Abstract
Background
Peripartum cardiomyopathy (PPCM) describes dilated cardiomyopathy (DCM) without known cause that occurs during the last month of pregnancy to 5 months postpartum. A related term, pregnancy associated cardiomyopathy (PACM), refers to DCM onset earlier in pregnancy. Multiple studies have focused on inflammatory, immunologic and environmental etiologies. An alternative hypothesis is that PPCM and PACM result, in part, from genetic cause. In this study, we sought to test the hypothesis that rare DCM-associated mutations underlie a proportion of pregnancy-associated or peripartum cardiomyopathy cases.
Methods and Results
A systematic search of our DCM database designed for family-based genetic studies was undertaken for cases associated with pregnancy and the postpartum period; in the identified cases, clinical and molecular genetic data, including exonic and near intron/exon boundaries of DCM genes were analyzed. Of 4110 females from 520 pedigrees in the Familial Dilated Cardiomyopathy (FDC) Research Project database, we identified 45 cases of PPCM/PACM. Evidence of familial clustering with DCM was present in 23 unrelated cases. Of the 45 cases, 19 had been resequenced for known DCM genes, and six carried mutations. Five had PPCM, of which 3 were familial with mutations found in MYH7, SCN5A, and PSEN2, and 2 were sporadic with mutations in MYH6 and TNNT2. One case had PACM and carried a mutation in MYBPC3.
Conclusion
These findings suggest that a proportion of PPCM/PACM results from genetic cause.
Keywords: genetics, peripartum cardiomyopathy, dilated cardiomyopathy
Introduction
In 1971, Demakis introduced the term peripartum cardiomyopathy (PPCM) and proposed diagnostic criteria: heart failure (HF) onset in the last month of pregnancy or within 5 months postpartum; no determinable etiology for the cardiac failure; and no heart disease before the last month of pregnancy.1, 2 Since then, additional diagnostic criteria have been added, including reduced left ventricular ejection fraction (LVEF) and left ventricular enlargement (LVE).3 A related term, early pregnancy associated cardiomyopathy (PACM), was used in a study to refer to onset occurring before the last month of pregnancy.4 In that study, the clinical characteristics of 23 women identified as PACM (earliest diagnosis reported at week 17 gestational age) were compared to those of 100 women with PPCM; no clinical differences were observed between groups.4 Phenotypically, PPCM and PACM are indistinguishable from dilated cardiomyopathy (DCM), which is characterized by systolic failure and LV enlargement. Thus, PPCM/PACM can be conceived as phenotypic descriptors for DCM occurring during or after pregnancy.
Considerable recent work has established a genetic basis for a proportion of DCM of unknown cause,5 but a genetic hypothesis for PPCM/PACM has not been formally tested. However, several lines of evidence suggest that some proportion of PPCM may result from genetic cause. Familial clustering of PPCM has been noted,6–11 from which possible genetic cause may be inferred. Also, a novel mutation in MYBPC3 in postpartum DCM,12 and a mutation in PDLIM3 in a woman with PACM13 have been reported. Further, female mice with a deletion of STAT3 develop PPCM, where STAT3 mediates hypertrophy, myocardial angiogenesis and protects the heart from oxidative stress.14 DCM, after identifiable causes have been excluded (otherwise known as idiopathic dilated cardiomyopathy, or IDC) is familial (familial dilated cardiomyopathy; FDC) in 20–35% of cases,5 and rare variant mutations in over 30 genes have been implicated in causing familial and some apparently sporadic cases.15
Therefore, to test the hypothesis that rare variant DCM mutations underlie a proportion of PPCM/PACM cases, a systematic search of our DCM database designed for family-based genetic studies was undertaken for cases associated with pregnancy and the postpartum period. In the identified cases, available clinical and molecular genetic data were analyzed.
Materials and Methods
Patient population
Subjects with IDC (with or without FDC) were enrolled, as previously described.16 Written, informed consent was obtained. A blood sample was obtained for genetic research. Medical family history was obtained and a pedigree was constructed. Medical records were obtained to confirm IDC and assign familial or sporadic status. IDC was defined as LV enlargement with systolic dysfunction, and coronary artery disease, cardiotoxic exposures and other known causes ruled out.16 FDC cases were defined as those in which the patient and at least one relative had IDC. Kindreds with confirmed and probable familial disease were classified as FDC. Individuals with a negative family history or possible FDC were classified as sporadic.16 Patient information was stored in Progeny, a relational database (Progeny Software, South Bend, IN).
Database Query and Medical Records Review
A database query among 4110 females whose data was part of the FDC study cohort, either from enrollment or reported family history, was conducted for DCM cases associated with pregnancy and the postpartum period. Medical records and family history were reviewed for each of the identified cases.
Selection Criteria
Cases were selected if medical records or family history intake indicated a diagnosis or history of PPCM or PACM. All available medical records were reviewed for onset during pregnancy or 5 months after pregnancy and cardiovascular data indicating an ejection fraction less than 50% and/or M-mode fractional shortening less than 30%, and a left ventricular end-diastolic dimension greater than the 97.5th percentile of a gender- and height-based method,17 as previously reported.16
Genetic Data
Genetic data from comprehensive resequencing studies were available from 19 unrelated subjects, where all coding exons and near intron/exon boundaries underwent bidirectional capillary-based Sanger sequencing, as previously described.15, 18–20 Fourteen cases were resequenced for 14 genes, including CSRP3, LDB3, MYH7, SCN5A, TCAP, TNNT2, LMNA, PSEN1 and PSEN2, as reported,15, 18, 19 as well as MYBPC3, MYH6, TNNIC, TNNI3, and TPM.20 An additional case was resequenced for the above mentioned genes except for LMNA, PSEN1 and PSEN2. One case was resequenced for all genes except for PSEN1 and PSEN2. One case was only sequenced for the LMNA gene. Another case was resequenced for LMNA, PSEN1, PSEN2, MYBPC3, MYH6, TNNIC, TNNI3, and TPM. Her father, who had IDC, was resequenced for the remaining six of 14 genes (CSRP3, LDB3, MYH7, SCN5A, TCAP, and TNNT2). One case was only sequenced for the SCN5A mutation identified in a relative with DCM.
Results
Clinical data
A search of 4110 females from 520 families enrolled in the Familial Dilated Cardiomyopathy Research Project cohort identified 45 cases with PPCM/PACM (Table 1). This group includes two first cousins, two sisters, and a half-aunt/half-niece pair for a total of 42 unrelated cases. Of the 42 unrelated cases, 23 had familial disease and 12 were apparently sporadic. The remaining 7 unrelated cases had insufficient family data to be categorized. Nineteen women met PPCM criteria. Two of these 19 PPCM cases were sisters; their mother had DCM (diagnosed at 60 years old). Eight other cases met criteria for PACM. For the remaining 18 cases, insufficient clinical data were available for cases to be categorized (Table 1).
Table 1.
Subject Demographics
| All cases | PPCM | PACM | ||
|---|---|---|---|---|
| A. Race, ethnicity, age and parity | ||||
| Number of cases | 45 | 19 | 8 | |
| Race and Ethnicity | ||||
| Non-Hispanic White | 24 | 12 | 4 | |
| Hispanic White | 5 | 2 | 1 | |
| Non-Hispanic African American | 10 | 5 | 2 | |
| Non-Hispanic American Indian/Alaska Native | 1 | 0 | 1 | |
| Unknown | 5 | 0 | 0 | |
| Age at diagnosis, median, (n) | 27 (33) | 28 (19) | 25 (8) | |
| Parity, mean, (n) | 1.58 (33) | 2 (19) | 0.625 (8) | |
| Parity, range | 0–7 | 0–7 | 0–3 | |
| B. Familial or sporadic disease in unrelated cases who had family data available for analysis | ||||
| All | 35 | 18 | 8 | |
| FDC | 23 | 11 | 6 | |
| IDC | 12 | 7 | 2 | |
Medical records were available for 32 and cardiovascular characteristics of these cases are provided (Table 2).
Table 2.
Clinical Characteristics of Cases with Cardiovascular Functional Data
| Clinical presentation, n | 32 |
| Heart Failure | 26 |
| Arrhythmia | 11 |
| Other (fever, infection, nausea) | 3 |
| Echocardiographic findings, n | 26 |
| Left ventricular end-diastolic dimension in mm, n, mean, SD | 24, 62.6 ± 7.3 |
| Left ventricular end-diastolic dimension Z score, n, mean, SD | 21, 4.8 ± 1.7 |
| Ejection fraction, n, mean, SD | 24, 23.9 ± 11.1 |
Pedigree and Genetic Data
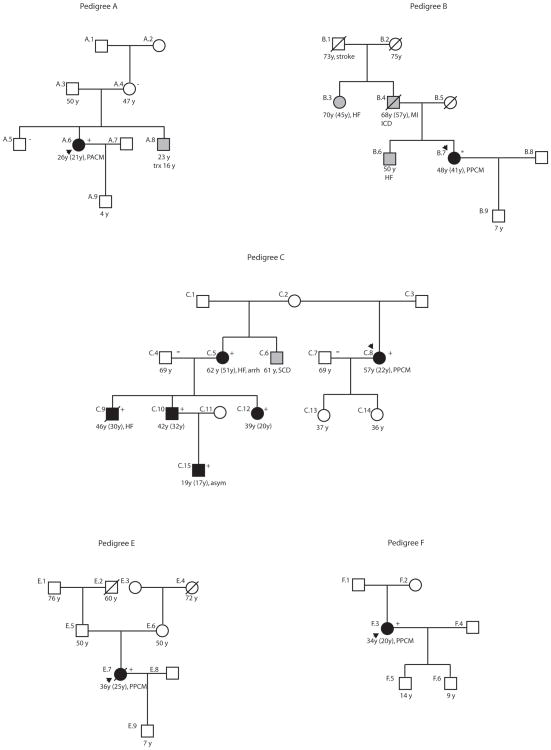
Genetic data were available for 19 (13 familial, 6 sporadic) of the 42 unrelated cases. The remaining cases were not sequenced either because the family’s proband was sequenced and a mutation was not found, or the subject was not enrolled in the FDC study so that the subject’s DNA was not available when the sequencing occurred. Nonsynonymous mutations were identified in six cases, each from different genes (Figure 1, Table 3 15, 19–21): myosin binding protein C (MYBPC3),20 β-myosin heavy chain 7 (MYH7),15 cardiac troponin T2 (TNNT2),15, 21 α– myosin heavy chain 6 (MYH6),20 sodium channel, voltage-gated, type V, alpha subunit (SCN5A),15 and presenilin 2 (PSEN2).19 Medical records confirmed a PPCM diagnosis in five of the six cases with mutations. These five cases included three with familial disease (Pedigree B, MYH7; Pedigree C, SCN5A; and Pedigree D, PSEN2) and two with sporadic disease (Pedigree E, MYH6, and Pedigree F, TNNT2). One, who carried a MYBPC3 mutation,20 met PACM criteria (Pedigree A) and had familial disease.
Figure 1. PPCM/PACM Pedigrees Associated with DCM Mutations.
Pedigrees have been labeled by letter, which correspond to their respective mutation as shown in Table 3. Pedigree D has been previously published and is not shown.19 Squares represent males, circles females. An arrowhead denotes the proband. A diagonal line marks deceased individuals. Solid symbols indicate confirmed or possible PPCM/PACM or IDC with or without heart failure; shaded symbols represent a family history suggestive of DCM that was not confirmed with medical records. Open symbols represent unaffected individuals. For PPCM/PACM or IDC cases, the current age or age at death, the age at diagnosis (in parenthesis), and the clinical presentation are shown. The presence or absence of a mutation is indicated by a + or − symbol, respectively. An asterisk represents a homozygous or hemizygous subject. Arrh=arrhythmia; asym=asymptomatic; HF= heart failure; ICD=implantable cardiac defibrillator; MI= myocardial infarction; PPCM=peripartum cardiomyopathy; PACM=pregnancy associated cardiomyopathy; SCD=sudden cardiac death; trx=heart transplant.
Table 3.
Clinical Characteristics of PPCM/PACM Cases and Family Members with DCM Mutations
| Subject | Gender | Diagnosis | Age at diagnosis | Gestational age at diagnosis (PPCM or PACM cases) | Parity | ECG/arrhythmia | LVEDD, mm (Z score) | Ejection fraction (%) | Comment |
|---|---|---|---|---|---|---|---|---|---|
| Pedigree A (FDC): MYBPC3 Arg272Cys 20 | |||||||||
| A.6 | F | PACM | 21 | 34 weeks | 0 | sinus tach, poor R wave progession | 66 (5.6) | 10 | |
| Pedigree B (FDC): MYH7 Gly1808Ala 15 | |||||||||
| B.7 | F | PPCM | 40 | 1 week post partum | 1 | tachycardia, IVCD, poor R wave progression, LAE, NSSTT | 64 (4.5) | 15 | homozygous or hemizygous mutation |
| Pedigree C (FDC): SCN5A Arg222Gln 15 | |||||||||
| C.8 | F | PPCM | 22 | post delivery | 2 | atrial ectopic rhythm | NA | 35 | |
| C.5 | F | IDC | 51 | NA | 3 | LAE, PACs, PVCs | 57 (3.7) | 30 | HF |
| C.9 | M | IDC | 30 | NA | NA | LBBB | 84 (7.1) | 15 | HF, ICD |
| C.10 | M | IDC | 32 | NA | NA | bigem | 74 (5.6) | 18 | HF, ICD |
| C.12 | F | IDC | 20 | NA | 0 | NA | 70 (6.6) | 15 | HF, ICD |
| C.15 | M | IDC | 17 | NA | NA | multifocal PVCs, bigem | 78 (6.3) | 13 | presented with arrhythmia |
| Pedigree D (FDC): PSEN2 Ser130Leu 19 | |||||||||
| F | PPCM | 37 | 2 weeks post partum | 7 | sinus tach NSSTT | 69 (5.8) | 18 | ||
| Pedigree E (IDC): MYH6 Arg568Cys 20 | |||||||||
| E.7 | F | PPCM | 25 | few days post partum | 1 | NA | 62.5 (4.9) | reduced | |
| Pedigree F (IDC): TNNT2 Arg159Gln 15, 21 | |||||||||
| F.3 | F | PPCM | 20 | 1 week post partum | 1 | NA | NA | NA | |
bigem, bigeminy; HF, heart failure; ICD, implantable cardiac defibrillator; IVCD, intraventricular conduction delay; LAE, left atrial enlargement; LBBB, left bundle branch block; NSSTT, non-specific ST-T wave changes; PAC, premature atrial complexes; PACM, pregnancy associated cardiomyopathy; PPCM, peripartum cardiomyopathy; PVC, premature ventricular complex; tach, tachycardia.
Pedigrees with Familial Disease
Pedigree A
The proband, of non-Hispanic white ethnicity, presented with shortness of breath (SOB) and pedal edema. She had advanced HF and required urgent cardiac transplant within one month postpartum. A diagnosis of PACM was made (Table 3). She carried a MYBPC3 Arg272Cys variant (absent in 246 controls) that was not detected in her unaffected mother or brother.20 This variant has been previously reported in one subject with DCM.22
Pedigree B
The proband, of non-Hispanic African American ethnicity, presented with progressive dyspnea that began in the last trimester of pregnancy. PPCM was diagnosed (Table 3). She was homozygous or hemizygous for a non-synonymous MYH7 Gly1808Ala variant not seen in 253 controls.15 According to pedigree intake, she had a family history consistent with FDC; however, DNA from her reportedly affected relatives was not available and thus segregation could not be assessed.
Pedigree C
The proband, of non-Hispanic white ethnicity, presented with palpitations, dyspnea on exertion and dizzy spells after the birth of her second child. PPCM was diagnosed (Table 3). She carried a SCN5A (Arg222Gln) variant absent in 253 controls.15 The variant segregated in family members with DCM.
Pedigree D
The proband, of non-Hispanic white ethnicity, presented with severe SOB and orthopnea two weeks postpartum. PPCM was diagnosed. Complaints of leg edema and intermittent chest pain noticed during the eighth month of pregnancy were attributed to normal pregnancy stress. A PSEN2 Ser130Leu variant that was absent in 413 controls was identified; it segregated with DCM in the family. The pedigree and molecular and functional data have been previously published.19
Pedigrees with Sporadic Disease
Pedigree E
The proband, a Hispanic white female, developed SOB and edema a few days after delivery of her first child. Chest X-ray showed cardiomegaly. She was diagnosed with PPCM (Table 3). A biventricular assist device was implanted and she eventually required heart transplantation. A novel MYH6 Arg568Cys variant not seen in 246 controls was detected.20 DNA from additional relatives was not available.
Pedigree F
The proband, a non-Hispanic white female, presented with paroxysmal nocturnal dyspnea, cardiomegaly on chest X-ray, and HF. PPCM was diagnosed one week postpartum (Table 3). A nonsynonymous TNNT2 Arg159Gln variant was identified that occurred at a conserved site and was absent in 253 controls.15 Functional studies demonstrated decreased calcium sensitivity, which indicated that this mutation was likely to be disease causing.21
Discussion
From our database designed for DCM genetic studies, we present cases of DCM onset occurring in pregnancy or the immediate postpartum period and their molecular genetic rare variant data, suggesting genetic causation. Among the 4110 females in the 520 pedigrees analyzed in our FDC cohort, we identified 45 PPCM or PACM cases. In six of the 19 cases for which resequencing data were available, mutations were identified in genes that have been previously shown to be associated with DCM. PPCM was diagnosed in five (Pedigrees B, C, D, E, F) and PACM in one (Pedigree A). To our knowledge, this is the first cohort of PPCM/PACM cases with sequencing data from genes that are relevant for genetic DCM.
Identifying the cause or causes of PPCM/PACM has been elusive, and no compelling data have previously supported any one central hypothesis.23 Numerous etiologies have been proposed, including autoimmune processes, myocarditis, abnormal hemodynamic responses to pregnancy, and selenium deficiency.23–26 Other proposed risk factors include maternal age >30 years old, twinning, hypertension, preeclampsia and tocolytic therapy.1, 4, 23, 24, 27 While African descent has also been proposed as a risk factor, this association may be confounded by socioeconomic status in some populations.27 Genetic cause has also been suggested in some studies, as noted above.
Because our 15+ year study has been primarily devoted to identifying patients with DCM who may have familial disease, it is not surprising that some of the participants who we identified, otherwise meeting criteria established for the diagnosis of PPCM or PACM, had familial disease. Previous reports6–11 of individual familial cases are at variance, as is this report, with several series of PPCM cases that have reported sporadic disease. For example, none of the PPCM subjects reported in two key studies2, 4 had positive family histories. One possible explanation for this difference is that the etiologic basis of the PPCM in those reports2, 4 may have been different (and non-hereditary) from that seen in our series of patients. Another possibility is that evidence of familial disease was not detected, as neither of the prior studies2, 4 specified that a family history had been obtained. That a careful, prospective 3–4 generation family history is essential to detect familial disease has been well established for DCM.5, 28, 29 It has also been well established that the family history is insensitive to detect familial DCM compared to clinical screening (history, exam, ECG and echocardiography) of closely related family members. This latter fact has led to the recent guideline recommending clinical screening for first-degree relatives of all patients newly diagnosed with DCM.29 We suggest that this clinical guideline recommendation also be considered for cases of suspected PPCM/PACM, and that a 3–4 generation family history (and consideration of clinical screening of first-degree family members) be integrated into all ongoing PPCM/PACM research study designs.
We also note that some have suggested that the PPCM diagnosis should be distinguished from FDC.30 However, we suggest the alternative possibility, that genetic DCM may underlie a significant proportion of PPCM cases, regardless of a positive family history. This point is illustrated by Pedigrees E and F, where both probands met PPCM criteria and were the only known affected individuals in their family, yet carried possibly or likely disease-causing DCM mutations, respectively. Prospective registries assessing the presence of DCM clinical findings and mutations in first-degree relatives of PPCM/PACM probands will be necessary to further evaluate this possibility.
Limitations
Due to the nature of our study design, we were unable to obtain all cardiovascular data from all subjects with a history of PPCM/PACM. However, we restricted our cardiovascular and clinical genetics assignments to those for whom requisite clinical data were available. The previously published data or the data presented herein to prove causation varied for each mutation. For mutations in two genes, TNNT2 and PSEN2, the previously published functional data19, 21 in concert with the segregation of the variant with DCM in multiple family members supported their role as highly likely disease-causing variants. For other mutations, segregation of DCM with the variant in SCN5A in multiple other family members (Pedigree C, Figure 1), or the prior report of the MYBPC3 variant in association with DCM,22 support their likely disease-causing roles. Mutations in two genes (MYH6, MYH7) had no functional or segregation data available. However, the MYH7 gene encodes the key sarcomeric protein beta myosin heavy chain that has had multiple previous mutations reported in association with DCM,12, 15, 31–33 and the MYH7 mutation identified Pedigree B met usual criteria (a rare, nonsynonymous variant) to be considered as possibly disease-causing. The MYH6 gene encodes alpha myosin heavy chain, another sarcomeric gene with variants that have been previously reported in association with DCM.20, 34 Although the MYH6 variant we identified in Pedigree E was also nonsynonymous and rare, because the role of MYH6 variants in DCM is overall less well established, the evidence that this variant is possibly disease-causing is not as strong as that of the MYH7 variant. We also note that our resequencing data were limited to those genes known to cause DCM in males and females of all ages without regard to the PPCM/PACM diagnosis, so it is possible that genes more relevant to cardiac function during pregnancy or the immediate postpartum period were missed; resequencing studies of additional genes will be required to assess this possibility. We also note that we are unable to specify how representative our cases for PPCM/PACM are, as our study was not specifically designed to identify PPCM/PACM cases per se, and the PPCM/PACM case series have not presented systematically obtained family history and pedigree data. However, our cases met usual criteria for PPCM or PACM, and are relevant to establish that genetic cause may underlie some proportion of PPCM/PACM.
Implications for Clinicians
This report has implications for clinicians caring for PPCM and PACM patients, as well as their families. For PPCM/PACM patients, the approach taken should be the same as that recommended for a new IDC diagnosis,29 which includes consideration of the possibility of FDC and of genetic cause. A genetic evaluation including family history, clinical screening, and genetic counseling and testing should be conducted for the proband and for first-degree relatives.29 Although PPCM/PACM are rare, these conditions may occur more frequently among relatives of patients with IDC, and therefore, reproductive risk counseling about PPCM/PACM is appropriate for female first-degree relatives of probands with IDC in the context of a genetic cardiomyopathy evaluation.29
Conclusion
Mutations associated with DCM were present in some subjects meeting formal criteria for PPCM/PACM, suggesting that a proportion of PPCM/PACM may result from genetic cause, and even in the absence of a disease-positive family history. These findings have implications for further research and may be of critical importance in the management of women with peripartum cardiomyopathy and their families.
Short Commentary/Clinical Summary
Peripartum cardiomyopathy (PPCM) describes dilated cardiomyopathy (DCM) without known cause that occurs during the last month of pregnancy to 5 months postpartum. A related term, pregnancy associated cardiomyopathy (PACM), refers to DCM onset earlier in pregnancy. Despite multiple studies focused on inflammatory, immunologic and environmental etiologies, no unifying hypothesis has been proven. An alternative hypothesis is that PPCM and PACM result from genetic cause. In an effort to identify preliminary support for this hypothesis, a systematic search of a large database, collected for family-based genetic DCM studies over the past 15 years, was undertaken for cases associated with pregnancy and the postpartum period. When cases were identified, available clinical and molecular genetic data were analyzed. Of the 4110 females from 520 pedigrees in the Familial Dilated Cardiomyopathy (FDC) Research Project database, 45 cases of PPCM/PACM were identified, 23 with familial clustering, of which 19 had been resequenced for known DCM genes. Six of these 19 carried mutations in genes previously shown to be associated with DCM. These data indicate that PPCM/PACM may have a genetic basis in some cases. Thus, we recommend that clinicians caring for PPCM/PACM patients be aware that PPCM/PACM may have a genetic basis, and that guidelines for evaluation of genetic cardiomyopathy be followed. Specifically, this includes clinical screening (family history, medical history, exam, ECG, echocardiography) of the patient and her first-degree relatives. Genetic counseling, including reproductive risk counseling about PPCM/PACM, is recommended, in addition to consideration of genetic testing.
Acknowledgments
Funding Sources. This work was supported by NIH award RO1-HL58626 (Dr Hershberger).
Footnotes
Disclosures: None.
References
- 1.Demakis JG, Rahimtoola SH. Peripartum cardiomyopathy. Circulation. 1971;44:964–8. doi: 10.1161/01.cir.44.5.964. [DOI] [PubMed] [Google Scholar]
- 2.Demakis JG, Rahimtoola SH, Sutton GC, Meadows WR, Szanto PB, Tobin JR, Gunnar RM. Natural course of peripartum cardiomyopathy. Circulation. 1971;44:1053–61. doi: 10.1161/01.cir.44.6.1053. [DOI] [PubMed] [Google Scholar]
- 3.Hibbard JU, Lindheimer M, Lang RM. A modified definition for peripartum cardiomyopathy and prognosis based on echocardiography. Obstet Gynecol. 1999;94:311–6. doi: 10.1016/s0029-7844(99)00293-8. [DOI] [PubMed] [Google Scholar]
- 4.Elkayam U, Akhter MW, Singh H, Khan S, Bitar F, Hameed A, Shotan A. Pregnancy-associated cardiomyopathy: clinical characteristics and a comparison between early and late presentation. Circulation. 2005;111:2050–5. doi: 10.1161/01.CIR.0000162478.36652.7E. [DOI] [PubMed] [Google Scholar]
- 5.Burkett EL, Hershberger RE. Clinical and genetic issues in familial dilated cardiomyopathy. J Am Coll Cardiol. 2005;45:969–81. doi: 10.1016/j.jacc.2004.11.066. [DOI] [PubMed] [Google Scholar]
- 6.Pierce JA, Price BO, Joyce JW. Familial occurrence of postpartal heart failure. Arch Intern Med. 1963;111:651–5. doi: 10.1001/archinte.1963.03620290117016. [DOI] [PubMed] [Google Scholar]
- 7.Pearl W. Familial occurrence of peripartum cardiomyopathy. Am Heart J. 1995;129:421–2. doi: 10.1016/0002-8703(95)90032-2. [DOI] [PubMed] [Google Scholar]
- 8.Fett JD, Sundstrom BJ, Etta King M, Ansari AA. Mother-daughter peripartum cardiomyopathy. Int J Cardiol. 2002;86:331–2. doi: 10.1016/s0167-5273(02)00357-1. [DOI] [PubMed] [Google Scholar]
- 9.Voss EG, Reddy CV, Detrano R, Virmani R, Zabriskie JB, Fotino M. Familial dilated cardiomyopathy. Am J Cardiol. 1984;54:456–7. doi: 10.1016/0002-9149(84)90223-6. [DOI] [PubMed] [Google Scholar]
- 10.Fett JD. Peripartum cardiomyopathy (PPCM) in both surrogate and biological mother. Hum Reprod. 2005;20:2666–8. doi: 10.1093/humrep/dei116. [DOI] [PubMed] [Google Scholar]
- 11.Ferguson JE, II, Harney KS, Bachicha JA. Peripartum maternal cardiomyopathy with idiopathic cardiomyopathy in the offspring. A case report. J Reprod Med. 1986;31:1109–12. [PubMed] [Google Scholar]
- 12.Moller DV, Andersen PS, Hedley P, Ersboll MK, Bundgaard H, Moolman-Smook J, Christiansen M, Kober L. The role of sarcomere gene mutations in patients with idiopathic dilated cardiomyopathy. Eur J Hum Genet. 2009;17:1241–9. doi: 10.1038/ejhg.2009.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arola AM, Sanchez X, Murphy RT, Hasle E, Li H, Elliott PM, McKenna WJ, Towbin JA, Bowles NE. Mutations in PDLIM3 and MYOZ1 encoding myocyte Z line proteins are infrequently found in idiopathic dilated cardiomyopathy. Mol Genet Metab. 2007;90:435–40. doi: 10.1016/j.ymgme.2006.12.008. [DOI] [PubMed] [Google Scholar]
- 14.Hilfiker-Kleiner D, Kaminski K, Podewski E, Bonda T, Schaefer A, Sliwa K, Forster O, Quint A, Landmesser U, Doerries C, Luchtefeld M, Poli V, Schneider MD, Balligand JL, Desjardins F, Ansari A, Struman I, Nguyen NQ, Zschemisch NH, Klein G, Heusch G, Schulz R, Hilfiker A, Drexler H. A cathepsin D-cleaved 16 kDa form of prolactin mediates postpartum cardiomyopathy. Cell. 2007;128:589–600. doi: 10.1016/j.cell.2006.12.036. [DOI] [PubMed] [Google Scholar]
- 15.Hershberger RE, Parks SB, Kushner JD, Li D, Ludwigsen S, Jakobs P, Nauman D, Burgess D, Partain J, Litt M. Coding sequence mutations identified in MYH7, TNNT2, SCN5A, CSRP3, LBD3, and TCAP from 313 patients with familial or idiopathic dilated cardiomyopathy. Clin Translational Science. 2008;1:21–6. doi: 10.1111/j.1752-8062.2008.00017.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kushner JD, Nauman D, Burgess D, Ludwigsen S, Parks S, Pantely G, Burkett EL, Hershberger R. Clinical characteristics of 304 kindreds evaluated for familial dilated cardiomyopathy. J Cardiac Failure. 2006;12:422–29. doi: 10.1016/j.cardfail.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 17.Vasan R, Larson M, Levy D, Evans J, Benjamin E. Distribution and categorization of echocardiographic measurements in relation to reference limits. The Framingham Heart Study: formulation of a height- and sex-specific classification and its prospective validation. Circ. 1997;96:1863–73. doi: 10.1161/01.cir.96.6.1863. [DOI] [PubMed] [Google Scholar]
- 18.Parks SB, Kushner JD, Nauman D, Burgess D, Ludwigsen S, Peterson A, Li D, Jakobs P, Litt M, Porter CB, Rahko PS, Hershberger RE. Lamin A/C mutation analysis in a cohort of 324 unrelated patients with idiopathic or familial dilated cardiomyopathy. Am Heart J. 2008;156:161–9. doi: 10.1016/j.ahj.2008.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li D, Parks SB, Kushner JD, Nauman D, Burgess D, Ludwigsen S, Partain J, Nixon RR, Allen CN, Irwin RP, Jakobs PM, Litt M, Hershberger RE. Mutations of presenilin genes in dilated cardiomyopathy and heart failure. Am J Hum Genet. 2006;79:1030–9. doi: 10.1086/509900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hershberger RE, Norton N, Morales A, Li D, Siegfried J, Gonzalez-Quintana J. Coding sequence rare variants identified in MYBPC3, MYH6, TPM1, TNNC1 And TNNI3 from 312 patients with familial or idiopathic dilated cardiomyopathy. Circ Cardiovasc Genet. 2010;3 doi: 10.1161/CIRCGENETICS.109.912345. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hershberger R, Pinto J, Parks S, Kushner J, Li D, Ludwigsen S, Cowan J, Morales A, Parvatiyar M, Potter J. Clinical and functional characterization of TNNT2 mutations identified in patients with dilated cardiomyopathy. Circ Genetics. 2009;2:306–13. doi: 10.1161/CIRCGENETICS.108.846733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ehlermann P, Weichenhan D, Zehelein J, Steen H, Pribe R, Zeller R, Lehrke S, Zugck C, Ivandic BT, Katus HA. Adverse events in families with hypertrophic or dilated cardiomyopathy and mutations in the MYBPC3 gene. BMC Med Genet. 2008;9:95. doi: 10.1186/1471-2350-9-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pearson GD, Veille JC, Rahimtoola S, Hsia J, Oakley CM, Hosenpud JD, Ansari A, Baughman KL. Peripartum cardiomyopathy: National Heart, Lung, and Blood Institute and Office of Rare Diseases (National Institutes of Health) workshop recommendations and review. Jama. 2000;283:1183–8. doi: 10.1001/jama.283.9.1183. [DOI] [PubMed] [Google Scholar]
- 24.Brown CS, Bertolet BD. Peripartum cardiomyopathy: a comprehensive review. Am J Obstet Gynecol. 1998;178:409–14. doi: 10.1016/s0002-9378(98)80034-3. [DOI] [PubMed] [Google Scholar]
- 25.Lampert MB, Lang RM. Peripartum cardiomyopathy. Am Heart J. 1995;130:860–70. doi: 10.1016/0002-8703(95)90089-6. [DOI] [PubMed] [Google Scholar]
- 26.Bhakta P, Biswas BK, Banerjee B. Peripartum cardiomyopathy: review of the literature. Yonsei Med J. 2007;48:731–47. doi: 10.3349/ymj.2007.48.5.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sliwa K, Fett J, Elkayam U. Peripartum cardiomyopathy. Lancet. 2006;368:687–93. doi: 10.1016/S0140-6736(06)69253-2. [DOI] [PubMed] [Google Scholar]
- 28.Morales A, Cowan J, Dagua J, Hershberger RE. Family history: an essential tool for cardiovascular genetic medicine. Congest Heart Fail. 2008;14:37–45. doi: 10.1111/j.1751-7133.2008.08201.x. [DOI] [PubMed] [Google Scholar]
- 29.Hershberger RE, Lindenfeld J, Mestroni L, Seidman CE, Taylor MR, Towbin JA. Genetic evaluation of cardiomyopathy--a Heart Failure Society of America practice guideline. J Card Fail. 2009;15:83–97. doi: 10.1016/j.cardfail.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 30.Fett JD, Carraway RD, Dowell DL, King ME, Pierre R. Peripartum cardiomyopathy in the Hospital Albert Schweitzer District of Haiti. Am J Obstet Gynecol. 2002;186:1005–10. doi: 10.1067/mob.2002.122423. [DOI] [PubMed] [Google Scholar]
- 31.Kamisago M, Sharma SD, DePalma SR, Solomon S, Sharma P, McDonough B, Smoot L, Mullen MP, Woolf PK, Wigle ED, Seidman JG, Seidman CE. Mutations in sarcomere protein genes as a cause of dilated cardiomyopathy. N Engl J Med. 2000;343:1688–96. doi: 10.1056/NEJM200012073432304. [DOI] [PubMed] [Google Scholar]
- 32.Daehmlow S, Erdmann J, Knueppel T, Gille C, Froemmel C, Hummel M, Hetzer R, Regitz-Zagrosek V. Novel mutations in sarcomeric protein genes in dilated cardiomyopathy. Biochem Biophys Res Commun. 2002;298:116–20. doi: 10.1016/s0006-291x(02)02374-4. [DOI] [PubMed] [Google Scholar]
- 33.Villard E, Duboscq-Bidot L, Charron P, Benaiche A, Conraads V, Sylvius N, Komajda M. Mutation screening in dilated cardiomyopathy: prominent role of the beta myosin heavy chain gene. Eur Heart J. 2005;26:794–803. doi: 10.1093/eurheartj/ehi193. [DOI] [PubMed] [Google Scholar]
- 34.Carniel E, Taylor MR, Fain P, Di Lenarda A, Sinagra G, Lascor J, Ku L, Feiger J, Slavov D, Zhu X, Dao D, Ferguson DA, Mestroni L. Molecular screening of α-myosin heavy chain in patients with dilated and hypertrophic cardiomyopathy. Circ. 2003;108:IV–263. [Google Scholar]