Abstract
Zoonotic pathogens including those transmitted by insect vectors are some of the most deadly of all infectious diseases known to mankind. A number of these agents have been further weaponized and are widely recognized as being potentially significant biothreat agents. We describe a novel method based on multiply-primed rolling circle in vitro amplification for profiling genomic DNAs to permit rapid, cultivation-free differential detection and identification of circular plasmids in infectious agents. Using Phi29 DNA polymerase and a two-step priming reaction we could reproducibly detect and characterize by DNA sequencing circular DNA from Borrelia burgdorferi B31 in DNA samples containing as little as 25 pg of Borrelia DNA amongst a vast excess of human DNA. This simple technology can ultimately be adapted as a sensitive method to detect specific DNA from both known and unknown pathogens in a wide variety of complex environments.
Keywords: Phi29 DNA polymerase, Multiply-primed rolling circle amplification
Zoonotic pathogens including those transmitted by insect vectors are some of the most deadly of all infectious diseases known to mankind. Many of these diseases remain endemic in various regions of the world and therefore pose serious threats to those who might enter endemic disease zones. A number of these agents have been further weaponized and are widely recognized as being potentially significant biothreat agents. With the threat of possible bioterrorist attacks in today’s society, methods to detect and recognize the pathogens used in these attacks quickly and efficiently are vital to a quick, successful response to the attack [1]. The goal of this project was to develop an improved DNA-based method that enables comprehensive, rapid, cultivation-free differential detection and identification of DNA plasmids present in biothreat infectious agents in clinical specimens.
As a characteristic feature, many of the category A and category B bioterrorism agents/diseases listed by the Centers for Disease Control and Prevention (CDC) including Anthrax (Bacillus anthracis), Botulism (Clostridium botulinum toxin), Plague (Yersinia pestis), and Tularemia (Francisella tularensis) (http://www.bt.cdc.gov/agent/agentlist-category.asp) have circular DNA molecules, either with circular chromosomes and/or plasmids. The size of these circular chromosomes/plasmids ranges from 9.6 Kbp (a plasmid from Yersinia pestis) to 4.7 Mbp (a chromosome from Yersinia pestis) (The Comprehensive Microbial Resource (CMR)—http://cmr.tigr.org/tigr-scripts/CMR/CmrHomePage.cgi).
Recently a technique has been described called multiplyprimed rolling circle amplification that uses Phi29 DNA polymerase and random primers to amplify DNA directly from various sources including whole cells [2], plasmids [3], virus [4,5] and plants [6]. The method takes advantage of the highly processive nature of Phi29 DNA polymerase as well as the enzymes exceptional strand displacement activity [3,7]. This single subunit, proofreading DNA polymerase, is able to promote efficient strand displacement coupled with the polymerization process and incorporates on average >70,000 nt per binding event with linear reaction kinetics at 30 °C for over 12 h [7]. Unlike PCR, DNA amplification does not appear to be limited by target length nor percent G+C content of the template [8]. Moreover, since amplification with Phi29 DNA polymerase is an isothermal procedure there is no need of special instrumentation for temperature cycling, which is required with PCR-based procedures. This is important for diagnostic reasons, as the amplification with Phi29 DNA polymerase could be done virtually anywhere, making Phi29-based procedures both useful and practical in the detection of certain pathogens.
Any new generation of diagnostic systems needs to be open and flexible to identify a broad array of agents, which may reside in complex clinical samples and may not be easily cultivatable. In the present study, we developed a rapid method using Phi29 DNA polymerase to detect the presence of a pathogen’s circular DNA in human DNA samples. We used Borrelia burgdorferi B31, the arthropod-borne etiologic agent of Lyme disease, as a source of total genomic pathogen DNA because B. burgdorferi B31 spirochetes is know to have a complex mixture of as many as 20 unique linear (lp) and circular (cp) plasmids [9,10]. Moreover, we have extensive experience in working with the spirochete in different complex environments including ticks, rodents and human samples.
Materials and methods
Reagent and enzyme
RepliPHI Phi29 DNA polymerase was purchased from Epicentre Biotechnologies (Madison, WI). The Illustra GenomiPhi V2 DNA amplification Kit was purchased from GE Healthcare (Piscataway, NJ) and SYBR Green JumperStart Taq ReadyMix was purchased from Sigma (St. Louis, MO). Dynabeads MyOne Streptavidin C1 were from Invitrogen (Carlsbad, CA). All other DNA purification reagents were from Qiagen (Valencia, CA).
DNA
Human genomic DNA was purified from the human cell line 293T using Qiagen DNeasy kit. B. Burgdorferi DNA was purified from a of low passage culture of strain B31 using Qiagen Genomictips. DNAs were quantified on agarose gel against serial dilutions of a known concentration of lambda phage DNA.
Phi29 DNA polymerase amplification
Fifty-microliter reactions contained 1× reaction buffer (40 mM Tris–HCl, pH 7.5–10 mM MgCl2–5 mM ammonium sulfate–4 mM DTT–50 mM KCl), 0.75 mMeach dNTP, 0.4 μMof each primer and 250 ng B. burgdorferi B31DNA alone or with 1.0 μg human DNA. DNA was denatured at 95 °C for 2 min followed by the addition of 100 U RepliPHI Phi29 DNA polymerase. Samples were incubated at 30 °C for 16 h. The primers used in Phi29 amplification reactions are listed in Table 1.
Table 1.
Primers used in Phi29 amplification.
| Primer name | Sequence | Comment |
|---|---|---|
| OC-F | AAAGAATACATTAAGTGC | Amplifies circular plasmid |
| OC-R | TGGACTTTCTGCCACAAC | cp26 |
| OA-F | TTCAGTAGATTTGCCTGGTGG | Amplifies linear plasmid |
| OA-R | ATTTCAACTGCTGACCCCTC | lp54 |
| lp36-F | ATAAAGTAAATATTAAATTAAGGAG | Amplifies linear plasmid |
| lp36-R | GCTTCTTTATTTTATTGAAATGTAG | lp36 |
| lp28-3-F | AATATTCTGTTAACAC | Amplifies linear plasmid |
| lp28-3-R | AACCCTAAATAAGTACC | lp28-3 |
| 5′cp32-6 | CTTTACATAGTATAAATGCTTTTGG | Amplifies circular plasmid |
| 3′cp32-6 | TCTCGTTATTATAAAATAAGTAGG | cp32-6 |
Real-time PCR
Real-time PCR was performed to monitor plasmid amplification following Phi29 amplification. One microliter of each Phi29 reaction was used as template in Real-time PCR reactions. Plasmid specific gene primers are listed in Table 2. Real-time PCR was carried out at the Stony Brook University DNA sequencing facility using an MJ Research DNA Engine Opticon 2 and SYBR green chemistry. Samples were analyzed in triplicate. A standard curve was generated using serial dilutions of a plasmid solution of known concentration.
Table 2.
Primers used in Real-time PCR
| Primer name | Sequence | Comment |
|---|---|---|
| 5′BBB08 | ATGAAAAAAAAGTTTAATTTTATTTTTC | Amplifies ORF BBB08 on |
| 3′BBB08 | ATTGTAAAAATTTTTCAATTGC | circular plasmid cp26 |
| 5′BBB18 | ATGCTAAGGATCCATGAAGTATAC | Amplifies ORF BBB18 on |
| 3′BBB18 | ATTATTCCCATTCTATGG | circular plasmid cp26 |
| 5′BBA03 | ATGAAAAAAACGATTATTG | Amplifies ORF BBA03 on linear |
| 3′BBA03 | GCTATATAGTGTCTTTAAG | plasmid lp54 |
| 5′BBA24 | ATAATGTTATGATTAAAT | Amplifies ORF BBA24 on linear |
| 3′BBA24 | TTAGTTATTTTTGCATTTTTC | plasmid lp54 |
| 5′BBM04 | ATGGCTTTAAAAGGC | Amplifies ORF BBM04 on |
| 3′BBM04 | TTAACCACCAGCTTC | circular plasmid cp32-6 |
| 5′BBH05 | ACTGGGGCACTATTTGG | Amplifies ORF BBH05/06 on |
| 3′BBH06 | AGATTTTAGCAGGGGAG | linear plasmid lp28-3 |
| 5′BBK21 | ATGGATCAAAAAAAACC | Amplifies ORF BBK21 on linear |
| 3′BBK21 | ATTTACAGTATATTTTTTAAAGCGC | plasmid lp36 |
Circular DNA amplification and sequencing
A series of reaction mixtures containing 1.0 μg of human DNA and B. burgdorferi B31 DNA ranging from 2.5 pg–250 ng were amplified with Phi29 DNA polymerase and biotinylated OC-F/OC-R primers specific to the cp26 of B. burgdorferi as described above. Reactions were 30 °C for 12 h. Reaction products were captured on streptavidin- coated beads and following extensive washing with 5 mM Tris–HCl, pH 7.5, 0.5 mM EDTA, pH 8.0, 1 M NaCl the bound material was subjected to universal amplification using the Illustra GenomiPhi V2 DNA Amplification Kit. The amplified DNA products were digested with HindIII, and the fragments cloned into the HindIII site of plasmid pBS/KS(+). Recombinant plasmids were purified and the inserts sequenced at the Stony Brook University DNA sequencing facility using ABI BigDye reaction mixture.
Results and discussion
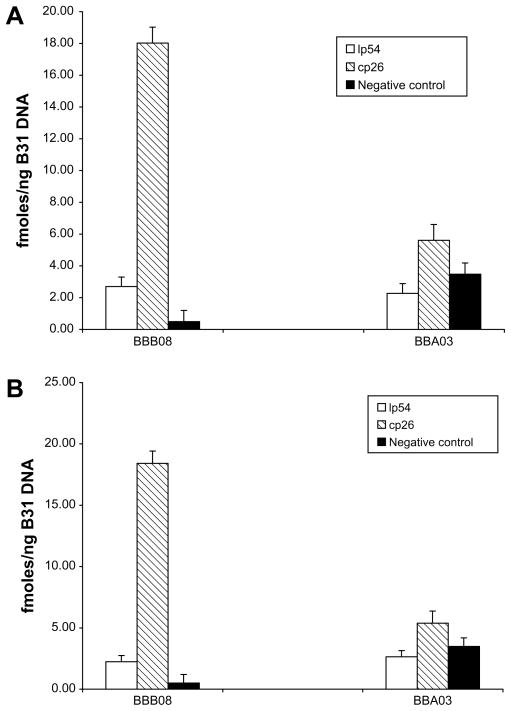
We first determined the ability of Phi29 DNA polymerase to selectively amplify a B. burgdorferi circular plasmid, p26 and a covalently-closed linear plasmid, lp54. B. burgdorferi B31 genomic DNA with or without human 293 T genomic DNA was amplified with Phi29 DNA polymerase and primers sets specific for either plasmid (Table 1). Real-time PCR was then performed on aliquots of amplified DNA using primers specific for genes located on each DNA (Table 2). As shown in Fig. 1, the concentration of gene BBB08, located on cp26, was increased dramatically in genomic DNA amplified with Phi29 DNA polymerase and primers specific for cp26. Importantly, specific amplification of BBB08 was observed in samples with or without human DNA (Fig. 1A vs. Fig. 1B). In contrast, little or no amplification was observed for lp54 DNA as evidenced by the failure to observe an increase in the amount of the lp54 gene BBA03 target region in samples amplified with primers specific for either linear plasmid lp54 or circular plasmid cp26. Furthermore, the BBB08 target was not amplified in Phi29DNA reactions which only included primers specific for the linear plasmid lp54. From these results we conclude that circular plasmids but not linear plasmids of B. burgdorferi DNA can be readily amplified with Phi29 DNA polymerase and that amplification occurs in the presence of a vast excess of human DNA.
Fig. 1.
Phi29 DNA polymerase selectively amplifies B. burgdorferi B31circular plasmid cp26. B. burgdorferi B31 genomic DNA (250 ng) with (A) or without (B) human genomic DNA (1 μg) was amplified with Phi29 DNA polymerase using primers specific for linear plasmid lp54 or circular plasmid cp26. The amount of specific plasmid amplified in each reaction was monitored by real-time PCR using primers specific for plasmid encoded genes (BBB08 for cp26 and BBA03 for lp54). Samples without added Phi29 DNA polymerase were used as a negative control.
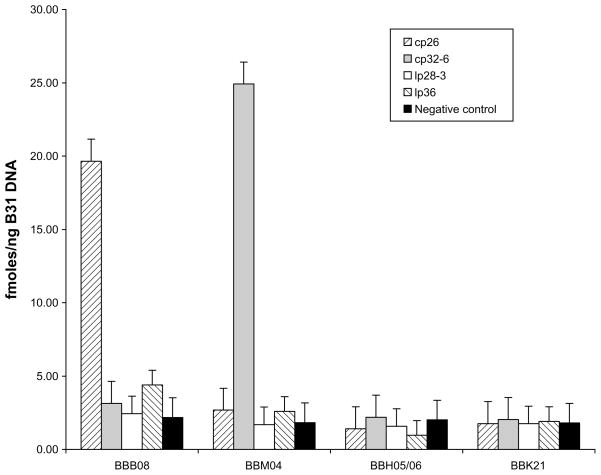
To verify that amplification of circular plasmid as opposed to linear plasmids was a general phenomenon and that the products are indeed primer specific, we tested other circular and linear plasmids in the B. burgdorferi B31 genome. A mixture of B. burgdorferi B31 genomic DNA and human genomic DNA was amplified with Phi29 DNA polymerase using primers specific for circular plasmids cp26 and cp32-6 as well as the linear plasmids lp28-3 and lp36. Relative amplification efficiency was monitored by real-time PCR using gene-specific primers (BBB08 for cp26, BBM04 for plasmid cp32-6, BBH05/06 for lp28-3, and BBK21 for lp36). These data (Fig. 2) confirm that Phi29 DNA polymerase specifically amplified circular plasmids cp26 and cp32-6 as evidenced by the accumulation of plasmid specific gene targets BBB08 and BBM04, respectively. As before, there was no amplification of the linear plasmids, lp36 and lp28-3 linear plasmids. Furthermore, the data also demonstrate that circular plasmid amplification by Phi29 DNA polymerase is primer specific, cp32-6 target gene BBM04 was not amplified in reactions using cp26 specific primers, and vise versa.
Fig. 2.
Phi29 DNA polymerase specifically amplifies B. burgdorferi B31circular plasmids. B. burgdorferi B31 genomic DNA with human genomic DNA was amplified with Phi29 DNA polymerase using primers specific for circular plasmids cp26 and cp32-6, and linear plasmids lp28-3 and lp36. The amount of specific plasmid amplified was monitored by real-time PCR using plasmid specific gene primers (BBB08 for plasmid cp26, BBM04 for plasmid cp32-6, BBH05/06 for plasmid lp28-3 and BBK21 for plasmid lp36). Samples without addition of Phi29 DNA polymerase were used as a negative control.
To develop a high-throughput sequence-based assay to detect pathogen DNA, we combined plasmid-specific primer amplification with subsequent random hexamer primer amplification. First, we amplified DNA mixtures containing a constant amount of human DNA spiked with decreasing amounts of Borrelia DNA (250 ng–2.5 pg) using biotinylated primers specific to cp26. Following amplification, the products were captured on streptavidin-coated magnetic beads. Bound material was subjected to universal amplification using the Illustra GenomiPhi V2 DNA Amplification Kit and the products digested with HindIII. After shot-gun cloning of the digests into Escherichia coli, 10 plasmid clones were randomly selected from each amplification reaction and sequenced.
Results revealed that the number of clones containing Borrelia cp26 DNA was directly related to the amount of Borrelia genomic DNA in the amplification reactions. Of the 10 clones isolated from each sample, 10 of 10 were positive for cp26 sequence in reactions containing 250 ng Borrelia DNA, 9 of 10 were positive in reactions containing 25 ng, 5 of 10 were positive in 2.5 ng reactions while 2 of 10 contained cp26 sequence in reactions with 250 pg. We could; moreover, reproducibly detect cp26 DNA in clones (1 of 10) obtained from reactions containing as little as 25 pg of Borrelia DNA which represents approximately 10,000 molecules of B. burgdorferi. Interestingly, the Borrelia positive clones only contained cp26 sequences and these were randomly distributed around the circular molecule; all non-positive clones contained human DNA. Thus, by performing an initial reaction with biotinylated primers, we were able to separate the amplified product from the original DNA mixture and re-amplify it with Phi29 DNA polymerase to generate sufficient material for cloning and DNA sequencing. The amplification reactions can be performed in several hours and has the advantage that only minimal quantities of starting material are needed for analysis.
In conclusion, we were able to specifically amplify pathogen circular DNA in the presence of human genomic DNA using Phi29 DNA polymerase mediated rolling-circle amplification. Our inability to amplify linear plasmids may be due to their having covalently closed ends which are know to promote rapid snap-back annealing following denaturation. Self-annealing would prevent primer binding and Phi29 DNA amplification. This technology can ultimately be adapted as a sensitive method to detect specific DNA from both known and unknown pathogens in a wide variety of complex clinical and environmental samples.
Acknowledgments
Supported by NIH Grant U01-AI56480. Studies performed at BNL were conducted under the auspices of the United States Department of Energy.
References
- 1.Knobler SL, Mahmoud AA, Pray LA, editors. Biological Threats and Terrorism: Assessing the Science and Response Capabilities: Workshop Summary. National Academy Press; Washington, DC: 2002. [PubMed] [Google Scholar]
- 2.Spits C, Le Caignec C, De Rycke M, Van Haute L, Van Steirteghem A, Liebaers I. Whole-genome multiple displacement amplification from single cells. Nat Protoc. 2006;1:1965–1970. doi: 10.1038/nprot.2006.326. [DOI] [PubMed] [Google Scholar]
- 3.Dean FB, Nelson JR, Giesler TL, Lasken RS. Rapid amplification of plasmid and phage DNA using Phi29 DNA polymerase and multiplyprimed rolling circle amplification. Genome Res. 2001;11:1095–1099. doi: 10.1101/gr.180501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biagini P, Uch R, Belhouchet M, Attoui H, Cantaloube JF, Brisbarre N, Micco P. Circular genomes related to Anelloviruses identified in human and animal samples by using a combined rolling-circle amplification/sequenceindependent single primer amplification approach. J Gen Virol. 2007;88:2696–2701. doi: 10.1099/vir.0.83071-0. [DOI] [PubMed] [Google Scholar]
- 5.Sugiyama M, Ito N, Minamoto N. Isothermal amplification of rabies virus gene. J Vet Med Sci. 2003;65:1063–1068. doi: 10.1292/jvms.65.1063. [DOI] [PubMed] [Google Scholar]
- 6.Knierim D, Maiss E. Application of Phi29 DNA Polymerase in identification and full-length clone inoculation of tomato yellow leaf curl Thailand virus and tobacco leaf curl Thailand virus. Arch Virol. 2007;152:941–954. doi: 10.1007/s00705-006-0914-9. [DOI] [PubMed] [Google Scholar]
- 7.Blanco L, Bernad A, Lazaro JM, Martin G, Garmendia C, Salas M. Highly efficient DNA synthesis by the phage 429 DNA polymerase. J Biol Chem. 1989;264:8935–8940. [PubMed] [Google Scholar]
- 8.Baner J, Nilsson JM, Mendel-Hartvig M, Landegren U. Signal amplification of padlock probes by rolling circle replication. Nucleic Acids Res. 1998;26:5073–5078. doi: 10.1093/nar/26.22.5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fraser C, Casjens S, Huang WM, Sutton GG, Clayton R, Lathigra R, White O, Fetchum KA, et al. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature. 1997;390:580–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- 10.Simpson WJ, Garon CF, Schwan TG. Analysis of supercoiled circular plasmids in infectious and non-infectious Borrelia burgdorferi. Microb Pathog. 1990;8:109–118. doi: 10.1016/0882-4010(90)90075-2. [DOI] [PubMed] [Google Scholar]