The genome continues to fascinate and humble its enthusiasts. Any major step in the ladder of scientific discoveries taken toward understanding the genome is soon overshadowed by newer discoveries. The large monotonous macromolecule that Friedrich Miescher, the discoverer of deoxyribonucleic acid (DNA), called the “nuclein,” has only 4 randomly repeating letters (1). This apparent simplicity misled scientists for about 75 years until the surprising discovery that the 4 randomly repeating nucleotides contained Johann Gregor Mendel's “elements of hereditary” (2). It was just as fascinating to discover that the “elements of hereditary” comprised <2% of over 3 billion nucleotide pairs in the genome. The rest was considered “junk DNA.” Then we were humbled to learn that there simply was no “junk DNA” but that signals were posted in every corner of the genome. The discovery of microribonucleic acids, transcribed from introns and intergenic regions, and demonstration of their role in regulating cardiac function are powerful evidence of the significance of the “junk DNA” (3–5). Moreover, the information conveyed by the genome is not restricted to its nucleotide sequence but also comprises an extra layer of complexity imparted by the epigenetic mechanisms (6).
The completion of the draft sequence of 2 sets of the human genome in 2001 (Human Genome Sequencing Consortium and Celera Genomics) was a huge step on the ladder of scientific discoveries (7,8). Yet, the triumph soon faced the challenge of deciphering the meaning of the sequence, i.e., annotation. Comparison of the draft sequence of the 2 genomes documented the abundance of DNA polymorphisms, primarily at the single nucleotide level. Since then, the number single nucleotide polymorphisms (SNPs) has increased to nearly 12 million. A plethora of studies have utilized the existing databases to correlate phenotypes with genotypes. The approach primarily has been SNP-centric, because until now SNPs were considered to be the primary genetic determinants of the interindividual variability in susceptibility to disease, response to treatment (pharmacogenetics), and clinical outcomes.
The genome surprised us once again when Dr. J. Craig Venter's DNA sequence—as a diploid genome sequence—was released in September 2007 (9). The distinction merits clarification, because the Human Genome Sequencing Consortium assembly was a composite sequence of haploids derived from several individuals, and the Celera Genome was a consensus sequence derived from 5 individuals. Surprisingly, 44% of Venter's annotated genes were heterozygous for one or more variants (9). Venter's DNA sequence had 4.1 million DNA sequence variations of which one third were novel. This is remarkable considering that there are already nearly 12 million SNPs in the database. More astonishing was the abundance of insertions, deletions, duplications and other rearrangements in Venter's genome. Importantly, the non-SNP DNA variations involved 74% of the variant nucleotides. The information changes the landscape and shifts the paradigm from an SNP-centric to a genome-centric approach in genetic studies of complex phenotypes. It is probably fair to suggest that the genome of each individual is “private.”
The genome, through the information imprinted in its DNA sequence and beyond, is an important determinant of biologic phenotypes. The magnitude of its contribution is context dependent but by and large is a lion's share of the burden. However, genomic information, although crucial for the development of the phenotype, is not usually complete. The underlying extra complexity of a phenotype arises from intertwined dynamic and nonlinear interactions between the genome—in the broader definition—and the host's personal environment. Therefore, biologic phenotypes, including all in health and disease, are for the most part the products of the genome interacting with environmental factors.
Advances in molecular genetic techniques have afforded the opportunity to identify the genetic determinants of the human phenotype. Accordingly, the molecular genetics of various Mendelian disorders have been delineated (10). In contrast, our understanding of the molecular genetic basis of complex diseases has remained rudimentary. Genome-wide association study (GWAS) has raised considerable enthusiasm for detecting, without prior acknowledge, the susceptibility alleles for complex traits such as coronary atherosclerosis and myocardial infarction (MI) (11–14). Unfortunately, the first set of GWAS was performed mostly in unmatched case and control subjects, and therefore the results are subject to the confounding effects of the covariates, such as the known risk factors for coronary atherosclerosis and MI.
A much more restrictive approach to genetic studies of complex traits is the candidate gene approach, where association of the phenotype with variants of a biologically plausible gene is analyzed. As in GWAS, the study design and robust data analysis are fundamental to gaining meaningful results. In all association studies, whether genome-wide or candidate-gene, replication is considered to be an important step toward validation (15). In the present issue of the Journal, 3 studies have attempted to replicate and extend the results of an earlier study showing a modest association between the nonsynonymous tryptophan719arginine (W719R) polymorphism in KIF6, encoding kinesin family member 6, and ischemic heart disease (16). Likewise, the R variant of KIF6 was more common in cases with MI than in control subjects in the CARE (Cholesterol and Recurrent Events) study, the WOSCOPS (West of Scotland Coronary Prevention Study), and the WHS (Women's Health Study) populations (17,18). On a similar theme, albeit a completely different phenotype, carriers of the R allele had a greater reduction in the coronary events rate in response to treatment with atorvastatin in the PROVE IT-TIMI 22 (Pravastatin or Atorvastatin Evaluation and Infection Therapy: Thrombolysis in Myocardial Infarction-22) study population, which was independent of changes in plasma low-density lipoprotein cholesterol (LDL-C) and C-reactive protein (CRP) levels (19).
These studies collectively raise interest in KIF6 as a possible candidate gene in susceptibility to MI, coronary atherosclerosis, and clinical response to statins. They have considerable strengths, including the relatively large sample size of the study populations, carefully phenotyped participants, and concordant findings in separate datasets. Nonetheless, the results at best should be considered provisional pending validation through experimentation. The possibility of a spurious association should be considered. In a candidate gene approach, first and foremost is to establish the biologic plausibility of the candidate gene. Unfortunately, there is a paucity of information on KIF6, which encodes a member of the kinesin-9 family with a yet to be defined function (20). The superfamily of motor protein kinesins is involved in the transportation of cellular cargo, such as protein complexes along the microtubules (20). The choices of KIF6 and the W719R SNP as the candidate gene were based on the results of an earlier study which reported a borderline association of the risk of ischemic heart disease with the W719R SNP (16). Like the previous study, the present studies also failed to establish the biologic plausibility of KIF6 in susceptibility to coronary atherosclerosis and MI. Likewise, the results of the pharmacogenetic study, which showed genotype-dependent differences in the primary clinical outcome rate in the atorvastatin but not in the pravastatin subgroups, require a plausible biologic explanation. It is equally desirable to understand the basis for the independence of the association between the clinical event rates and the KIF6 genotypes from the response of the plasma LDL-C and CRP levels to statin therapy. These shortcomings are further compounded by the fact that KIF6 is not known to be expressed in the vasculature, the primary site of atherosclerosis. It is expressed at relatively low levels in the brain, connective tissue, colon, eye, pharynx, skin, and testes, organs not known to play a direct role in susceptibility to atherosclerosis.
Thus, there is insufficient evidence for the biologic plausibility of KIF6 as a candidate gene for risk of atherosclerosis and MI or the response of clinical outcome to statins. Nevertheless, biologic plausibility, although an integral part in the candidate gene approach, is only based on existing knowledge and alone is not sufficient to refute the results. Another common and largely arbitrary approach is the pooling of 2 of the 3 genotypes in data analysis in the absence of supportive biologic evidence. It increases the chance of a spurious association and could be justified when the risk allele is rare, the sample size of the study population is small, and the supportive biologic evidence exists. However, W719R is a common SNP, and the sample sizes of the study populations are large enough to afford independent analysis of the genotypes. The lack of a gene dose effect also needs biologic support and is a concern. Finally, the studies are restricted to analysis of a single SNP with a yet-to-be-defined function. The approach should have been complemented with the comprehensive genetic screening of the KIF6 locus, which is known to contain several additional SNPs, including common and uncommon nonsynonymous SNPs. Despite these serious deficiencies, the results, if validated through experimentation, could portend a new pathway for the pathogenesis of coronary atherosclerosis and MI and possibly pharmacogenetics of statins.
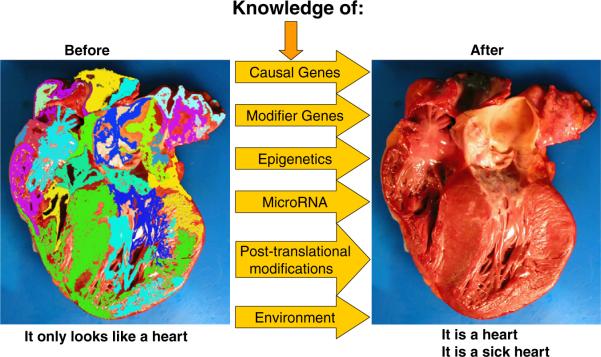
The current SNP-centric approach to genetic studies of complex traits when complemented with mechanistic studies could provide valuable insight into the pathogenesis of disease and lead to identification of new prognosticators, diagnostic markers, and therapeutic targets. The SNP-centric approach, however, at best could encompass only one-fourth of the variant nucleotides in the genome (9). A complete paradigm shift from the current SNP-centric to a more comprehensive genome-centric approach is necessary to capture the full potential of the genome. Given the rapid pace of evolution of sequencing techniques, a genome-centric approach is becoming a reality. Moreover, given the intricacy of factors that determine complex phenotypes, only a comprehensive pluralistic approach that integrates all of the constituents of the phenotype (Fig. 1), such as genomic, epigenomic, transcriptomic, proteomic, and metabolomic profiles, could propel us toward “personalized” medicine. Otherwise, the clinical utility of genetic association studies in individualization of diagnosis, risk stratification, prevention, and treatment will fall short of Dr. Koshland's cha-cha-cha theory of scientific discoveries (21).
Figure 1. Determinants of a Complex Phenotype.
RNA = ribonucleic acid.
Acknowledgments
Supported by grants from the National Heart, Lung, and Blood Institute, a Clinical Scientist Award in Translational Research from Burroughs Wellcome Fund, and the TexGen Fund from the Greater Houston Community Foundation.
Footnotes
Editorials published in the Journal of the American College of Cardiology reflect the views of the authors and do not necessarily represent the views of JACC or the American College of Cardiology.
REFERENCES
- 1.Miersky AE. The discovery of DNA. Sci Am. 1968;218:78–88. doi: 10.1038/scientificamerican0668-78. [DOI] [PubMed] [Google Scholar]
- 2.Hershey AD, Chase M. Independent function of viral protein and nucleic acid in growth of bacteriophage. J Gen Physiol. 1952;36:39–56. doi: 10.1085/jgp.36.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Care A, Catalucci D, Felicetti F, et al. MicroRNA-133 controls cardiac hypertrophy. Nat Med. 2007;13:613–8. doi: 10.1038/nm1582. [DOI] [PubMed] [Google Scholar]
- 4.Zhao Y, Ransom JF, Li A, et al. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1–2. Cell. 2007;129:303–17. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]
- 5.van Rooij E, Sutherland LB, Qi X, Richardson JA, Hill J, Olson EN. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science. 2007;316:575–9. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- 6.Jiang YH, Bressler J, Beaudet AL. Epigenetics and human disease. Annu Rev Genomics Hum Genet. 2004;5:479–510. doi: 10.1146/annurev.genom.5.061903.180014. [DOI] [PubMed] [Google Scholar]
- 7.Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 8.Venter JC, Adams MD, Myers EW, et al. The sequence of the human genome. Science. 2001;291:1304–51. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 9.Levy S, Sutton G, Ng PC, et al. The diploid genome sequence of an individual human. PLoS Biol. 2007;5:e254. doi: 10.1371/journal.pbio.0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marian AJ, Brugada R, Roberts R. In: Cardiovascular diseases due to genetic abnormalities. Fuster V, editor. Hurst's The Heart. McGraw-Hill; New York, NY: 2004. pp. 1747–83. [Google Scholar]
- 11.Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–78. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McPherson R, Pertsemlidis A, Kavaslar N, et al. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316:1488–91. doi: 10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Samani NJ, Erdmann J, Hall AS, et al. Genomewide association analysis of coronary artery disease. N Engl J Med. 2007;357:443–53. doi: 10.1056/NEJMoa072366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Helgadottir A, Thorleifsson G, Manolescu A, et al. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007;316:1491–3. doi: 10.1126/science.1142842. [DOI] [PubMed] [Google Scholar]
- 15.NCI-NHGRI Working Group on Replication in Association Studies Replicating genotype phenotype associations. Nature. 2007;447:655–60. doi: 10.1038/447655a. [DOI] [PubMed] [Google Scholar]
- 16.Morrison AC, Bare LA, Chambless LE, et al. Prediction of coronary heart disease risk using a genetic risk score: the atherosclerosis risk in communities study. Am J Epidemiol. 2007;166:28–35. doi: 10.1093/aje/kwm060. [DOI] [PubMed] [Google Scholar]
- 17.Iakoubova OA, Tong CH, Rowland CM, et al. Association of the Trp719Arg polymorphism in kinesin-like protein 6 with myocardial infarction and coronary heart disease in 2 prospective trials: the CARE and WOSCOPS trials. J Am Coll Cardiol. 2008;51:435–43. doi: 10.1016/j.jacc.2007.05.057. [DOI] [PubMed] [Google Scholar]
- 18.Shiffman D, Chasman DJ, Zee RY, et al. A kinesin family member 6 variant is associated with coronary heart disease in the Women's Health Study. J Am Coll Cardiol. 2008;51:444–8. doi: 10.1016/j.jacc.2007.09.044. [DOI] [PubMed] [Google Scholar]
- 19.Iakoubova OA, Sabatine MS, Rowland CM, et al. Polymorphism in KIF6 gene and benefit from statins after acute coronary syndromes: results from the PROVE IT–TIMI 22 study. J Am Coll Cardiol. 2008;51:449–55. doi: 10.1016/j.jacc.2007.10.017. [DOI] [PubMed] [Google Scholar]
- 20.Miki H, Okada Y, Hirokawa N. Analysis of the kinesin superfamily: insights into structure and function. Trends Cell Biol. 2005;15:467–76. doi: 10.1016/j.tcb.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 21.Koshland J. Philosophy of science: the cha-cha-cha theory of scientific discovery. Science. 2007;317:761–2. doi: 10.1126/science.1147166. [DOI] [PubMed] [Google Scholar]