Abstract
Expression strains of Escherichia coli BL21(DE3) overproducing the E. coli m5C McrA restriction protein were produced by cloning the mcrA coding sequence behind a T7 promoter. The recombinant mcrA minus BL21 (DE3) host produces active McrA as evidenced by its acquired ability to selectively restrict the growth of T7 phage containing DNA methylated in vitro by HpaII methylase. The mcrA coding region contains several non-optimal E. coli triplets. Addition of the pACYC-RIL tRNA encoding plasmid to the BL21(DE3) host increased the yield of recombinant McrA (rMcrA) upon induction about 5- to 10-fold. McrA protein expressed at 37 °C is insoluble but a significant fraction is recovered as soluble protein after autoinduction at 20 °C. rMcrA protein, which is predicted to contain a Cys4-Zn2+ finger and a catalytically important histidine triad in its putative nuclease domain, binds to several metal chelate resins without addition of a poly-histidine affinity tag. This feature was used to develop an efficient protocol for the rapid purification of nearly homogeneous rMcrA. The native protein is a dimer with a high α-helical content as measured by circular dichroism analysis. Under all conditions tested purified rMcrA does not have measurable nuclease activity on HpaII methylated (Cm5CGG) DNA, although the purified protein does specifically bind HpaII methylated DNA. These results have implications for understanding the in vivo activity of McrA in “restricting” m5C-containing DNA and suggest that rMcrA may have utility as a reagent for affinity purification of DNA fragments containing m5C residues.
Keywords: McrA, Zn2+ finger, DNA methylation, C5-methylcytosine (m5C), DNA binding, CpG island affinity purification
Epigenetic factors, and in particular methylation at the 5-position in cytosine residues in CpG dinucleotides to form m5CpG1, regulate the function of vertebrate genomes by controlling gene expression and chromatin folding. Aberrant hypermethylation of CpG islands near promoters of human tumor suppressor and other genes is now recognized as an important contributing factor in cancer, aging and several other pathological states. Detecting these aberrantly methylated regions and accurately determining their methylation profile is an area of considerable interest primarily because of their potential use as diagnostic and prognostic biomarkers for cancer [1–4]. Reagents, such as m5CpG binding proteins, which preferentially bind to methylated CpG dinucleotides and enzymes that cleave specifically at methylated CpG dinucleotides, are useful tools for identification and characterization of m5C-containing DNA regions [1–3,5–8]. In this study we set out to characterize McrA, an Escherichia coli protein purported to be a nuclease with specificity towards m5C-containing DNA to evaluate its usefulness as a reagent for the identification of m5C residues at single nucleotide resolution.
Wild-type E. coli K-12 strains possess several restriction systems in addition to the classical EcoK hsdR/M/S host-specificity restriction–modification mechanism. One of these, Mar (for methylated adenine restriction), is directed only against DNA containing N6-methyladenine residues while another Mrr (for methylated adenine recognition and restriction) has been reported to restrict DNA containing N6-methyladenine and also DNA with C5-methyl-cytosine residues [9,10]. Neither system restricts DNA methylated by the E. coli enzymes encoded by dam, which methylates the A residue in the sequence GATC, or by dcm, which modifies the internal cytosine in CCWGG (W is A or T) sequences at the C5 position [9,11].
DNA containing C5-methylcytosine (m5C) is also restricted by the Mcr (for modified cytosine restriction) system which is identical to the previously described Rgl (for restricts glucose-less phage) restriction system that blocks the growth of T-even phages, but only when they contain 5-hydroxymethylcytosine in their DNA, i.e., when their 5-hydroxymethylcytosine residues are not glucosylated [12]. Later work further subdivided the Mcr system into two genetically distinct regions: McrA (equal to RglA) on an easily excisable but defective lambdoid prophage element e14 located at 25 min on the E. coli K-12 chromosome and McrB (or RglB) at map position 99 min in a region that includes the EcoK restriction/modification and Mrr systems [13,14]. McrA recognizes DNA containing C5-methylcytosine or C5-hydroxymethylcytosine while McrB also recognizes DNA containing N4-methylcytosine. The mcrB locus encodes two polypeptides McrB and C which together function as a nuclease recognizing in cis two half sites 5′-G/A 5mC (N40–3000) G/A 5mC-3′. Cleavage requires GTP hydrolysis and occurs at a non-fixed distance between the two methylated half sites [15].
Early studies showed that DNAs methylated by M. HpaII (Cm5CGG), M. Eco1831I (Cm5CSGG where S is C or G) and M. SssI (m5CG) are restricted by the McrA system and further studies demonstrated that clones expressing the McrA open reading frame conferred both McrA and RglA phenotypes on a mcr minus host [14,16]. However, since the McrA protein has never been purified its precise sequence preferences and its mode of action remain unclear although it is generally believed to be a member of the ββα-Me finger superfamily of nucleases acting specifically on m5C-containing DNA. McrA also contains a HNH motif common to homing endonucleases as well as many restriction and DNA repair enzymes. The core ββα-Me domain of McrA (residues 159–272 of the 277 amino acid long polypeptide) was modeled by Bujnicki [17] and coworkers using a protein sequence threading approach. This region contains three histidine residues (H-228, 252, and 256) predicted to coordinate a Mg2+ ion, as well as four cysteine residues (C-207, 210, 248, and 251) which form a putative zinc finger, most likely involved in coordinating Zn2+ or some other divalent metal ion to help stabilize the protein’s structure.
While McrA is predicted to function as a nuclease this has never been demonstrated and to date the mechanism for biological restriction of modified phage or plasmid DNAs by McrA is not known. Furthermore, although a slightly N-terminal truncated form of the polypeptide has been cloned in an expression vector [18,19], McrA protein has not been purified. Here, we report the cloning, expression, purification and initial characterization of full-length, biologically active rMcrA. All attempts to demonstrate that rMcrA is a nuclease acting on m5C-containing DNA have failed but electrophoretic mobility shift analysis demonstrates that purified rMcrA interacts specifically with DNA fragments containing Cm5CGG sequences. The production of the recombinant McrA protein in good yield opens up the possibility of obtaining its 3D-structure and will help further investigations into its genuine mode of action in vivo.
Material and methods
Materials
All enzymes are from (New England Biolabs (NEB), Ipswich, MA, USA) if not stated otherwise. Colored protein SDS–PAGE markers were purchased from Lonza Rockland, Inc., Rockland, ME. T7 DNA [20] was isolated by phenol extraction of CsCl purified virus grown in E. coli Bl21, a dcm minus host.
Cloning of McrA
Escherichia coli K-12 W3110 (mcrA+) genomic DNA was used as template to PCR amplify the 833 bp [16] McrA coding sequence with McrA-F 5′-GACGTCTCCCATGCATGTTTTTGAT-3′ and McrA-R 5′-AGAGGATCCCTATTATTTCTGTAATC-3′ as the forward and reverse primers, respectively, and PfuTurbo Cx Hotstart DNA polymerase (Stratagene, San Diego, CA). Bases in the primers complementary to the McrA coding sequence are underlined. McrA-F contains a unique BsmBI recognition sequence while McrA-R contains a unique site for BamHI (shown in italics) which were added to facilitate directional cloning of the digested DNA into the unique NcoI and BamHI sites of a pET28 expression plasmid. The initial amplicons were purified and blunt end ligated into pZERO-2 cut with EcoRV. Following electroporation into E. coli TOP10 (F-mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80lacZΔM15 ΔlacX74 nupG recA1 araD139 Δ (ara-leu)7697 galE15 galK16 rpsL(StrR) endA1 λ−) cells (Stratagene) several pZERO-McrA (KanR) plasmids were purified and the correct sequence of the inserts verified by DNA sequencing using standard M13 primers flanking the cloning site. One of the correct recombinant plasmid was digested with BsmBI and BamHI to release a full-length McrA fragment having ends compatible with those of NcoI + BamHI digested pET28. Following gel purification and elution, the McrA containing fragment was directionally cloned into pET28, previously digested with NcoI and BamHI, using standard ligation conditions and subsequently electroporated into Top10 cells. The resulting pET-rMcrA plasmid DNA was used to transform BL21(DE3) (FompT gal dcm lon hsdSB(rB− mB−) λ(DE3 [lacI lacUV5-T7 gene 1 ind1 sam7 nin5]) cells with and without the chloramphenicol resistant pACYC-RIL tRNA encoding plasmid. All transformants were plated on 2 × YT agar plates supplemented with 50 μg/ml Kan or with 50 μg/ml Kan and 25 μg/ml chloramphenicol as needed. The pRIL tRNA plasmid: argU(AGA, AGG), ileY(AUA) and leuW(CUA), was isolated from Stratagene BL21-CodonPlus McrA+ cells (Strategies Newsletter 14.2, pp. 50–53) and then moved into BL21(DE3) to place the plasmid in a McrA− background. This BL21(DE3) mcr−/pRIL+ strain was kindly provided by F.W. Studier (BNL).
McrA expression in E. coli
Expression of McrA was initially tested in BL21(DE3) with and without the pRIL tRNA plasmid by addition of 0.5 mM IPTG to mid log phase cells in 2 × YT medium or by growth to saturation in ZYM 5052 autoinduction medium at 37 and 20 °C [21]. These media contained 100 μg/ml Kan and 25 μg/ml chloramphenicol as needed and were supplemented with 30 μM ZnSo4. Induction and solubility of the expressed McrA protein was followed by SDS–PAGE. Expression of rMcrA was increased ≥ 5-fold (data not shown) by the presence of the pRIL plasmid. This strain was used for all future experiments. Recombinant McrA expressed at 37 °C is insoluble but a sizeable fraction is soluble if expression is done at 20 °C. We therefore chose to induce protein expression at 20 °C and for ease we also used autoinducing conditions [21]. Shaking cultures (100–200 ml in 500 or 1000 ml flasks) were initially started at 37 °C by addition of 1 ml of an overnight culture in 2 × YT and moved to 20 °C once growth became visible. Cells were harvested after 48 h by centrifugation at 5000g for 10 min at 4 °C, washed with 1/10 vol. 1 × PBS, recentrifuged and the cell pellets (~1.2 g/50 ml culture) stored frozen at −20 °C.
rMcrA purification
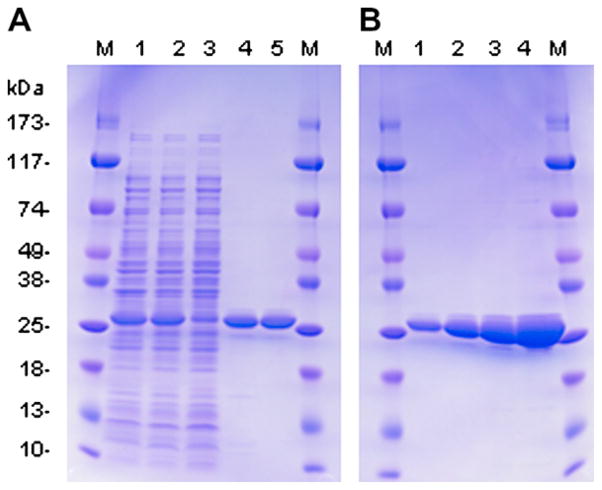
Cell pellets were thawed and resuspended in 10 ml of LEW buffer (50 mM NaPO4, pH 8.0; 300 mM NaCl). Lysozyme (20 mg/ml) was added to a final concentration of 100 μg/ml and the cells were frozen and thawed 3× using a dry ice–ethanol slurry to promote lysis. The extract was sonicated 4–6 times in 30 s bursts alternated with chilling on ice to reduce viscosity and then centrifuged at 10,000 rpm for 10 min at 4 °C. The pellet was resuspended in 5 ml of LEW and recentrifuged at 10,000 rpm for 10 min at 4 °C. The soluble fractions were pooled and mixed for 30 min at 4 °C with 500 mg of PrepEase High-Yield Ni-chelate IDA resin (USB #78806) pre-equilibrated with LEW. The resin was pelleted by centrifugation (5000 rpm for 5 min) and batch washed consecutively with 15 ml LEW, LEW + 700 mM NaCl (1 M final NaCl concentration), and LEW + 2 mM imidazole before pouring into a small 1 cm i.d. column. The settled resin was washed with 25 ml of LEW + 2 mM imidazole then with LEW + 100 mM imidazole to elute rMcrA collecting 2 ml fractions. All chromatography steps were carried out at RT. Peak fractions were pooled, diluted with an equal vol. of 10 mM NaPO4, pH 8.0, and passed through a 5-ml bed of SP-Sepharose Fast Flow (Pharmacia Biotech, Uppsala, Sweden) pre-equilibrated with 0.5 × LEW. The 1 cm i.d. column was washed with ~25 ml 0.5 × LEW and the bound rMcrA eluted with LEW. Peak fractions were pooled and stored at 4 °C. At this stage the protein is ≥99% pure as estimated by Coomassie Blue staining following SDS–PAGE (Fig. 1B).
Fig. 1.
(A) Purification of McrA from E. coli BL21(DE3)/pRIL. The following samples were loaded onto 4%–20% SDS–PAGE: lane 1, total extract prior to centrifugation; lane 2, supernatant after centrifugation; lane 3, protein not bound to the IDA resin; lane 4, pool of IDA 100 mM imidazole eluant; lane 5, pool of SP-Fast Flow Sepharose eluant. (B) SDS–PAGE of increasing amounts of purified rMcrA: lane 1, 1.5 μg; lane 2, 3.5 μg; lane 3 μg; 7.0 μg; lane 4, 15 μg. M indicates lanes with ProSieve color protein molecular weight markers.
Analytical methods
Analytical size exclusion chromatography
Analytical size exclusion chromatography (20 μl injection) was performed on a TSK-GEL G3000SWXl (7.8 mm i.d. × 30 cm, TosoHaas) column equilibrated in 25 mM MES, 300 mM NaCl, pH 6.5. The column was calibrated with thyroglobulin (669 k Da), apoferritin (443 k Da), β-amylase (200 k Da), alcohol dehydrogenase (150 k Da), bovine serum albumin (66 k Da), ovalbumin (45 k Da), carbonic anhydrase (29 k Da) and sperm whale myoglobin (17.8 k Da) in the same buffer. The size of rMcrA was determined from its elution time relative to those of the protein standards plotted against the logarithm of their molecular weights.
Mass determination
Mass spectrometry (TOF-MS) was used to determine the molecular mass of McrA. Samples were desalted and mixed (1:1 dilution) with the matrix sinapinic acid (10 mg/ml in 50% CH3CN, 0.3% TFA) immediately prior to analysis on a Perseptive Biosystems Voyager-DE Linear Mass Spectrometer.
Circular dichroism
Far UV circular dichroism spectra (CD) of rMcrA (180–280 nm) was measured at NSLS beamline U11 using a quartz cell with a 20 μ path length. rMcrA was 1.6 mg/ml in 50 mM HEPES, 500 mM NaCl, pH 7.2 and the data were corrected by subtraction of a blank spectrum obtained using only buffer. The secondary structure content of rMcrA from the CD spectrum was calculated using the software analysis program CDSSTR from DICHROWEB [22,23].
In vitro activity assays
The ability of purified rMcrA to digest or bind to methylcytosine containing DNA was assayed using restriction fragments of normal, unmethylated T7 DNA or DNA incubated with different methyltransferases and S-adenosylmethionine as methyl donor using conditions provided by the supplier. Completeness of the methylation step was confirmed by testing the modified DNA’s resistance to digestion with the appropriate restriction enzymes as well as acquired sensitivity, where appropriate, to digestion by McrBC. Digestion was monitored by agarose electrophoresis. These same conditions were used for electrophoretic mobility shift assays (EMSA). EMSA reactions (10 μl) typically contained a mixture of 50 ng NarI digested unmethylated T7 DNA, 50 ng KpnI digested HpaII methylated T7 DNA, 100 μg/ml BSA and 250 ng sonicated E. coli ER2925 as non-specific competitor in various binding buffers with and without added Mg2+. Reactions were started by adding 1 μl of rMcrA diluted in the appropriate binding buffer plus 100 μg/ml BSA. After 15–20 min at RT the samples were loaded on 0.7% agarose Tris–borate/EDTA (1 × TBE; 89 mM Tris, 89 mM boric acid and 2 mM EDTA.) gels which were electrophoresed at RT. DNA was detected by staining with ethidium bromide (0.5 μg/ml) and illumination with UV light.
In vitro packaging
Unmodified and HpaII methylated T7 DNAs were packaged into virions in vitro using an extract obtained from Novagen (San Diego, CA) using 1 μg DNA/25 μl extract for 30 min at room temperature. Reactions were stopped by dilution with 2 × YT medium and stored at 4 °C. Cells used as indicator were grown overnight at 37 °C in non-inducing 2 × YT medium supplemented with antibiotics as required.
Results and discussion
Expression of recombinant McrA in E. coli
Prior to this study, expression of full-length recombinant McrA (rMcrA) protein has never been reported. Our approach was to place, by PCR, a unique recognition sequence, for BsmBI at an appropriate distance upstream of the McrA start codon and another unique site, BamHI, just past the McrA stop codon. BsmBI is a Type IIS restriction endonuclease that cleaves the double-stranded DNA outside of its recognition site. Cutting this amplicon with BsmBI and BamHI leaves 4 base cohesive ends complementary to the overhangs produced by cutting a standard pET28 cloning/T7-based expression vector with NcoI and BamHI. After directional cloning and sequence verification, rMcrA expression conditions were optimized for reproducible purification of soluble protein. Routine expression was carried out by autoinduction in ZYM 5052 medium [21] at 20 °C in a mcrA− BL21(DE3) host harboring the pRIL tRNA plasmid. ZnSO4 (30 μM) was included in the medium as McrA is thought to have a C4-Zn finger motif [17]. Since we wanted to determine whether the cloned mcrA gene was active we did not use the similar expression host provided by Strategene as it has a mcrA+ genotype.
To test for biological activity we titered T7 phage containing normal, non-methylated or in vitro HpaII methylated, T7 DNA on BL21(DE3) pRIL cells with and without pET-rMcrA. The presence of pET-rMcrA reduced the efficiency of plating (EOP) of the phage containing methylated DNA approximately a 1000-fold compared to their EOP (see Table 2) on this same host containing a similar pET28 vector with a non-related recombinant gene insert at the some position. This reduction in EOP was selective and not seen with in vitro packaged T7 phages containing non-methylated DNA. From these results we concluded that rMcrA is biologically active.
Table 2.
Biological properties of rMcrA
| Methylase | Sequence | # Of sites in T7 | rMcrA binding | T7 inhibitiona |
|---|---|---|---|---|
| AluI | 5′…AGm5CT…3′ | 140 | No | ND |
| Dam | 5′…Gm6ATC…3′ | 6 | No | ND |
| HaeIII | 5′…GGm5CC…3′ | 68 | No | ND |
| HhaI | 5′…Gm5CGC…3′ | 103 | No | ND |
| HpaII | 5′…Cm5CGG…3′ | 58 | Yes | Yes ~103-fold |
| MspI | 5′…m5CCGG…3′ | 58 | No | ND |
| MspI + HpaII | 5′…m5C m5CGG…3′ | 58 | Yes | ND |
| TaqI | 5′…TCGm6A…3′ | 111 | No | ND |
Determined by plating T7 phage containing in vitro methylated DNA on BL21(DE3)/pRIL with and without the pET28-rMcrA plasmid relative to plating of in vitro packaged phage without methylated DNA on these same hosts.
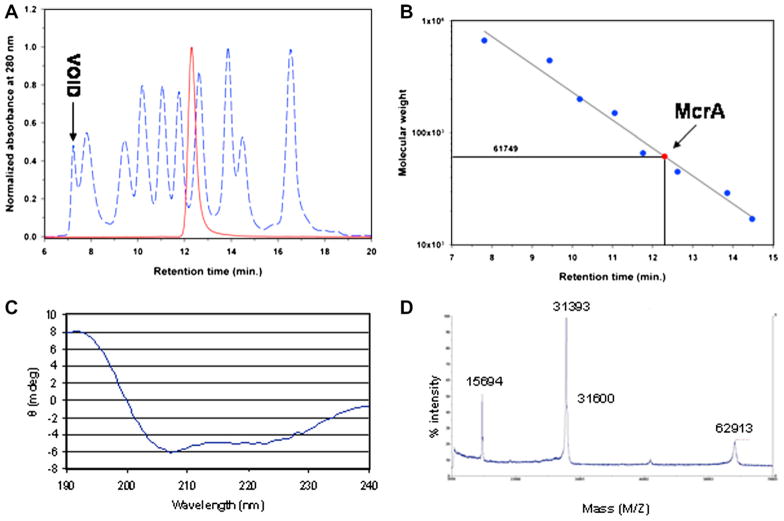
McrA contains three histidines in a ββα-Me domain predicted to coordinate a Mg2+ ion which is believed to be essential for the phosphodiester bond cleavage [17]. This prediction suggested to us that rMcrA might bind directly to matrices typically used for affinity purification of poly-His tagged proteins. Preliminary experiments demonstrated that fairly high purity rMcrA, as visualized by Coomassie blue stained SDS–PAGE gels, could be obtained by affinity chromatography of crude extracts on Ni, Zn or Co charged NTA beads followed by elution with buffers containing 100–250 mM imidazole. No binding was observed to uncharged NTA beads. Presumably binding to these metal resins is via one or more histidine residues in the ββα-Me domain as the rMcrA protein lacks a poly-His tag. Several different metal-ion-charged resins were tried but we chose the IDA resin from USB for routine use because of its ease of preparation, high capacity and purity of the eluted rMcrA protein. Batch chromatography on SP-Sepharose Fast Flow was used to further purify rMcrA (Fig. 1A). This step also removes imidazole from the buffer. The concentration of rMcrA was determined by spectrophotometry assuming a calculated molar extinction coefficient of 35,870 from its amino acid composition (http://encorbio.com/protocols/Prot-MW-Abs.htm) ignoring the contribution of the protein’s seven cysteine residues. The absorbance of a 1 mg/ml solution at 280 nm is calculated to be 1.14. Approximately 7–8 mg of rMcrA is obtained from 50 ml of autoinduced cells. Size exclusion chromatography (Fig. 2A) indicates that rMcrA is a dimer (62 k Da) under native conditions with a high α-helical content (~60%) as determined from its CD spectrum (Fig. 2C) [24]. ESI mass spectrometric analysis (Fig. 2D) of non-reduced rMcrA is in good agreement with the calculated mass of the monomeric protein (31,389 expected vs 31,393 observed). Furthermore, the relatively small peak detected at the expected mass position of an rMcrA dimer is consistent with non-covalent associations forming the rMcrA dimer.
Fig. 2.
(A) Analytical size exclusion chromatography of rMcrA. rMcrA eluted as a single peak (solid red line). The elution peaks for the void and marker proteins are indicated by dashed lines. (B) The mass of native rMcrA calculated relative to its elution time and that of the molecular mass standards (C) CD spectrum of rMcrA. The shape of the spectrum indicates a high content of α-helix in the secondary structure (see Table 1). (D) TOF-MS analysis of native (non-reduced) rMcrA.
rMcrA does not appear to be a m5C specific nuclease
Numerous attempts to find conditions under which rMcrA would digest HpaII methylated T7 DNA or pGEM3 plasmid DNA in vitro were unsuccessful. Among the parameters tested were different buffers (NEB #1, #2, #3 and #4; 50 mM K glutamate, pH 6.0; LEW; and 20 mM HEPES, pH 7.2, 300 mM NaCl) and addition of 1.5 or 3 mM Ca2+; Cd2+; Cu2+, Fe2+; Mg2+; Mn2+, Ni2+ or Zn2+ ions; as well as addition 3 mM ATP or GTP or 10 μM S-adenosylmethionine to the standard NEB restriction buffers. While we cannot rule out the possibility that rMcrA was inactivated during purification this seems unlikely given the ability of the protein to selectively bind HpaII methylated DNA (see below).
rMcrA selectively binds HpaII methylated DNA
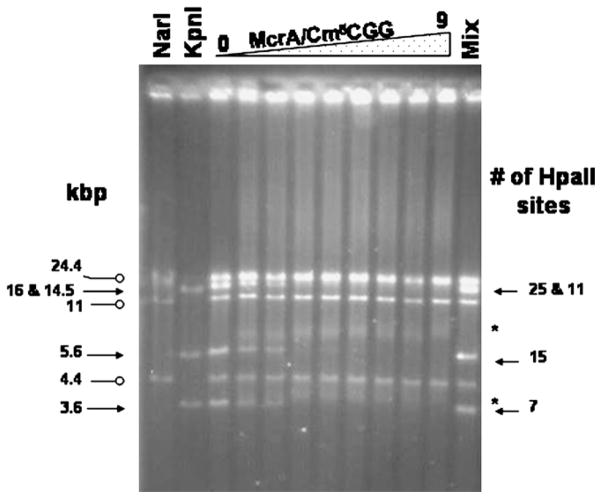
The DNA binding properties of rMcrA were examined using fragments of unmethylated and HpaII methylated T7 DNA by EMSA. The results of a typical EMSA carried out in the absence of added Mg2+ are shown in Fig. 3 and similar results were obtained if Mg2+ was present. The addition of rMcrA had no effect on the mobility of the non-methylated fragments until much higher ratios of moles of protein to CCGG sites in the DNA were used (T7 DNA contains 58 CCGG sites); however, the motilities of the fragments with methylated HpaII sites were significantly retarded in the presence of even small amounts of rMcrA. Furthermore, the addition of rMcrA resulted in the appearance of two distinct shifted products for each of the smaller HpaII methylated KpnI fragments. Separate experiments demonstrated that binding of rMcrA to HpaII methylated T7 DNA does not modify the m5cytosine ring as does binding of the Arabidopsis thaliana proteins DEMETER (DME) and repressor of silencing (ROS1) [25–27]. These closely related DNA glycosylase domain containing proteins remove 5-methylcytosine from the DNA backbone and then their lyase activities cleave the resulting abasic site by successive β- and δ-elimination reactions. If McrA has a similar activity it could account for its known restriction of methylated DNA and initiation of a SOS response following in vivo induction of HpaII in mcrA+ cells. To rule out this possibility we treated rMcrA reacted DNA with proteinase K, EDTA and SDS followed by phenol/chloroform extraction and demonstrated that the DNA was still resistant to cutting by HpaII but sensitive to cutting with MspI which cleaves the same sequence even when the internal cytosine is 5-methylated.
Fig. 3.
Electrophoretic mobility shift assay (EMSA) showing the DNA binding activity of rMcrA to HpaII methylated (solid arrows) but not unmethylated (open circles) T7 DNA fragments. Unmodified and HpaII methylated (Cm5CGG) T7 DNA was digested with NarI and KpnI, respectively. Increasing amounts of rMcrA were added to a 1:1 mixture of the digested T7 DNAs. The molar ratio of added rMcrA to total Cm5CGG sites is: 0; 0.23; 0.3; 0.45; 0.56; 1.14; 2.3; 4.5 and 9.0. The sizes of the fragments (kbp) and number of Cm5CGG sites in each is indicated on the left and right, respectively [20]. The positions of the EMSA shifted smaller HpaII methylated KpnI fragments are labeled with asterisks.
We next determined by EMSA whether rMcrA binds to other methylated DNA sequences. For these assays we used T7 DNA modified by HpaII and the other six methyltransferases listed in Table 2. Most of these enzymes with the exception of dam methyltransferase have more than 50 sites in T7 DNA. Interestingly no shift products were observed for any of these non-HpaII methylation patterns nor did dual methylation by HpaII and Msp I (m5Cm5CGG) prevent binding (data not shown). These observations are consistent with and extend the finding about the specificity of McrA based solely on in vivo studies [9,11,13]. However, they still do not identify conclusively the minimal sequence or number of sites needed for McrA binding nor explain its mode of action in vivo. Presumably, McrA acting simply as a DNA binding protein could interfere with methylated phage development or plasmid maintenance [28]. It is possible that bound McrA interacts with some other E. coli protein(s) to cause cleavage in vivo at methylated HpaII sites. Our attempts to demonstrate that rMcrA can act in concert with McrBC to cause cleavage at methylated HpaII sites have been unsuccessful.
In this study, the smallest number of methylated sites observed to cause a mobility shift was seven although preliminary data (EAM) indicates that as few as two sites may be sufficient for efficient binding. Such detailed studies will require the use of synthetic oligonucleotides.
Our data further indicate that since rMcrA does not bind to HhaI methylated DNA it should only interact with a subset of the sequences recognized by the methyl binding domains of the eukaryotic proteins MeCP2 and MBD2 which together with a monoclonal antibody to m5cytosine are used routinely to affinity purify methylated CpG islands from total genomic sonicates or restriction digests [1,2,5–7]. Fusing the McrA to an appropriate, presumably non-poly His, affinity tag might permit its use in matrix assisted binding of fragments containing only Cm5CGG sequences which are frequently found in aberrantly methylated CpG islands having diagnostic value.
In summary, purified rMcrA does not by itself appear to be a nuclease. Although we cannot fully exclude the trivial explanation that the protein was damaged during purification or that it requires a hitherto unknown co-factor or untried reaction conditions, we favor the idea that its role in vivo is more elusive and complicated than initially proposed.
Table 1.
Secondary structure analysis of rMcrA from CD spectrum
| Secondary structure type | |
|---|---|
| α-Helix Type 1–regular (%) | 38 |
| α-Helix Type 2–distorted (%) | 22 |
| β-Sheet Type 1–regular (%) | 5 |
| β-Sheet Type 2–distorted (%) | 3 |
| Turns (%) | 11 |
| Unordered/random coil (%) | 21 |
| Total | 100 |
Acknowledgments
This work was supported by a Laboratory Directed Research and Development Award at Brookhaven National Laboratory, the Low Dose Radiation Research Program of the Office of Biological and Environmental Research (BER) program of the U.S. Department of Energy (DOE). J.J.D. was also supported by NIH Grant U01-AI56480. We thank John Trunk for CD analysis using beamline U11 at the NSLS which is supported by BER-DOE. We also acknowledge the help of Mike Blewitt, Ed Whittle and Vito Graziano in analytical analysis of rMcrA and the technical assistance of Barbara Lade, Laura-Li Loffredo and Judi Romeo in this work.
Footnotes
Abbreviations used: m5C, C5-methylcytosine; CD, CpG, cytosine-guanosine dinucleotide; circular dichroism; SDS–PAGE, sodium dodecyl sulfate polyacrylamide gel electrophoresis; RT, room temperature; ORF, open reading frame; YT, yeast-tryptone medium; PBS, phosphate buffered saline; Kan, kanamycin; EMSA, electrophoretic mobility shift assay.
References
- 1.Cross SH, Charlton JA, Nan X, Bird AP. Purification of CpG islands using a methylated DNA binding column. Nat Genet. 1994;6:236–244. doi: 10.1038/ng0394-236. [DOI] [PubMed] [Google Scholar]
- 2.Gebhard C, Schwarzfischer L, Pham TH, Andreesen R, Mackensen A, Rehli M. Rapid and sensitive detection of CpG-methylation using methyl-binding (MB)-PCR. Nucleic Acids Res. 2006;34:e82. doi: 10.1093/nar/gkl437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gebhard C, Schwarzfischer L, Pham TH, Schilling E, Klug M, Andreesen R, Rehli M. Genome-wide profiling of CpG methylation identifies novel targets of aberrant hypermethylation in myeloid leukemia. Cancer Res. 2006;66:6118–6128. doi: 10.1158/0008-5472.CAN-06-0376. [DOI] [PubMed] [Google Scholar]
- 4.Baylin SB, Esteller M, Rountree MR, Bachman KE, Schuebel K, Herman JG. Aberrant patterns of DNA methylation, chromatin formation and gene expression in cancer. Hum Mol Genet. 2001;10:687–692. doi: 10.1093/hmg/10.7.687. [DOI] [PubMed] [Google Scholar]
- 5.Rauch T, Li H, Wu X, Pfeifer GP. MIRA-assisted microarray analysis, a new technology for the determination of DNA methylation patterns, identifies frequent methylation of homeodomain-containing genes in lung cancer cells. Cancer Res. 2006;66:7939–7947. doi: 10.1158/0008-5472.CAN-06-1888. [DOI] [PubMed] [Google Scholar]
- 6.Rauch T, Pfeifer GP. Methylated-CpG island recovery assay: a new technique for the rapid detection of methylated-CpG islands in cancer. Lab Invest. 2005;85:1172–1180. doi: 10.1038/labinvest.3700311. [DOI] [PubMed] [Google Scholar]
- 7.Zilberman D, Gehring M, Tran RK, Ballinger T, Henikoff S. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat Genet. 2007;39:61–69. doi: 10.1038/ng1929. [DOI] [PubMed] [Google Scholar]
- 8.Tarasova GV, Nayakshina TN, Degtyarev SK. Substrate specificity of new methyl-directed DNA endonuclease GlaI. BMC Mol Biol. 2008;9:7. doi: 10.1186/1471-2199-9-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heitman J, Model P. Site-specific methylases induce the SOS DNA repair response in Escherichia coli. J Bacteriol. 1987;169:3243–3250. doi: 10.1128/jb.169.7.3243-3250.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Waite-Rees PA, Keating CJ, Moran LS, Slatko BE, Hornstra LJ, Benner JS. Characterization and expression of the Escherichia coli Mrr restriction system. J Bacteriol. 1991;173:5207–5219. doi: 10.1128/jb.173.16.5207-5219.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raleigh EA, Wilson G. Escherichia coli K-12 restricts DNA containing 5-methylcytosine. Proc Natl Acad Sci USA. 1986;83:9070–9074. doi: 10.1073/pnas.83.23.9070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fleischman RA, Cambell JL, Richardson CC. Modification and restriction of T-even bacteriophages. In vitro degradation of deoxyribonucleic acid containing 5-hydroxymethylctosine. J Biol Chem. 1976;251:1561–1570. [PubMed] [Google Scholar]
- 13.Raleigh EA, Murray NE, Revel H, Blumenthal RM, Westaway D, Reith AD, Rigby PW, Elhai J, Hanahan D. McrA and McrB restriction phenotypes of some E. coli strains and implications for gene cloning. Nucleic Acids Res. 1988;16:1563–1575. doi: 10.1093/nar/16.4.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raleigh EA, Trimarchi R, Revel H. Genetic and physical mapping of the mcrA (rglA) and mcrB (rglB) loci of Escherichia coli K-12. Genetics. 1989;122:279–296. doi: 10.1093/genetics/122.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stewart FJ, Raleigh EA. Dependence of McrBC cleavage on distance between recognition elements. Biol Chem. 1998;379:611–616. [PubMed] [Google Scholar]
- 16.Hiom K, Sedgwick SG. Cloning and structural characterization of the mcrA locus of Escherichia coli. J Bacteriol. 1991;173:7368–7373. doi: 10.1128/jb.173.22.7368-7373.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bujnicki JM, Radlinska M, Rychlewski L. Atomic model of the 5-methylcytosine-specific restriction enzyme McrA reveals an atypical zinc finger and structural similarity to ββα-Me endonucleases. Mol Microbiol. 2000;37:1280–1281. doi: 10.1046/j.1365-2958.2000.02010.x. [DOI] [PubMed] [Google Scholar]
- 18.Ramalingam R, Prasad R, Shivapriya R, Dharmalingam K. Molecular cloning and sequencing of mcrA locus and identification of McrA protein in Escherichia coli. J Biosci. 1992;17:217–232. [Google Scholar]
- 19.Shivapriya R, Prasad R, Narayanan IL, Krishnaswamy S, Dharmalingam K. Expression of the mcrA gene of Escherichia coli is regulated posttranscriptionally, possibly by sequestration of the Shine–Dalgarno region. Gene. 1995;157:201–207. doi: 10.1016/0378-1119(94)00746-f. [DOI] [PubMed] [Google Scholar]
- 20.Dunn JJ, Studier FW. Complete nucleotide sequence of bacteriophage T7 DNA and the locations of T7 genetic elements. J Mol Biol. 1983;166:477–535. doi: 10.1016/s0022-2836(83)80282-4. [DOI] [PubMed] [Google Scholar]
- 21.Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 22.Whitmore L, Wallace BA. DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res. 2004;32:W668–W673. doi: 10.1093/nar/gkh371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Whitmore L, Wallace BA. Protein secondary structure analyses from circular dichroism spectroscopy: methods and reference databases. Biopolymers. 2008;89:392–400. doi: 10.1002/bip.20853. [DOI] [PubMed] [Google Scholar]
- 24.Sreerama N, Woody RW. A self-consistent method for the analysis of protein secondary structure from circular dichroism. Anal Biochem. 1993;209:32–44. doi: 10.1006/abio.1993.1079. [DOI] [PubMed] [Google Scholar]
- 25.Agius F, Kapoor A, Zhu JK. Role of the Arabidopsis DNA glycosylase/lyase ROS1 in active DNA demethylation. Proc Natl Acad Sci USA. 2006;103:11796–11801. doi: 10.1073/pnas.0603563103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu J, Kapoor A, Sridhar VV, Agius F, Zhu JK. The DNA glycosylase/lyase ROS1 functions in pruning DNA methylation patterns in Arabidopsis. Curr Biol. 2007;17:54–59. doi: 10.1016/j.cub.2006.10.059. [DOI] [PubMed] [Google Scholar]
- 27.Morales-Ruiz T, Ortega-Galisteo AP, Ponferrada-Marin MI, Martinez-Macias MI, Ariza RR, Roldan-Arjona T. DEMETER and REPRESSOR OF SILENCING 1 encode 5-methylcytosine DNA glycosylases. Proc Natl Acad Sci USA. 2006;103:6853–6858. doi: 10.1073/pnas.0601109103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anton BP, Raleigh EA. Transposon-mediated linker insertion scanning mutagenesis of the Escherichia coli McrA endonuclease. J Bacteriol. 2004;186:5699–5707. doi: 10.1128/JB.186.17.5699-5707.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]