Abstract
Disorder prediction for short peptides is important and difficult. All modern predictors have to be optimized on a preselected dataset prior to prediction. In the succeeding prediction process, the predictor works on a query sequence or its short segment. For implementing the prediction smoothly and obtaining sound prediction results, a specific length of the sequence or segment is usually required. The need of the preselected dataset in the optimization process and the length limitation in the prediction process restrict predictors’ performance. To minimize the influence of these limitations, we developed a method for the prediction of intrinsic disorder in short peptides based on large dataset sampling and statistics. As evident from the data analysis, this method provides more reliable prediction of the intrinsic disorder status of short peptides.
Introduction
The concepts of intrinsic disorder (ID) and intrinsically disordered proteins (IDPs) are being accepted by the scientific community (Wright & Dyson 1999; Uversky et al. 2000; Dunker et al. 2001; Tompa 2003). IDPs do not have unique 3D structures in their native states under physiological conditions. However, they play important roles in living organisms, being frequently involved in crucial biological processes, such as signaling, recognition and regulation. Often, the function of IDPs relies on the large-scale conformational changes of corresponding intrinsically disordered regions (IDRs) (Wright & Dyson 1999; Dunker et al. 2002a, b; Minezaki et al. 2006). The disordered residues and regions can be identified by experiments as regions of missing electron density in X-ray crystallography maps (Ringe & Petsko 1986) or as highly dynamic regions in nuclear magnetic resonance (NMR) spectroscopy (Dyson & Wright 2002b), or by computational predictions (Ferron et al. 2006; He et al. 2009). IDRs are highly abundant in nature. Approximately 70% of proteins in protein data bank (PDB) have regions of missing electron density (Obradovic et al. 2003), of which approximately 40% have regions of missing density corresponding to fragments of 10–20 residues. Over 10% of proteins in PDB have long segments of missing electron density consisting of at least 30 amino acids (Le Gall et al. 2007). Computational studies at the genome level revealed that typically 7– 30% prokaryotic proteins contain long disordered regions of more than 30 consecutive residues, whereas in eukaryotes the amount of such proteins reaches 45–50% (Romero et al. 1997, 2001; Dunker et al. 2001; Oldfield et al. 2005a, b).
Inferences from the earlier observations are more interesting. Over half of the proteins in PDB have short disordered regions of 30 or fewer residues. The vast majority of proteins in various genomes may have short disordered regions (<30 consecutive residues). These facts immediately raised several interesting questions: Why are the short disordered regions so abundant in nature? What functions do they have? How can we identify them? Recent experimental studies have confirmed the functional importance of short IDRs. They can mediate protein–protein interaction (Vershon & Johnson 1993), facilitate multimerization and proceed membrane binding (Liang et al.2003). Computational analyses identified a group of protein segments called molecular recognition features (MoRFs) (Oldfield et al. 2005b; Mohan et al. 2006), which are a short protein fragment undergoing disorder-to-order transition during the protein recognition and binding processes. In other words, short IDRs often help proteins to interact with other molecules or facilitate such interactions. Actually, as estimated by our computational studies, over 40% of proteins in eukaryotes genomes are predicted to contain at least one α-helical MoRF (Oldfield et al. 2005b; Mohan et al. 2006). Based on their fundamental biological roles, short biologically active peptides were collected and classified into various databases, such as ELM (Puntervoll et al. 2003), MnM (Balla et al. 2006) and SLiMDISC (Davey et al. 2006). Pharmaceutical industries have also begun to use more and more peptides in their drug design (Marasco et al. 2008).
Knowing that the short IDRs are related to many biological functions, it is of great importance to identify them with high accuracy. However, this is not a trivial task. Experimental methods are both time and cost consuming. Computational methods, although fast, are less accurate and have many application restrictions.
All state-of-art computational predictors of intrinsic disorder are knowledge-based, meaning that predictor training depends on a collection of examples exhibiting and not exhibiting features of interest. First, a set of proteins is selected in advance. Next, the predictor is optimized by training on these proteins of known features. When the query proteins are very similar to proteins in the training set with regard to the features adopted by the predictor, high accuracy predictions are typically the result. However, when the query protein is different from the training set proteins in chosen features, the prediction accuracy would be subject to many factors. The variability of the prediction accuracy in this case is essentially a sampling problem in the phase space of features adopted by the predictor. The inappropriate selection of true positive and true negative samples in the training set will definitely reduce the generality of the predictor. Hence, selecting proper representation for the phase space of features is of key importance in improving the reliability of the predictor. However, the available structural information obtained from experiments is still limited in comparison with the number of known sequences. Only a small portion of known sequences have resolved structures. In PDB, the number of known disordered residues/regions is noticeably smaller than the number of structured residues. This limitation restricts the sampling of the entire phase space of features. Besides, each predictor can be simulated by a specific algorithm. Mathematical approximation in the algorithm can cause new problems, such as artificial multiple minima and over-fitting. These problems become more critical when the phase space is sparsely sampled.
A second issue for all the disorder predictors currently used comes from the input requirement of protein sequence as a consecutive segment. Amino acids and their sequence on that consecutive segment provide various features as inputs for predictor. These inputs are transformed into the disorder index by the predictor. The accuracy of the disorder index depends on the selection of features, as well as on the length of the consecutive segment. The reasons for this limitation are as follows. The predictor is trained on segments of a particular length. Therefore, the query segments of appropriate length can imitate the local interactions, which are important for local structure and function. If the length is too short, the imitation of the local interactions may create errors. Usually, the length of this chosen segment is approximately 20–30 residues. From this starting point, shorter peptides with only a few residues could not be properly predicted. Many predictors cannot even be applied to short peptides.
The third problem for current disorder predictors is the low prediction accuracy for short IDRs (He et al. 2009; Xue et al. 2010). This problem is strongly dependent upon the first and second problems. By definition, IDRs are flanked sequentially by structured regions. Because a consecutive segment is required for the prediction, the prediction of disordered region boundaries will be influenced by neighboring structured residues. When the disordered region becomes short, the entire predictions for all residues in such an IDR will be influenced by the flanking structured regions. More practically, residues in short IDRs need to be more disorder-prone to maintain the disordered status. That is the reason why the composition profile of short disordered regions is very different from that of longer ones (Peng et al. 2006). Predictors, which take composition profile as the input and are trained on datasets of long segments, will have low accuracy in predicting short segments. As shown by earlier studies, although the prediction accuracy for longer disordered regions is 75–95%, the accuracy for short disordered regions is only 25–66% (Obradovic et al.2003; Xue et al. 2010).
Previously, to predict short peptides, the predictors were built using the datasets of known short peptides. In this article, we proposed a different methodology to deal with the disorder prediction in short peptides. The new computational tool, Short Peptides Analyzer (SPA), first extends the query peptide by embedding it into a preselected segment of 30 residues and then analyzes the disorder status of this extended fragment by one of the previously developed disorder predictors, PONDR-VLXT. The purposes of this study were (i) to develop a specific tool for the accurate disorder prediction of short peptides; and (ii) to improve the prediction accuracy of short disordered regions inside longer sequences. Because the boundary in defining a given segment as short or long region is usually set at 30 residues, and because the most of the PONDR family predictors work well for sequences with 28 or more residues, we restricted our studies to short peptides of 28 or fewer residues.
Results
SPA prediction scheme
The short query peptide is embedded inside a preselected protein segment of 30 amino acids to create a longer combined peptide. Embedding is carried out in such a way that each side of the query peptide is extended by 15 amino acids fragments from this preselected 30-residue-long protein segment. Therefore, the combined peptide has at least 31 amino acids and is obviously above the minimal length limitation posted by all PONDR predictors. Hence, this combined peptide can be predicted by any PONDR predictor. The predicted results corresponding to the central region of the combined peptide are extracted as the prediction for the original query short peptide. Apparently, there are two problems to be solved: Which predictor shall be used? How to choose the 30 a segment?
Each predictor in PONDR family has its own specialty. For example, PONDR-VLXT is very sensitive to local amino acid composition (Romero et al. 1997, 2001). Because of this sensitivity, PONDR-VLXT is able to identify the subtle difference between various short peptides. That is why this predictor is one of the major components of the MoRF identifiers (Oldfield et al. 2005b; Cheng et al. 2007). By definition, MoRF is a specific short protein segment that undergoes disorder-to-order transition during protein–ligand binding. Based on these considerations, PONDR-VLXT was chosen to predict combined peptides in this study.
Obviously, because the amino acid compositions and physicochemical properties of two preselected protein segments may be enormously different, the preselected peptide may have very large influence on the results of final prediction. A common solution to this problem is the ensemble average. Here, a large number of protein segments are selected to make an ensemble. The short query peptide is inserted into every protein segment in this ensemble. The disorder propensities are predicted for all these combined peptides. The disorder predictions corresponding to the short query peptide from all the combined peptides are averaged and are taken as the final disorder scores. Such ensemble averaging helps to reduce the random influence of single preselected protein segment.
Statistics of various short segment datasets
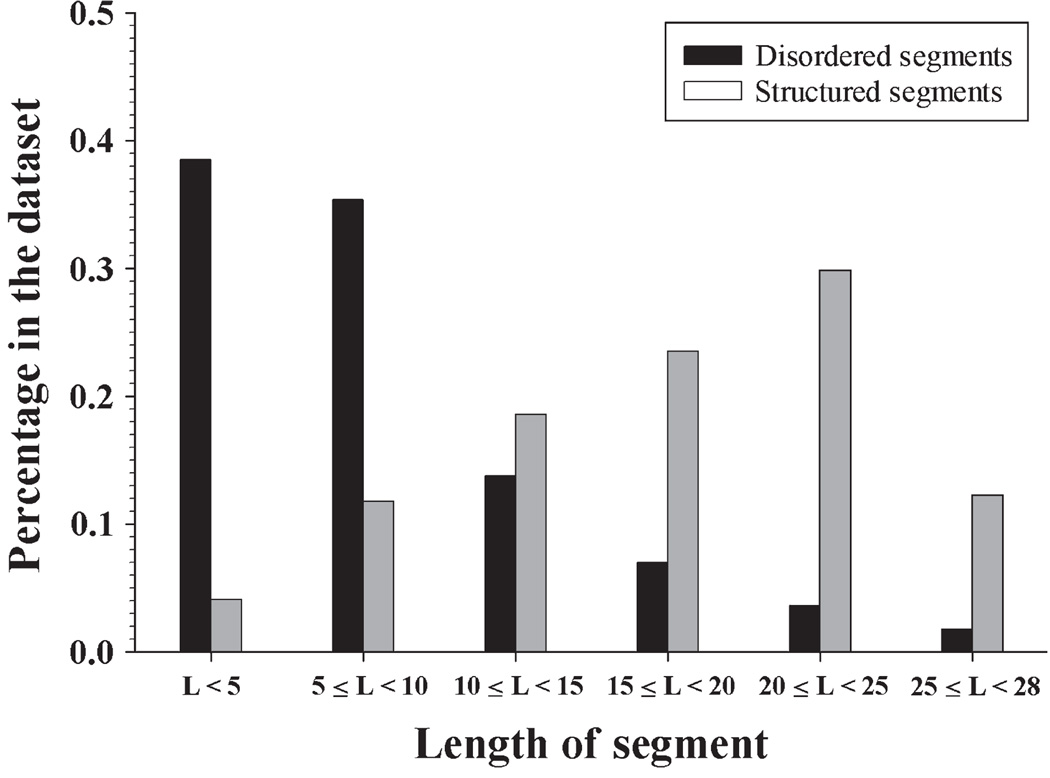
The accuracy of the previously described computational tool for the disorder status analysis of the short peptides, SPA, was tested using the disordered segments of partially disordered proteins (DSP) and ordered segments of partially disordered proteins (OSP) datasets, containing DSP and OSP, respectively. Both datasets originated from the previously generated partially disordered dataset (PDD) (Xue et al. 2010), and the protocol for their development is described in the Materials and Methods section. Fig. 1 shows the length distribution of short disordered and structured segments in the DSP and OSP datasets. The length distributions of these two types of segments were completely different: the DSP dataset mostly contained short segments (5–10 residues), whereas the majority of segments in the OSP dataset were noticeably longer. More specifically, ~75% of disordered segments in DSP were shorter than 10 residues, and only 16% of structured segments in OSP had 10 or fewer residues. Only 5% of disordered segments were longer than 20 residues, whereas over 40% of ordered segments had more than 20 residues. This distribution reflects the natural (PDB-based) abundance of short disordered and ordered segments of various lengths, because both DSP and OSP were directly extracted from PDB. Apparently, because of the overpopulation of short segments, the overall prediction accuracy on these two datasets will be dominated by these short segments.
Figure 1.
Length distribution of short disordered and short ordered segments from DSP and OSP.
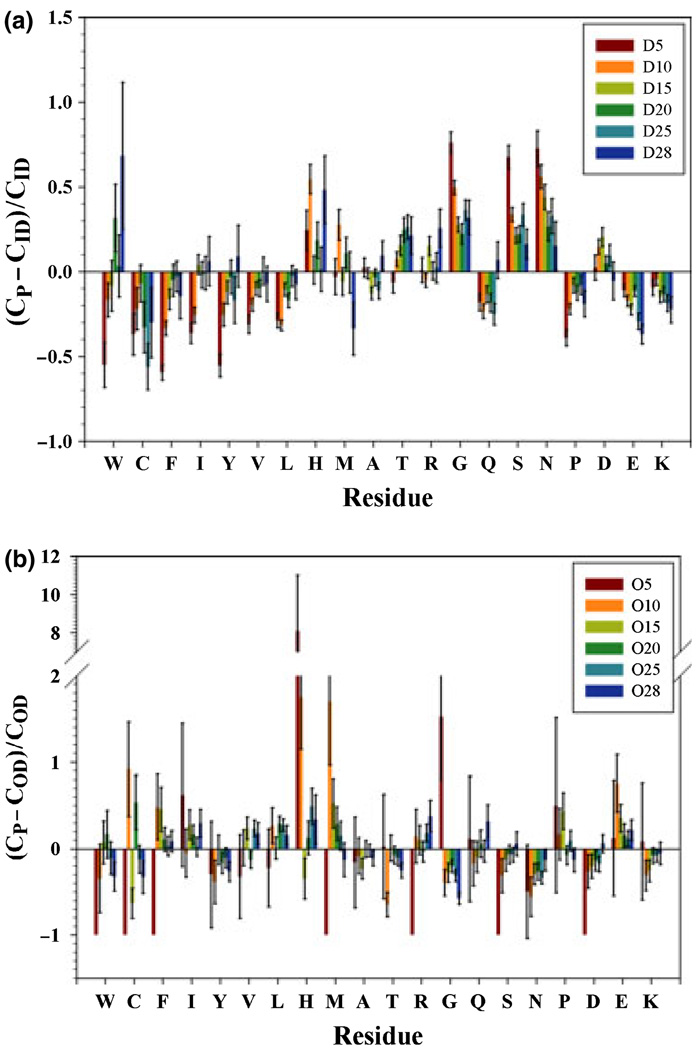
The physicochemical properties of protein chains of different lengths could be different. These differences may eventually invalidate predictors optimized under dissimilar environments. The composition profiling (Vacic et al. 2007b) and the balanced Kullback-Leibler (KL) divergence (Kullback 1987) were employed to compare the datasets. Figure 2(a) illustrates the composition profiles of disordered segments of various lengths compared to sequences in fully disordered dataset (FDD). When the DSP segment length became short, the abundance of order-promoting residues (W, C, F, I, Y, V, and L) clearly decreased, and the content of major disorder-promoting residues (G, S, N, D, E, and K) increased with except for R, Q and P. In essence, these data suggested that short disordered segments in DSP were noticeably more disorder prone than fully disordered proteins in FDD. This outcome was expected because short disordered segment is flanked by ordered segments at both sides. Therefore, there should be more disorder-promoting residues and less order-promoting residues in the short disordered segments to counteract the influence of ordered segments at both sides. Alternatively, these short disordered segments do not have as much potential to interact with other proteins (because they are typically low in the order-promoting residues such as aromatic amino acids) and could therefore have evolved to promote solvation. More interestingly, short disordered segments had much more G, S and N and fewer P, E and K than FDD. The fraction of R and D residues in both datasets was not too different. Hence, the disorder status of short disordered segments was mainly dictated by the high abundance of polar residues rather than by prevalence of charged residues and proline.
Figure 2.
Relative composition profile of DSP segments vs FDD (a) and that of OSP segments vs fully ordered dataset (FOD) (b). On x-axis, amino acids are arranged in ascending disorder tendency. CP is the absolute composition of one amino acid in the query dataset; CID is the absolute composition of the same amino acid in FDD; COD is the same amino acid composition in FOD. Error bars are from 200 times of bootstrapping sampling. ‘D’ indicates subsets from DSP while ‘O’ is for subsets from OSP. D5 includes all the segments with segment length less than 5; D10 is for segments longer than or equal to 5 but <10; D15 corresponds to segments with 10 ≤ L < 15; D20 is 15 ≤ L < 20; D25 is 20 ≤ L < 25; D28 is 25 ≤ L ≤ 28. The same nomenclature is applied to subsets obtained from OSP.
Figure 2(b) represents the relative composition profiles of short structured segments when compared to sequences of proteins in a fully ordered dataset (FOD). Because of the limited number of samples in the dataset, the bootstrapping errors were relatively large. Although the overall trend was still recognizable, the individual content of each amino acid varied greatly among the datasets of short ordered segments. In general, short ordered segments had more aliphatic residues (I, V and L) and less polar and charged residues (G, Q, S, N, K, and D). The content of aromatic residues W, Y and F, as well as that of H, R, P and M fluctuated in a very wide range. Another/interesting observation is the extremely large abundance of histidine residues in the O5 and O10 datasets. This fact is also expected because the histidine residues often have a significant contribution to the protein structural stability.
The KL divergence between various datasets analyzed in this study is shown in Table 1. In our previous study, a KL value <0.01 was used as the indication of two similar datasets (Xue et al. 2009a). By applying this rule of thamb, almost all the segments of various lengths in different datasets were very distinct from each other, except to D10, D15 and D20. Furthermore, difference between ordered subsets was much bigger than the difference between disordered subsets. This observation was in line with the composition variations shown in Fig. 2. In addition, subsets containing very short segments (<10 residues) always had larger KL distance from other subsets than subsets of longer segments. This finding clearly showed that short segments constitute a unique entity and therefore should be considered separately.
Table 1.
Balanced Kullback-Leibler divergence between various subsets
| D5 | D10 | D15 | D20 | D25 | D28 | O5 | O10 | O15 | O20 | O25 | O28 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. | 1105 | 1014 | 394 | 200 | 104 | 44 | 9 | 26 | 41 | 52 | 66 | 27 |
| D5 | 0 | 0.02 | 0.05 | 0.06 | 0.06 | 0.08 | 2.18 | 0.43 | 0.34 | 0.28 | 0.28 | 0.30 |
| D10 | – | 0 | 0.02 | 0.02 | 0.02 | 0.04 | 2.06 | 0.31 | 0.23 | 0.19 | 0.19 | 0.22 |
| D15 | – | – | 0 | 0.01 | 0.01 | 0.02 | 2.11 | 0.27 | 0.15 | 0.13 | 0.13 | 0.14 |
| D20 | – | – | – | 0 | 0.01 | 0.02 | 2.03 | 0.25 | 0.14 | 0.12 | 0.12 | 0.14 |
| D25 | – | – | – | – | 0 | 0.02 | 2.10 | 0.29 | 0.14 | 0.14 | 0.13 | 0.15 |
| D28 | – | – | – | – | – | 0 | 1.94 | 0.27 | 0.16 | 0.12 | 0.11 | 0.14 |
| O5 | – | – | – | – | – | – | 0 | 2.11 | 2.06 | 1.97 | 1.89 | 2.13 |
| O10 | – | – | – | – | – | – | – | 0 | 0.14 | 0.10 | 0.10 | 0.13 |
| O15 | – | – | – | – | – | – | – | – | 0 | 0.05 | 0.04 | 0.06 |
| O20 | – | – | – | – | – | – | – | – | – | 0 | 0.02 | 0.05 |
| O25 | – | – | – | – | – | – | – | – | – | – | 0 | 0.03 |
| O28 | – | – | – | – | – | – | – | – | – | – | – | 0 |
The meaning of abbreviations is explained in the caption of Fig. 2.
Prediction accuracy
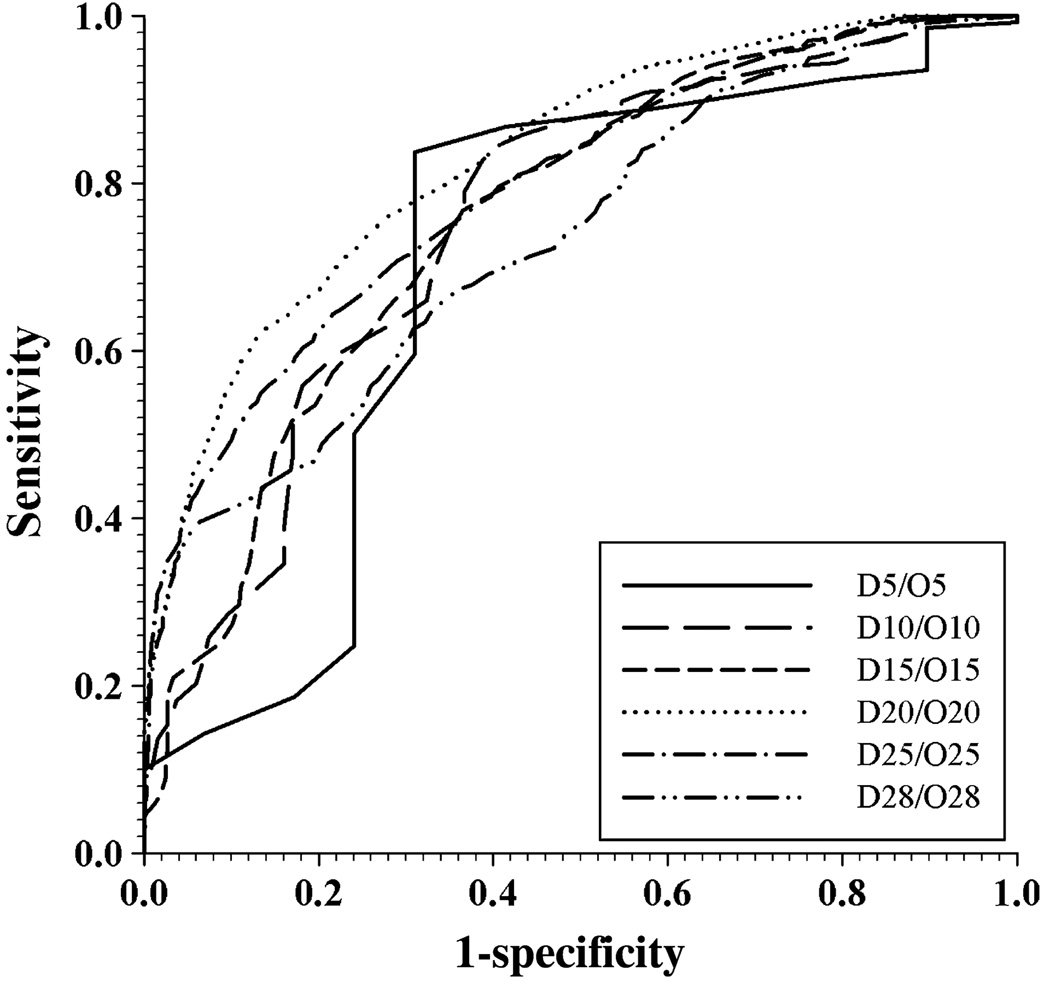
The receiver operating characteristic (ROC) curve for the SPA performance on various subsets is shown in Fig. 3. The corresponding values of area under curve (AUC), breakeven point and accuracy at breakeven point are listed in Table 2. It is clear that the prediction of disorder status in shorter segments was less accurate than the prediction of disorder in middle-sized segments. The accuracy of prediction of longer fragments was also reduced. This probably was because of the insufficient number of samples in the dataset. The datasets D20/O20 achieve the highest accuracy with the AUC of 0.83 and the breakeven accuracy of 74%.
Figure 3.
Receiver operating characteristic (ROC) curve of SPA in DSP/OSP datasets. These two datasets are furthermore grouped into subsets according to the length of segments in them. ‘D’ indicates subsets from DSP while ‘O’ is for subsets from OSP. D5 includes all the segments with segment length less than 5; D10 is for segments longer than or equal to five but <10; D15 corresponds to segments with 10 ≤ L < 15; D20 is 15 ≤ L < 20; D25 is 20 ≤ L < 25; D28 is 25 ≤ L ≤ 28. The same nomenclature is applied to subsets obtained from OSP. Each pair of subsets with the same range of length, originated from DSP and OSP, respectively, are put together to calculate the ROC curve for segments of that length.
Table 2.
Area under ROC curve of various subsets and corresponding prediction accuracy
| AUC | Breakeven accuracy |
Breakeven point |
|
|---|---|---|---|
| D5/O5 | 0.704 | 67.8% | 0.51 |
| D10/O10 | 0.754 | 66.0% | 0.49 |
| D15/O15 | 0.757 | 69.8% | 0.44 |
| D20/O20 | 0.829 | 74.1% | 0.37 |
| D25/O25 | 0.794 | 70.7% | 0.37 |
| D28/O28 | 0.733 | 66.0% | 0.36 |
In comparison, the AUC of PONDR-VLXT in PDD dataset is only 0.71 (Xue et al. 2009a). Hence, it was interesting to compare the accuracy of PONDR-VLXT and SPA performance on various subsets analyzed in this study. The results of this comparison are summarized in Table 3. In datasets of short disordered segments, the accuracy of SPA remarkably exceeded the PONDR-VLXT accuracy, especially for segments shorter than 20 residues. Even in the datasets of short ordered segments, the accuracy of SPA was noticeably better than that of PONDR-VLXT in half of the cases, which represent 65% of the segments. These data clearly show that SPA not only provided a methodology of accurate prediction of disorder status in short segments, which cannot be predicted by traditional disorder predictors because of their limitation on the length of analyzed sequences, but also presented a new way of improving the prediction accuracy. As an illustration of the SPA performance, Table 4 represents several examples of short intrinsically disordered and intrinsically ordered regions (containing 16–20 amino acids) with known crystal or NMR structures, which were correctly or incorrectly predicted by SPA. Here, the experimentally validated disorder status was established from the corresponding PDB entries, with regions with missing electron density being identified as disordered, and segments flanked by the disordered regions being considered as structured. In SPA prediction, a segment was considered as disordered if the content of disordered residues was equal to or higher than 50%. On the contrary, if segment contained <50% of predicted disordered residues, it was assigned as structured. Table 4 shows that in addition to this arbitrary classification of segments as wholly disordered or structured, SPA can provide a mean disorder propensity score for a given peptide, which then can be used for a more accurate disorder status assignment.
Table 3.
Comparison of accuracy of PONDR-VLXT and SPA in various datasets. Acc is the true positive rate of each prediction
| D5 | D10 | D15 | D20 | D25 | D28 | |
|---|---|---|---|---|---|---|
| No. of segments | 1105 | 1014 | 394 | 200 | 104 | 44 |
| Acc – VLXT | 12.7% | 23.6% | 33.8% | 42.5% | 55.2% | 50.0% |
| Acc – SPA | 75.1% | 62.8% | 60.7% | 59.8% | 59.2% | 52.0% |
| O5 | O10 | O15 | O20 | O25 | O28 | |
| No. of segments | 9 | 26 | 41 | 52 | 66 | 27 |
| Acc – VLXT | 81.8% | 69.3% | 81.0% | 79.4% | 78.6% | 83.1% |
| Acc – SPA | 69.0% | 72.3% | 75.5% | 88.0% | 82.7% | 76.4% |
Table 4.
Examples of correctly and incorrectly predicted short segments in O20/D20 datasets
| Dataset | PDB id | Protein name | Short segment | Location | ID status |
SPA prediction (Avg. ID score) |
|---|---|---|---|---|---|---|
| D20 | 5EAU | 5-EPI-aristolochene Synthase | MASAAVANYEEEIVRPVADF | 1–20 | D | D⋆(0.64) |
| 3GJY | Spermidine Synthase | SDTPQHPAETPEHSNTQP | 300–317 | D | D⋆(0.86) | |
| 3FTO | Selenomethionine | MGSNQSSSTSTKKLKAG | 1–17 | D | D⋆(0.71) | |
| 3C96 | Flavin-containing Monooxygenase phzS | KTEKSAALEAITGSYRNQV | 380–398 | D | D⋆(0.49) | |
| 2G5D | MltA from Neisseria gonorrhoeae Monoclinic | GSQSRSIQTFPQPDTSVING | 1–20 | D | D⋆(0.49) | |
| 1ACC | Anthrax protective Antigen | HGNAEVHASFFDIGGS | 304–319 | D | S⋆(0.25) | |
| 1B8X | Glutathione S-transferase | ATRYHTYLPPPYPGEFIVID | 261–280 | D | S⋆(0.23) | |
| 1DYK | Laminin Alpha 2 Chain LG4–5 Domain PAIR | APLASVPTPAFPFPVPTMV | 1–19 | D | S⋆(0.29) | |
| 1Q79 | Poly(A) Polymerase | SHVLQKKKKHSTEGVK | 499–514 | D | S⋆(0.36) | |
| 1SR8 | Cobalamin Biosynthesis Protein | EDDMDSWVWDVQGTDH | 283–298 | D | S⋆(0.19) | |
| O20 | 1HAR | HIV-1 Reverse Transcriptase N-terminal | WAKLVDFRELNKRTQDFWEV | 71–90 | S | S⋆(0.26) |
| 1B9D | HIV-1 integrase | GYSAGERIVDIIATDIQT | 144–161 | S | S⋆(0.41) | |
| 1DHY | KKS102 BPHC Enzyme | WTVARHSRTAMWGHKSV | 272–288 | S | S⋆(0.44) | |
| 2GA8 | YFH7 | EECTAVVARGGTANAIRIAA | 119–138 | S | S⋆(0.51) | |
| 1HU3 | Middle Domain of Human EIF4GII | NFRKLLLNRCQKEFEKDKA | 89–107 | S | S⋆(0.46) | |
| 3E0C | DDB1 | ALRPSASTQALSSSVS | 751–766 | S | D⋆(0.81) | |
| 2QQH | C8alpha-MACPF | RKAMAVEDIISRVRGGSSG | 250–268 | S | D⋆(0.64) | |
| 2IQC | Human FANCF | EDSLMKTQAELLLERLQEV | 12–30 | S | D⋆(0.69) | |
| 1VZW | PRIA | SKLELLPAVDVRDGQAVR | 2–19 | S | D⋆(0.51) | |
| 1PWA | Fibroblast Growth Factor 19 | LPLSHFLPMLPMVPEEP | 126–142 | S | D⋆(0.58) |
‘ID status’ is from PDB database, segments with missing electron density are identified as disordered ‘D’, while segments flanked by disordered regions are assigned as structured ‘S’; In SPA prediction, ‘D⋆’ refers to ratio of disordered residue equal to or higher than 50%; ‘S⋆’ is for ratio of predicted disordered residues less than 50%. Avg. ID score represents the mean intrinsic disorder propensity value estimated by SPA for a given fragment.
Applications
Recently, a concept of MoRFs was introduced to characterize a specific structural element that mediates many of the binding events of IDPs (Oldfield et al.2005b; Mohan et al. 2006; Cheng et al. 2007; Vacic et al. 2007a). These structural elements consist of short regions – on the order of 20 residues – that undergo disorder-to-order transitions upon binding to their partners. Furthermore, these regions are typically flanked with regions of intrinsic disorder (Oldfield et al. 2005b; Cheng et al. 2007). The search of PDB for proteins that fit the general MoRF model of disorder-mediated protein interactions revealed a dataset of 372 short fragments that are very likely to be disordered prior to binding their protein partners, as shown by both sequence- and structure-based predictions (Mohan et al. 2006; Vacic et al. 2007a). These MoRFs were separated into four major groups based on their secondary structure content (Mohan et al. 2006): α-MoRFs, which form α-helices; β-MoRFs, which form β-strands or β-sheets; ι-MoRFs, which have irregular, nonrepeating psi- and phi- angles; and complex-MoRFs, which have two or more secondary structure types of approximately equal abundance (see Fig. 5). Subsequent analyses revealed that MoRFs are very common in various proteomes and occupy a unique structural and functional niche in which function is a direct consequence of intrinsic disorder (Oldfield et al. 2005b; Mohan et al. 2006; Cheng et al. 2007; Vacic et al. 2007a). The functional capacities of MoRFs were shown to be exploited in many molecular settings suggesting that MoRFs may play crucial roles in many different functions (Mohan et al. 2006; Vacic et al. 2007a). MoRFs clearly exemplify a molecular recognition mechanism, which is coupled to the folding process, and which confers exceptional specificity and versatility (Dunker et al. 2001, 2005, 2008a, b; Dyson & Wright 2002a, 2005; Gunasekaran et al. 2003; Uversky et al. 2005; Radivojac et al.2007; Dunker & Uversky 2008; Uversky & Dunker 2008; Wright & Dyson 2009).
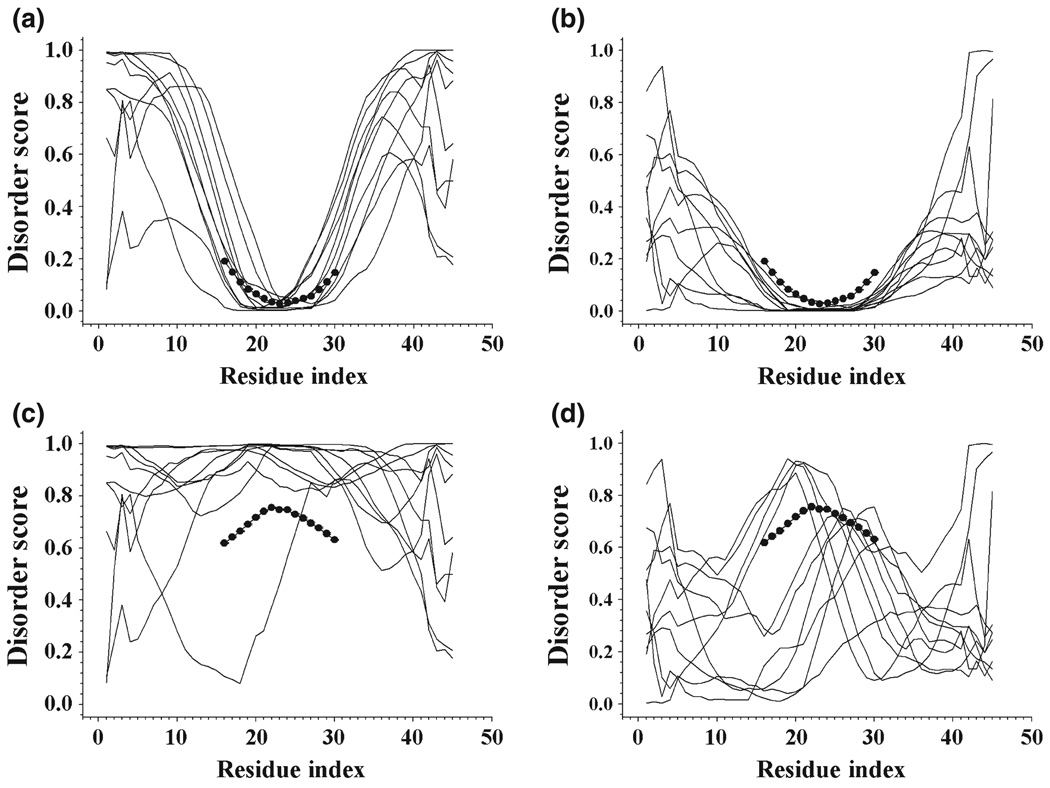
Figure 5.
Application of SPA on two peptides P1 (PFVVSDIAFMGLFYD) and P2 (PLSHGSVVYPRSSLG). P1 is experimentally identified as ordered while P2 is disordered. All the slim curves are PONDR-VLXT predictions for the combined peptides by inserting the query peptides into disordered and ordered segments selected from fully disordered segments (FDS) and fully ordered segments (FOS). The large connected dots are predictions and error bars from SPA. (a) Predictions of 10 randomly selected combined peptides by embedding P1 on disordered protein segments from FDS. (b) Predictions of 10 randomly combined peptides by implanting P1 into ordered segments of FOS. (c) Predictions for peptides generated by inserting P2 into 10 segments used in (a). (d) Combining P2 onto 10 segments shown in (b).
Because α- and β-MoRFs are basically structured peptides flanked by disordered regions at both side, and because ι-MoRFs look like segments ‘frozen’ in the irregular configurations because of their interaction with binding partner, we decided to use our SPA tool to evaluate the predicted disorder status of various MoRFs. Almost all the MoRF segments analyzed so far were predicted to be either fully disordered or fully structured (data not shown). Therefore, we simply considered the segments with 50% or more disordered residues as disordered and counted the number of disordered MoRF segments in each case. Results of this analysis are summarized in Table 5, which shows that only three of 12 α-MoRFs were predicted to be disordered, whereas for ι-MoRFs the disorder/order ratio was close to four out 10. These data are in agreement with earlier observations that MoRFs tend to maintain higher net charge than ordered monomers and although they show lower proportions of aromatic residues, the vast majority of MoRF regions were shown to contain at least one aromatic amino acid (Mohan et al. 2006). The presence of aromatic residues in MoRFs was expected because the side chains of aromatic amino acids tend to make strong and specific interactions (Burley & Petsko 1985), which are expected to exist in regions involved in molecular recognition (Mohan et al. 2006). All these features are shown in Fig. 4 for various MoRF complexes.
Table 5.
Percentage of disordered residues in various molecular recognition feature (MoRF) datasets of different length
| Length | <5 | 5–10 | 10–15 | 15–20 | 20–25 | 25–28 | |
|---|---|---|---|---|---|---|---|
| Alpha-MoRF | No. | – | 2 | 5 | 4 | 1 | – |
| ID Segment | – | 0 | 2 | 1 | 0 | – | |
| Beta-MoRF | No. | – | 1 | – | – | – | – |
| ID Segment | – | 0 | – | – | – | – | |
| Coil-MoRF | No. | 6 | 14 | 11 | 7 | – | – |
| ID Segment | 5 | 6 | 2 | 1 | – | – |
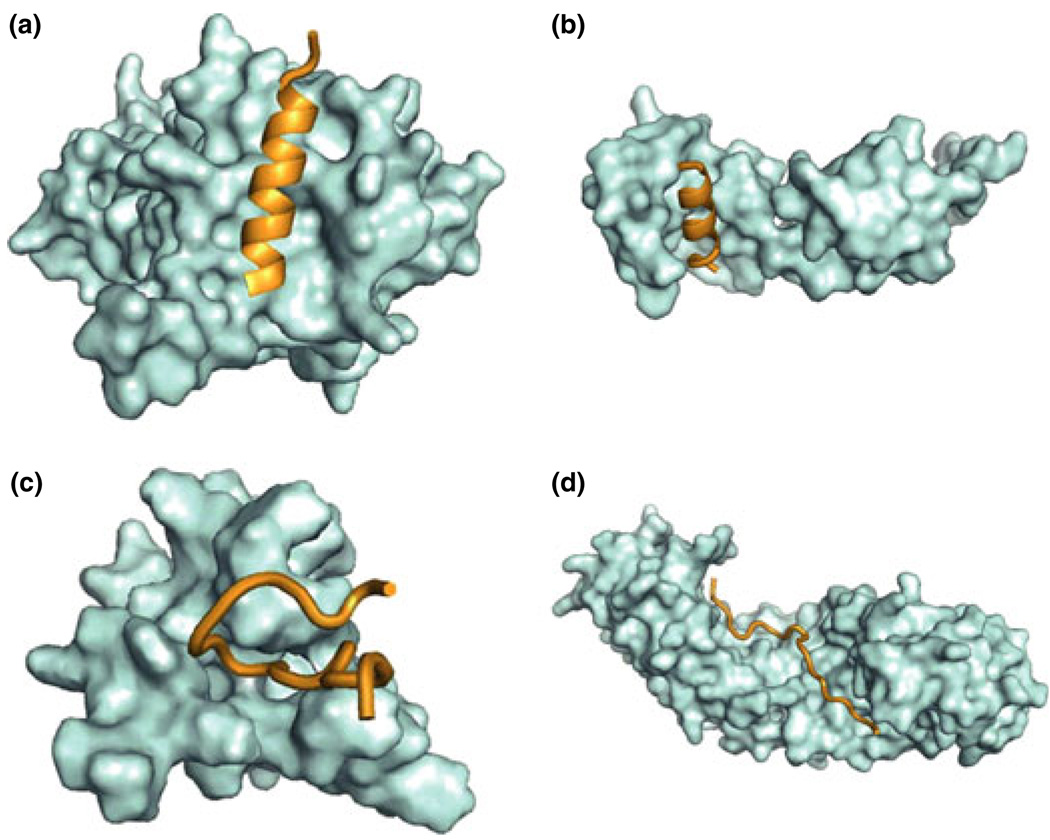
Figure 4.
3D Structure of molecular recognition features (MoRFs) with their substrates. (a) (PDBid:2NM1) and (b) (PDBid:2AUC) are alpha-MoRFs. (a) is predicted to be structured while (b) is disordered. (c) (PDBid:1LXH) and (d) (PDNid: 1PJM) are coil-MoRFs with (c) structured and (d) disordered.
Discussion
The reliable disorder prediction on short protein segments is a difficult task (He et al. 2009; Xue et al. 2010). For a short peptide, the information on long range interactions, which may contribute to its stability and dynamics, is always insufficient. Besides, the majority of modern disorder predictors are typically trained on a set of relatively long sequences. As a result, their prediction accuracies on short segments are relatively poor. All the predictors need a consecutive segment of certain length as the input of the prediction and segments which are shorter than a chosen threshold are rejected by the predictor. Actually, it is a wise strategy in designing predictor to reject the prediction if its accuracy cannot be assured. To the best of our knowledge, PONDR-VSL2 has the shortest threshold of nine residues, whereas the thresholds of the majority of disorder predictors are set at approximately 30 residues. Therefore, there is an urgent need to develop a method for an accurate evaluation of disorder status in such short peptides. Described in this article is a novel computational tool, the short peptide analyzer or SPA, which applies the method of artificial extension of the length of a short query peptide, implements traditional disorder prediction on the extended peptide, and adopts an ensemble average approach to reduce the random error.
To illustrate the performance of SPA analyzer on known ordered and disordered fragments, Fig. 5 represents the SPA prediction for an ordered peptide P1, PFVVSDIAFMGLFYD, and a disordered peptide P2, PLSHGSVVYPRSSLG. Both P1 and P2 peptides have 15 residues, and their order and disorder, respectively, have been experimentally identified. These two peptides were identified as potential sites of protein–protein interactions by the phage display experiments. All the curves in Fig. 5 can be divided into three regions: AA1-AA15, AA16-AA30 and AA31-AA45. Here, AA16-AA30 is the query peptide P1 or P2, whereas flanking segments AA1-AA15 and AA31-45 correspond to the N-terminal and C-terminal halves of fully disordered or fully ordered segments (FOS) selected from fully disordered segments (FDS) and FOS, respectively. Because query sequences were embedded into the preselected ordered or disordered fragments, the resulting flanking regions were predicted as ordered or disordered depending on the nature of the preselected sequences. However, the AA16-AA30 region corresponding to the query sequence was much less affected by the order/disorder status of the flanking regions. By taking the ensemble average as a final step, the results were furthermore consolidated. From Fig. 5, it is clear that the SPA predictions were in a good agreement with the experimental results.
As shown in Fig. 5, the disordered status of terminal residues is heavily influenced by the boundaries. This is one of the reasons why boundary residues usually have less prediction accuracies (He et al. 2009; Xue et al. 2010). We believe that the strategies proposed in SPA can be helpful in improving the accuracy for boundary residues.
Experimental procedures
Datasets of preselected segments
To reduce the fluctuation of the averaged prediction because of insufficient samples, it is important to select as many protein segments as possible. If the ergodicity in the space of combined peptides is satisfied, then the final averaged prediction over all the possible combinations should be highly reliable. However, this exhaustive sampling involves 2030 possible combinations. For the simplicity and feasibility, the number of preselected protein segments has to be reduced to a computationally acceptable level. Such a reduced set of protein segments should provide a sound representation of the original phase space of the combined peptides. By try-and-error approach, two datasets of preselected protein segments were chosen to compose the ensemble, the dataset of FDS and the dataset of FOS.
These two segment datasets were extracted from the previously generated datasets of ordered and disordered proteins, FDD and FOD (Xue et al. 2009b). The set of fully ordered proteins, FOD, was extracted from PDB by choosing X-ray structures of single-chain nonmembrane proteins, which were characterized by unit cell and primitive space groups. Structures with ligand, disulfide bonds and missing electron density were removed from the dataset. The sequence identity of 25% was applied in the BLASTClust (basic local alignment search tool with clustering) from NCBI to find redundant sequences, and shorter sequences in the same cluster were removed. The final dataset has 554 chains and 113,895 residues. The dataset of fully disordered proteins, FDD, was extracted from the Dis-Prot database (Sickmeier et al. 2007) by selecting proteins, which were experimentally shown to be wholly disordered. The final version of FDD has 84 proteins and 17,420 residues.
Next, all the proteins from FOD and FDD were analyzed by PONDR-VLXT (Romero et al. 1997, 2001) and PONDR-VSL2 predictors (Obradovic et al. 2005; Peng et al. 2005, 2006) to evaluate the disorder propensity distribution in their sequences. Segments having consistent ‘ordered’ or ‘disordered’ status among the experiment and the results of these two predictions were selected. Segments shorter than 30 residues were filtered away. Segments longer than 30 residues were chopped down to 30 residues starting from their C-termini. Finally, all segments, which were predicted and experimentally verified as ordered, were grouped into FOS. There were a total of 1470 such segments. By applying the BLASTCLUST and 25% threshold value to filter redundancy, the final FOS dataset of 1439 segments was created. Similarly, 197 disordered segments were classified into FDS. The application of BLASTCLUST did not reduce the number of sequences in this dataset.
Independent prediction and bootstrapping
For each short query peptide, there were 1636 combined peptides, among which 1439 resulted from embedding a query peptide into the FOS sequences, and 197 combined peptides originated from the insertion of a query sequence into the FDS sequences. Because the sizes of FOS and FDS are very different, a simple mathematical average over all these 1636 predictions will undoubtedly bias to the predictions from FOS-embedded fragments. To avoid this bias, a balanced bootstrapping procedure was applied 1000 times to represent the final disorder score for each residue of the query sequence. At each bootstrapping step, to calculate the disorder score for each residue, an equal number of predictions made for ordered-segment-combined peptides and for disorder-segment-combined peptides were randomly selected from the original set of 1439 and 197 predictions. This process was repeated 1000 times to provide mega average values, as well as the statistical error of the prediction.
Test datasets
To evaluate the accuracy of the proposed method, two additional datasets were created. The first dataset, DSP, contained disordered segments of partially disordered proteins, whereas ordered segments of partially disordered proteins were included in OSP dataset. Both datasets originated from the previously generated PDD (Xue et al. 2010), which was created by selecting from PDB the X-ray structures of single chain protein with resolutions higher than 3.0 Å and without prosthetic groups. These sequences were then clustered by using BLASTCLUST with a 30% cut-off of sequence identity. In the case, there were multiple sequences in the same cluster, the longest one was selected. The resulting sequences were furthermore filtered by removing histidine tags and initial methionines, as well as sequences having only 20 or less disordered residues totally in the entire sequence by applying xml2pdb (http://dunbrack.fccc.edu/Guoli/s2c/index.php). The purpose of removing sequences with low number of disordered residues was to keep the reasonable size of the dataset. There were 647 sequences with totally 230,314 residues, in which 16,011 disordered residues were located in 1376 disordered regions. After removing the segments longer than 28 residues, there were 2861 and 221 short disordered and short structured segments in DSP and OSP, respectively.
The performance of SPA was also evaluated on a dataset of multi-partner MoRF segments and illustrated for two biologically active short peptides with known disorder status. The multi-partner MoRF dataset was also extracted from PDB by following procedures: First, select all the complex structures in PDB that have short nonglobular protein fragments (5–25 residues) bound to large globular structural partner (>70 residues). Then, remove all the complexes that have solvent surface area difference (ΔASA) of <400 square angstroms from unbound to bound state. Third, by aligning each short protein segment in the complexes back onto all the sequences in UniProt, extract the sequences that contain multiple protein segments, and these multiple segments overlap (at least one residue) with each other. Followed by this, the overlapped common regions are taken as multi-partner MoRFs. Finally, after applying BlastCluster to remove the redundancy, there are 150 multi-partner MoRFs in 51 clusters. Two 15-mer peptides were identified through a screening for protein–protein interaction using the phage display technology.
Acknowledgements
This work was supported in part by the grants R01 LM007688-01A1 (to A.K.D and V.N.U.), R01 GM071714-01A2 (to A.K.D and V.N.U.), the grant EF 0849803 from the National Science Foundation (to A.K.D and V.N.U.), and the Program of the Russian Academy of Sciences for the ‘Molecular and Cellular Biology’ (to V.N.U.). We gratefully acknowledge the support of the IUPUI Signature Centers Initiative.
References
- Balla S, Thapar V, Verma S, Luong T, Faghri T, Huang CH, Rajasekaran S, del Campo JJ, Shinn JH, Mohler WA, Maciejewski MW, Gryk MR, Piccirillo B, Schiller SR, Schiller MR. Minimotif Miner: a tool for investigating protein function. Nat. Methods. 2006;3:175–177. doi: 10.1038/nmeth856. [DOI] [PubMed] [Google Scholar]
- Burley SK, Petsko GA. Aromatic-aromatic interaction: a mechanism of protein structure stabilization. Science. 1985;229:23–28. doi: 10.1126/science.3892686. [DOI] [PubMed] [Google Scholar]
- Cheng Y, Oldfield CJ, Meng J, Romero P, Uversky VN, Dunker AK. Mining alpha-helix-forming molecular recognition features with cross species sequence alignments. Biochemistry. 2007;46:13468–13477. doi: 10.1021/bi7012273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey NE, Shields DC, Edwards RJ. SLiMDisc: short, linear motif discovery, correcting for common evolutionary descent. Nucleic Acids Res. 2006;34:3546–3554. doi: 10.1093/nar/gkl486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunker AK, Brown CJ, Lawson JD, Iakoucheva LM, Obradovic Z. Intrinsic disorder and protein function. Biochemistry. 2002a;41:6573–6582. doi: 10.1021/bi012159+. [DOI] [PubMed] [Google Scholar]
- Dunker AK, Brown CJ, Obradovic Z. Identification and functions of usefully disordered proteins. Adv. Protein Chem. 2002b;62:25–49. doi: 10.1016/s0065-3233(02)62004-2. [DOI] [PubMed] [Google Scholar]
- Dunker AK, Cortese MS, Romero P, Iakoucheva LM, Uversky VN. Flexible nets. The roles of intrinsic disorder in protein interaction networks. FEBS J. 2005;272:5129–5148. doi: 10.1111/j.1742-4658.2005.04948.x. [DOI] [PubMed] [Google Scholar]
- Dunker AK, Lawson JD, Brown CJ, et al. Intrinsically disordered protein. J. Mol. Graph. Model. 2001;19:26–59. doi: 10.1016/s1093-3263(00)00138-8. [DOI] [PubMed] [Google Scholar]
- Dunker AK, Oldfield CJ, Meng J, Romero P, Yang JY, Chen JW, Vacic V, Obradovic Z, Uversky VN. The unfoldomics decade: an update on intrinsically disordered proteins. BMC Genomics. 2008a;9 Suppl 2:S1. doi: 10.1186/1471-2164-9-S2-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunker AK, Silman I, Uversky VN, Sussman JL. Function and structure of inherently disordered proteins. Curr. Opin. Struct. Biol. 2008b;18:756–764. doi: 10.1016/j.sbi.2008.10.002. [DOI] [PubMed] [Google Scholar]
- Dunker AK, Uversky VN. Signal transduction via unstructured protein conduits. Nat. Chem. Biol. 2008;4:229–230. doi: 10.1038/nchembio0408-229. [DOI] [PubMed] [Google Scholar]
- Dyson HJ, Wright PE. Coupling of folding and binding for unstructured proteins. Curr. Opin. Struct. Biol. 2002a;12:54–60. doi: 10.1016/s0959-440x(02)00289-0. [DOI] [PubMed] [Google Scholar]
- Dyson HJ, Wright PE. Insights into the structure and dynamics of unfolded proteins from nuclear magnetic resonance. Adv. Protein Chem. 2002b;62:311–340. doi: 10.1016/s0065-3233(02)62012-1. [DOI] [PubMed] [Google Scholar]
- Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- Ferron F, Longhi S, Canard B, Karlin D. A practical overview of protein disorder prediction methods. Proteins. 2006;65:1–14. doi: 10.1002/prot.21075. [DOI] [PubMed] [Google Scholar]
- Gunasekaran K, Tsai CJ, Kumar S, Zanuy D, Nussinov R. Extended disordered proteins: targeting function with less scaffold. Trends Biochem. Sci. 2003;28:81–85. doi: 10.1016/S0968-0004(03)00003-3. [DOI] [PubMed] [Google Scholar]
- He B, Wang K, Liu Y, Xue B, Uversky VN, Dunker AK. Predicting intrinsic disorder in proteins: an overview. Cell Res. 2009;19:929–949. doi: 10.1038/cr.2009.87. [DOI] [PubMed] [Google Scholar]
- Kullback S. The Kullback-Leibler distance. Am. Stat. 1987;41:340–341. [Google Scholar]
- Le Gall T, Romero PR, Cortese MS, Uversky VN, Dunker AK. Intrinsic disorder in the Protein Data Bank. J. Biomol. Struct. Dyn. 2007;24:325–342. doi: 10.1080/07391102.2007.10507123. [DOI] [PubMed] [Google Scholar]
- Liang C, Hu J, Whitney JB, Kleiman L, Wainberg MA. A structurally disordered region at the C terminus of capsid plays essential roles in multimerization and membrane binding of the gag protein of human immunodeficiency virus type 1. J. Virol. 2003;77:1772–1783. doi: 10.1128/JVI.77.3.1772-1783.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marasco D, Perretta G, Sabatella M, Ruvo M. Past and future perspectives of synthetic peptide libraries. Curr. Protein Pept. Sci. 2008;9:447–467. doi: 10.2174/138920308785915209. [DOI] [PubMed] [Google Scholar]
- Minezaki Y, Homma K, Kinjo AR, Nishikawa K. Human transcription factors contain a high fraction of intrinsically disordered regions essential for transcriptional regulation. J. Mol. Biol. 2006;359:1137–1149. doi: 10.1016/j.jmb.2006.04.016. [DOI] [PubMed] [Google Scholar]
- Mohan A, Oldfield CJ, Radivojac P, Vacic V, Cortese MS, Dunker AK, Uversky VN. Analysis of molecular recognition features (MoRFs) J. Mol. Biol. 2006;362:1043–1059. doi: 10.1016/j.jmb.2006.07.087. [DOI] [PubMed] [Google Scholar]
- Obradovic Z, Peng K, Vucetic S, Radivojac P, Brown CJ, Dunker AK. Predicting intrinsic disorder from amino acid sequence. Proteins. 2003;53 Suppl 6:566–572. doi: 10.1002/prot.10532. [DOI] [PubMed] [Google Scholar]
- Obradovic Z, Peng K, Vucetic S, Radivojac P, Dunker AK. Exploiting heterogeneous sequence properties improves prediction of protein disorder. Proteins. 2005;61 Suppl 7:176–182. doi: 10.1002/prot.20735. [DOI] [PubMed] [Google Scholar]
- Oldfield CJ, Cheng Y, Cortese MS, Brown CJ, Uversky VN, Dunker AK. Comparing and combining predictors of mostly disordered proteins. Biochemistry. 2005a;44:1989–2000. doi: 10.1021/bi047993o. [DOI] [PubMed] [Google Scholar]
- Oldfield CJ, Cheng Y, Cortese MS, Romero P, Uversky VN, Dunker AK. Coupled folding and binding with alpha-helix-forming molecular recognition elements. Biochemistry. 2005b;44:12454–12470. doi: 10.1021/bi050736e. [DOI] [PubMed] [Google Scholar]
- Peng K, Radivojac P, Vucetic S, Dunker AK, Obra-dovic Z. Length-dependent prediction of protein intrinsic disorder. BMC Bioinformatics. 2006;7:208. doi: 10.1186/1471-2105-7-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng K, Vucetic S, Radivojac P, Brown CJ, Dunker AK, Obradovic Z. Optimizing long intrinsic disorder predictors with protein evolutionary information. J Bioinform Comput Biol. 2005;3:35–60. doi: 10.1142/s0219720005000886. [DOI] [PubMed] [Google Scholar]
- Puntervoll P, Linding R, Gemund C, et al. ELM server: A new resource for investigating short functional sites in modular eukaryotic proteins. Nucleic Acids Res. 2003;31:3625–3630. doi: 10.1093/nar/gkg545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radivojac P, Iakoucheva LM, Oldfield CJ, Obradovic Z, Uversky VN, Dunker AK. Intrinsic disorder and functional proteomics. Biophys. J. 2007;92:1439–1456. doi: 10.1529/biophysj.106.094045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringe D, Petsko GA. Study of protein dynamics by X-ray diffraction. Methods Enzymol. 1986;131:389–433. doi: 10.1016/0076-6879(86)31050-4. [DOI] [PubMed] [Google Scholar]
- Romero P, Obradovic Z, Kissinger CR, Villafranca JE, Dunker AK. Identifying Disordered regions in proteins from amino acid sequences. IEEE Int. Conf. Neural Networks. 1997;1:90–95. [Google Scholar]
- Romero P, Obradovic Z, Li X, Garner EC, Brown CJ, Dunker AK. Sequence complexity of disordered protein. Proteins. 2001;42:38–48. doi: 10.1002/1097-0134(20010101)42:1<38::aid-prot50>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- Sickmeier M, Hamilton JA, LeGall T, Vacic V, Cortese MS, Tantos A, Szabo B, Tompa P, Chen J, Uversky VN, Obradovic Z, Dunker AK. DisProt: the Database of Disordered Proteins. Nucleic Acids Res. 2007;35:D786–D793. doi: 10.1093/nar/gkl893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tompa P. The functional benefits of protein disorder. Journal of Molecular Structure-Theochem. 2003;666:361–371. [Google Scholar]
- Uversky VN, Dunker AK. Biochemistry. Controlled chaos. Science. 2008;322:1340–1341. doi: 10.1126/science.1167453. [DOI] [PubMed] [Google Scholar]
- Uversky VN, Gillespie JR, Fink AL. Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins. 2000;41:415–427. doi: 10.1002/1097-0134(20001115)41:3<415::aid-prot130>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- Uversky VN, Oldfield CJ, Dunker AK. Showing your ID: intrinsic disorder as an ID for recognition, regulation and cell signaling. J. Mol. Recognit. 2005;18:343–384. doi: 10.1002/jmr.747. [DOI] [PubMed] [Google Scholar]
- Vacic V, Oldfield CJ, Mohan A, Radivojac P, Cortese MS, Uversky VN, Dunker AK. Characterization of molecular recognition features, MoRFs, and their binding partners. J. Proteome Res. 2007a;6:2351–2366. doi: 10.1021/pr0701411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacic V, Uversky VN, Dunker AK, Lonardi S. Composition Profiler: a tool for discovery and visualization of amino acid composition differences. Bmc Bioinformatics. 2007b;8:211. doi: 10.1186/1471-2105-8-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vershon AK, Johnson AD. A short, disordered protein region mediates interactions between the homeodomain of the yeast alpha 2 protein and the MCM1 protein. Cell. 1993;72:105–112. doi: 10.1016/0092-8674(93)90054-t. [DOI] [PubMed] [Google Scholar]
- Wright PE, Dyson HJ. Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. J. Mol. Biol. 1999;293:321–331. doi: 10.1006/jmbi.1999.3110. [DOI] [PubMed] [Google Scholar]
- Wright PE, Dyson HJ. Linking folding and binding. Curr. Opin. Struct. Biol. 2009;19:31–38. doi: 10.1016/j.sbi.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue B, Dunbrack RL, Williams RW, Dunker AK, Uversky VN. PONDR-FIT: a meta-predictor of intrinsically disordered amino acids. Biochim. Biophys. Acta. 2010;1804:996–1010. doi: 10.1016/j.bbapap.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue B, Li L, Meroueh SO, Uversky VN, Dunker AK. Analysis of structured and intrinsically disordered regions of transmembrane proteins. Mol Biosyst. 2009a;5:1688–1702. doi: 10.1039/B905913J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue B, Oldfield CJ, Dunker AK, Uversky VN. CDF it all: consensus prediction of intrinsically disordered proteins based on various cumulative distribution functions. FEBS Lett. 2009b;583:1469–1474. doi: 10.1016/j.febslet.2009.03.070. [DOI] [PMC free article] [PubMed] [Google Scholar]