159/000326445. [DOI] [PubMed] [Google Scholar]
- 65.Wentzensen N, Bergeron C, Cas F, Eschenbach D, Vinokurova S, von Knebel Doeberitz M. Evaluation of a nuclear score for p16INK4a-stained cervical squamous cells in liquid-based cytology samples. Cancer. 2005 Dec 25;105(6):461–7. doi: 10.1002/cncr.21378. [DOI] [PubMed] [Google Scholar]
- 66.Wentzensen N, Bergeron C, Cas F, Vinokurova S, von Knebel Doeberitz M. Triage of women with ASCUS and LSIL cytology: use of qualitative assessment of p16INK4a positive cells to identify patients with high-grade cervical intraepithelial neoplasia. Cancer. 2007 Feb 25;111(1):58–66. doi: 10.1002/cncr.22420. [DOI] [PubMed] [Google Scholar]
- 67.Negri G, Moretto G, Menia E, et al. Immunocytochemistry of p16INK4a in liquid-based cervicovaginal specimens with modified Papanicolaou counterstaining. J Clin Pathol. 2006 Aug;59(8):827–30. doi: 10.1136/jcp.2005.030726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Baak JP, Kruse AJ, Robboy SJ, Janssen EA, van DB, Skaland I. Dynamic behavioural interpretation of cervical intraepithelial neoplasia with molecular biomarkers. J Clin Pathol. 2006 Oct;59(10):1017–28. doi: 10.1136/jcp.2005.027839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bose S, Evans H, Lantzy L, Scharre K, Youssef E. p16(INK4A) is a surrogate biomarker for a subset of human papilloma virus-associated dysplasias of the uterine cervix as determined on the Pap smear. Diagn Cytopathol. 2005 Jan;32(1):21–4. doi: 10.1002/dc.20175. [DOI] [PubMed] [Google Scholar]
- 70.Ekalaksananan T, Pientong C, Sriamporn S, et al. Usefulness of combining testing for p16 protein and human papillomavirus (HPV) in cervical carcinoma screening. Gynecol Oncol. 2006 Oct;103(1):62–6. doi: 10.1016/j.ygyno.2006.01.033. [DOI] [PubMed] [Google Scholar]
- 71.Meyer JL, Hanlon DW, Andersen BT, Rasmussen OF, Bisgaard K. Evaluation of p16INK4a expression in ThinPrep cervical specimens with the CINtec p16INK4a assay: correlation with biopsy follow-up results. Cancer. 2007 Apr 25;111(2):83–92. doi: 10.1002/cncr.22580. [DOI] [PubMed] [Google Scholar]
- 72.Pientong C, Ekalaksananan T, Swadpanich U, et al. Immunocytochemical detection of p16INK4a protein in scraped cervical cells. Acta Cytol. 2003 Jul;47(4):616–23. doi: 10.1159/000326578. [DOI] [PubMed] [Google Scholar]
- 73.Pientong C, Ekalaksananan T, Kongyingyoes B, et al. Immunocytochemical staining of p16INK4a protein from conventional Pap test and its association with human papillomavirus infection. Diagn Cytopathol. 2004 Oct;31(4):235–42. doi: 10.1002/dc.20122. [DOI] [PubMed] [Google Scholar]
- 74.Saqi A, Pasha TL, McGrath CM, Yu GH, Zhang P, Gupta P. Overexpression of p16INK4A in liquid-based specimens (SurePath) as marker of cervical dysplasia and neoplasia. Diagn Cytopathol. 2002 Dec;27(6):365–70. doi: 10.1002/dc.10205. [DOI] [PubMed] [Google Scholar]
- 75.Yoshida T, Fukuda T, Sano T, Kanuma T, Owada N, Nakajima T. Usefulness of liquid-based cytology specimens for the immunocytochemical study of p16 expression and human papillomavirus testing: a comparative study using simultaneously sampled histology materials. Cancer. 2004 Apr 25;102(2):100–8. doi: 10.1002/cncr.20046. [DOI] [PubMed] [Google Scholar]
- 76.Sahebali S, Depuydt CE, Boulet GA, et al. Immunocytochemistry in liquid-based cervical cytology: Analysis of clinical use following a cross-sectional study. Int J Cancer. 2005 Sep 13; doi: 10.1002/ijc.21489. [DOI] [PubMed] [Google Scholar]
- 77.Akpolat I, Smith DA, Ramzy I, Chirala M, Mody DR. The utility of p16INK4a and Ki-67 staining on cell blocks prepared from residual thin-layer cervicovaginal material. Cancer. 2004 Jun 25;102(3):142–9. doi: 10.1002/cncr.20258. [DOI] [PubMed] [Google Scholar]
- 78.Chen SF, Yang SF, Chu TY, et al. Which test is a better strategy to determine the outcome of atypical glandular cell-categorized Pap smears? Immunocytochemical p16INK4A expression or human papillomavirus test--a retrospective cohort study. Gynecol Oncol. 2005 Dec;99(3):578–84. doi: 10.1016/j.ygyno.2005.06.060. [DOI] [PubMed] [Google Scholar]
- 79.Negri G, Egarter-Vigl E, Kasal A, Romano F, Haitel A, Mian C. p16INK4a is a useful marker for the diagnosis of adenocarcinoma of the cervix uteri and its precursors: an immunohistochemical study with immunocytochemical correlations. Am J Surg Pathol. 2003 Feb;27(2):187–93. doi: 10.1097/00000478-200302000-00006. [DOI] [PubMed] [Google Scholar]
- 80.Nieh S, Chen SF, Chu TY, Lai HC, Fu E. Expression of p16INK4A in Pap smears containing atypical glandular cells from the uterine cervix. Acta Cytol. 2004 Mar;48(2):173–80. doi: 10.1159/000326312. [DOI] [PubMed] [Google Scholar]
- 81.Bibbo M, DeCecco J, Kovatich AJ. P16INK4A as an adjunct test in liquid-based cytology. Anal Quant Cytol Histol. 2003 Feb;25(1):8–11. [PubMed] [Google Scholar]
- 82.Nieh S, Chen SF, Chu TY, Lai HC, Fu E. Expression of p16 INK4A in Papanicolaou smears containing atypical squamous cells of undetermined significance from the uterine cervix. Gynecol Oncol. 2003 Oct;91(1):201–8. doi: 10.1016/s0090-8258(03)00479-7. [DOI] [PubMed] [Google Scholar]
- 83.Nieh S, Chen SF, Chu TY, et al. Is p16(INK4A) expression more useful than human papillomavirus test to determine the outcome of atypical squamous cells of undetermined significance-categorized Pap smear? A comparative analysis using abnormal cervical smears with follow-up biopsies. Gynecol Oncol. 2005 Apr;97(1):35–40. doi: 10.1016/j.ygyno.2004.11.034. [DOI] [PubMed] [Google Scholar]
- 84.Ziemke P. Immunocytochemistry (p16INK4a) as an additional examination for cytological specimens Class III D (Munich Classification II) Zentralbl Gynakol. 2005 Jun;127(3):140–5. doi: 10.1055/s-2005-836406. [DOI] [PubMed] [Google Scholar]
- 85.Carozzi F, Cecchini S, Confortini M, et al. Role of P16((INK4a)) expression in identifying CIN2 or more severe lesions among HPV-positive patients referred for colposcopy after abnormal cytology. Cancer. 2006 Jan 12; doi: 10.1002/cncr.21713. [DOI] [PubMed] [Google Scholar]
- 86.Wentzensen N, Hampl M, Herkert M, et al. Identification of high-grade cervical dysplasia by the detection of p16(INK4a) in cell lysates obtained from cervical samples. Cancer. 2006 Nov 1;107(9):2307–13. doi: 10.1002/cncr.22247. [DOI] [PubMed] [Google Scholar]
- 87.Mao C, Balasubramanian A, Yu M, et al. Evaluation of a new p16(INK4A) ELISA test and a high-risk HPV DNA test for cervical cancer screening: results from proof-of-concept study. Int J Cancer. 2007 Jun 1;120(11):2435–8. doi: 10.1002/ijc.22612. [DOI] [PubMed] [Google Scholar]
- 88.Wentzensen N, von Knebel Doeberitz M. Biomarkers in cervical cancer screening. Dis Markers. 2007;23(4):315–30. doi: 10.1155/2007/678793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kraus I, Molden T, Holm R, et al. Presence of E6 and E7 mRNA from human papillomavirus types 16, 18, 31, 33, and 45 in the majority of cervical carcinomas. J Clin Microbiol. 2006 Apr;44(4):1310–7. doi: 10.1128/JCM.44.4.1310-1317.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]