Abstract
Missense PTPN11 mutations cause Noonan and LEOPARD syndromes (NS and LS), two developmental disorders with pleiomorphic phenotypes. PTPN11 encodes SHP2, an SH2 domain-containing protein tyrosine phosphatase functioning as a signal transducer. Generally, different substitutions of a particular amino acid residue are observed in these diseases, indicating that the crucial factor is the residue being replaced. For a few codons, only one substitution is observed, suggesting the possibility of specific roles for the residue introduced. We analyzed the biochemical behavior and ligand-binding properties of all possible substitutions arising from single-base changes affecting codons 42, 139, 279, 282 and 468 to investigate the mechanisms underlying the invariant occurrence of the T42A, E139D and I282V substitutions in NS and the Y279C and T468M changes in LS. Our data demonstrate that the isoleucine-to-valine change at codon 282 is the only substitution at that position perturbing the stability of SHP2's closed conformation without impairing catalysis, while the threonine-to-alanine change at codon 42, but not other substitutions of that residue, promotes increased phosphopeptide-binding affinity. The recognition specificity of the C-SH2 domain bearing the E139D substitution differed substantially from its wild-type counterpart acquiring binding properties similar to those observed for the N-SH2 domain, revealing a novel mechanism of SHP2's functional dysregulation. Finally, while functional selection does not seem to occur for the substitutions at codons 279 and 468, we point to deamination of the methylated cytosine at nucleotide 1403 as the driving factor leading to the high prevalence of the T468M change in LS.
INTRODUCTION
Germline mutations in PTPN11 (MIM# 176876) account for ∼50% of cases of Noonan syndrome (NS; MIM# 163950) (1,2) and the vast majority of cases of LEOPARD syndrome (LS; MIM# 151100) (3,4), two clinically related developmental disorders with features that include short stature, facial dysmorphism, congenital heart defects, as well as skeletal and hematologic anomalies (5,6). A distinct class of missense mutations in the same gene has been identified to occur as somatic events contributing to leukemogenesis (7). PTPN11 encodes SHP2, a widely expressed cytoplasmic protein tyrosine phosphatase functioning as a signal transducer downstream of growth factor and cytokine receptors with roles in signaling pathways controlling developmental processes, hematopoiesis and metabolism (8–10). SHP2 is composed of two tandemly arranged Src homology 2 (N-SH2 and C-SH2) domains at the N-terminus, a catalytic domain (PTP), and a C-terminal tail containing two tyrosyl phosphorylation sites and a proline-rich stretch. Its function is controlled by a complex autoinhibitory mechanism mediated by the interaction of the N-SH2 with the PTP domain (11).
Available records based on more than 800 germline and somatic defects indicate that PTPN11 mutations are almost always missense changes and are not randomly distributed throughout the gene (12). Most of the NS- and leukemia-causing mutations affect residues of the N-SH2 domain located in or close to the PTP-interacting surface. Experimental evidence indicates that these mutations upregulate SHP2's function by destabilizing the catalytically inactive conformation of the protein (12,13). A number of mutations affect residues located within the PTP domain and are predicted to perturb SHP2's function through opposite mechanisms, either promoting dissociation of the N-SH2/PTP domain interaction, and/or perturbing substrate specificity or affecting the phosphatase activity, as observed for the catalytically impaired LS-causing SHP2 mutants (12,14,15). Finally, a few mutations involve residues that are located within the phosphopeptide-binding cleft of each SH2 domain. These lesions are expected to promote SHP2's gain of function by altering the binding affinity and/or specificity for pY-containing signaling partners controlling SHP2's translocation to the membrane and its activation (12,13). Biochemical and molecular data also indicate that the specificity in the amino acid change is relevant to the functional dysregulation of the enzyme and disease pathogenesis. Of note, while the nature of the substitution does not seem to be critical in most cases, suggesting that the amino acid residue being replaced occupies a crucial role in SHP2's function, a largely invariant amino acid change is observed for a number of recurrently affected codons, possibly indicating a specific role for the introduced residue (12).
Here, we analyzed the biochemical behavior and ligand-binding properties of all possible substitutions arising from single-base changes affecting codons 42, 139, 279, 282 and 468 and performed molecular dynamics (MD) simulations to determine the structural effects of selected mutants, showing that different forces drive the specificity of the invariant NS-causing T42A, E139D and I282V and LS-causing T468M lesions. The results of those analyses provide a picture of the complex pathogenic mechanisms involved in PTPN11-related NS and LS.
RESULTS
T42A and E139D amino acid substitutions in the N-SH2 and C-SH2 domains
Available published records of PTPN11 mutations document that codons 42 and 139 are the preferential target of germline mutations involving the phosphopeptide-binding cleft of the N- and C-SH2 domains, respectively. These mutations recur among individuals with NS and have not been documented in LS. At the nucleotide level, the mutations are invariably 124A>G (T42A), and 417G>C or 417G>T (E139D), neither altering a CpG dinucleotide (12). The recurrence rate of these amino acid substitutions is statistically significantly higher than expected by chance (T42A, p = 2.8 × 10−9; E139D, P = 2.0 × 10−9). Both the residues are highly conserved among SHP2 orthologs and paralogs (Supplementary Material, Fig. S1).
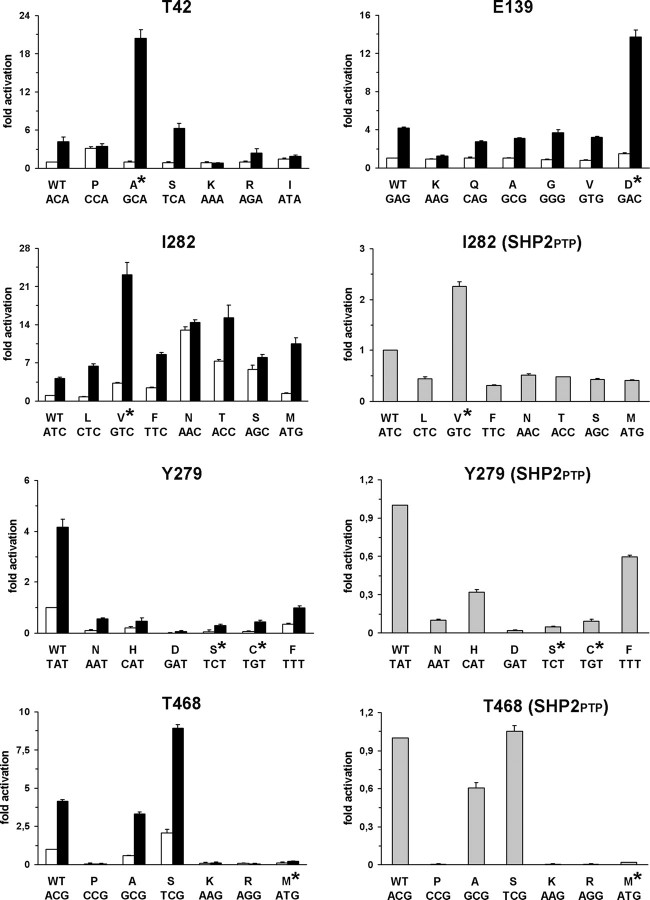
To explore the effects of the T42A and E139D substitutions on protein function and provide an explanation for their invariant occurrence, all possible SHP2 mutants arising from single-base changes affecting those codons (Supplementary Material, Table S1) as well as wild-type enzyme were expressed in bacteria, purified and their phosphatase activities were determined in vitro basally and following stimulation with the protein tyrosine phosphatase non-receptor type substrate 1 (PTPNS1) bisphosphotyrosyl-containing activation motif (BTAM peptide, hereafter) (16) (Fig. 1). Under basal conditions, all but one SHP2 mutant exhibited a relatively low catalytic activity, which was comparable to that of the wild-type protein, the exception being SHP2P42 that showed a threefold increase in substrate dephosphorylation. Following BTAM peptide stimulation, mutants not associated with NS either showed an increase in phosphatase activity comparable to that observed for wild-type SHP2 (SHP2S42, SHP2R42, SHP2A139, SHP2V139, SHP2Q139 and SHP2G139) or were unresponsive to the phosphopeptide (SHP2I42, SHP2P42, SHP2K42 and SHP2K139). In contrast, the NS-causing SHP2A42 and SHP2D139 mutants exhibited dramatically enhanced catalytic activation (Student's t-test, P < 0.001 in all the comparisons), strongly suggesting a higher ligand affinity for both proteins.
Figure 1.
Biochemical characterization of SHP2 mutants. In vitro phosphatase assay of wild-type SHP2 and all possible mutants arising from a single-base change at codons 42, 139, 282, 279 and 468. Catalytic activity was measured as pmoles of phosphate released using pNPP as substrate, basally (white bars) and following stimulation with the PTPNS1 BTAM peptide (black bars). Activities of recombinant wild-type SHP2PTP encoding for the isolated PTP domain and mutants at positions 282, 279 and 468 are also reported (gray bars). Values are expressed as mean ± standard deviation of at least three independent experiments and are normalized to the basal activity of the wild-type enzyme.
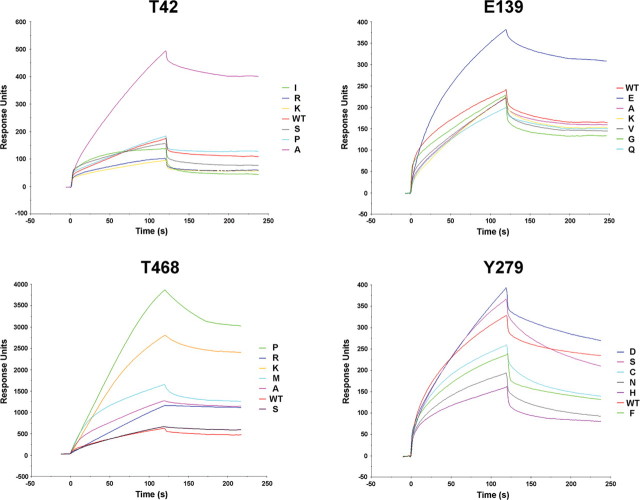
To confirm this hypothesis, the phosphopeptide binding properties of wild-type SHP2 and each of the 12 mutants were assayed by surface plasmon resonance (SPR). Following injection of biotinylated BTAM peptide over a streptavidin-coated sensor chip, recombinant proteins were applied at a concentration of 125 nM, and the amount of protein bound was monitored by the change in refractive index as a function of time at 25°C. SPR analyses demonstrated that T42A and E139D were the only substitutions leading to a significant increase in phosphopeptide binding affinity (Fig. 2), further supporting the proposed molecular mechanism of their pathogenicity.
Figure 2.
SPR analysis. Sensorgrams of the interaction of recombinant wild-type SHP2 or mutants at codons 42, 139, 279 and 468 with the biotinylated PTPNS1 BTAM peptide. Following binding of the phosphopeptide over a streptavidin-coated sensor chip, recombinant proteins were applied, and the amount of interacting protein was monitored by the change in arbitrary response units as a function of time at 25°C.
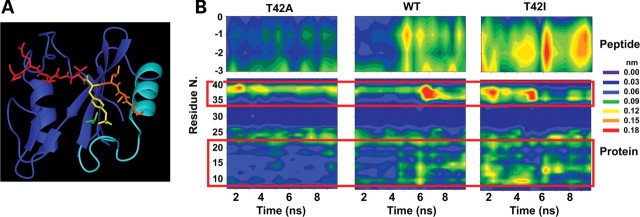
To investigate the structural bases underlying the increased binding affinity of the SHP2A42 mutant, MD simulations of the complex consisting of the N-SH2 domain bound to the PDGFRB's SVLpYTAVQP phosphopeptide were performed for the wild-type protein, and the SHP2A42 and SHP2I42 mutants, the latter included as a representative of the unresponsive proteins (Fig. 3A). On the basis of the crystal structure of the complex, the side chain of T42 forms a hydrogen bond with the pY phosphate, which contributes to the stability of the interaction between the phosphopeptide and the N-SH2 domain (17) and is lost upon substitution of the residue. Other salt bridge, H-bonding and hydrophobic interactions stabilize the binding of the pY to the N-SH2 domain, and MD analyses documented that these interactions were stably retained in all simulations (Supplementary Material, Table S2). This suggested that the major effect of both mutations did not involve a direct perturbation of the binding of the SH2 domain to the pY phosphate. Notably, the total mobility of the peptide Cα atoms during the trajectories, which provides an indication of the overall stability of the protein–peptide complex, differed significantly among complexes (total variance of the coordinates of peptide Cα atoms: SHP2I42: 18.3 Å2; wild-type SHP2: 15.0 Å2; SHP2A42: 8.3 Å2). These differences mapped specifically to the N-terminal segment of the peptide (residues −1 to −3) (Fig. 3B). Similar differences were consistently observed for the total variance of the coordinates of the N-SH2 Cα atoms (SHP2I42: 157 Å2; wild-type SHP2: 119 Å2; SHP2A42: 85 Å2), implying that the higher peptide mobility was correlated to a higher level of protein fluctuations, particularly in the BC loop (residues 34–40), N-terminal helix (residues 13–22) and the loop preceding it (residues 9–12) (Fig. 3B). These protein regions, which constitute the binding site for the N-terminal portion of the phosphopeptide, are spatially contiguous to the mutated residue. These data suggest that T42A and T42I mutations lead, respectively, to a structural stabilization and destabilization of the protein region surrounding the N-terminal portion of the phosphopeptide, which, in turn, affects the stability of the protein–peptide complex.
Figure 3.
MD simulations. (A) Structure of the N-SH2 domain complexed with the PDGFRB SVLpYTAVQP phosphopeptide (pdb entry 1AYA). The peptide is shown in red with the N-terminal residues (−3 to −1) and the phosphorylated tyrosine (pY) highlighted in orange and yellow, respectively. The N-SH2 domain is displayed in blue with the BC loop, N-terminal helix and adjacent loop highlighted in cyan. The side chain of T42 is shown in green. (B) Mobility of Cα atoms in selected protein and phosphopeptide regions during the simulations of wild-type SHP2, SHP2A42 and SHP2I42. Plots report the atom's root mean square fluctuations as a function of time and residue number. Phosphopeptide Cα atoms are numbered with respect to the pY residue (indicated as 0). Red boxes highlight protein regions where significant mobility differences are observed, corresponding to the BC loop (residues 34–40), the N-terminal helix (residues 13–22) and the loop preceding it (residues 9–12).
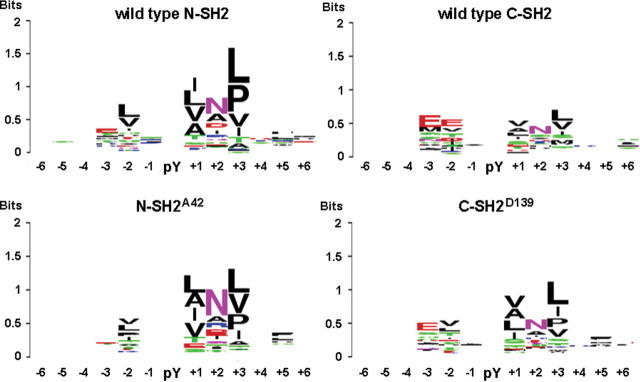
Aside from their effect on binding affinity, mutations involving residues located within the phosphopeptide-binding cleft of each SH2 domain might also perturb the recognition specificity for pY-containing signaling partners. To explore this possibility, the isolated wild-type and NS-causing mutated forms of each SH2 domain were expressed in bacteria and challenged against an array of 6400 pY-containing peptides known or predicted to be phosphorylated in vivo (see Materials and Methods). Consistent with the available published records based on combinatorial phosphopeptide libraries (18–20), wild-type N-SH2 and C-SH2 domains displayed unique features, notwithstanding some overlap in recognition specificity. Of the 73 and 78 phosphopeptides that were identified as high-affinity binders of the N-SH2 and C-SH2 domains, respectively, only 24 were shared between the two domains (Table 1 and Supplementary Material, Table S3). Such a difference in binding specificity is graphically represented by the sequence logo generated for each domain, which summarizes the residue enrichment at the positions flanking the phosphotyrosine in the best ligand peptides (Fig. 4). As indicated by the height of vertical stacks in each logo, the positions with the highest information content, i.e. residues mediating binding specificity, were those at positions +1, +2 and +3 from the phosphotyrosine. Residues on the N-terminal side of the phosphotyrosine (positions –2 and –3) also were found to contribute significantly to specificity, a notable difference from most SH2 domains. As is apparent from the sequence logos, the two domains displayed different binding preferences with the C-SH2 domain being characterized by more promiscuous behavior (Fig. 4). The analysis performed on the N-SH2A42 mutant domain documented that it retained a recognition specificity similar to that of its wild-type counterpart, as summarized by the relatively well-conserved logo (Fig. 4) and high percentage of shared binders (Table 1 and Supplementary Material, Table S3). Consistent with the SPR data, this mutant was found to bind to 156 phosphopeptides with intensity higher than the median plus 1 standard deviation, which was significantly greater compared with the number of binders observed for the wild-type domain, indicating a higher overall affinity (Table 1). In contrast, the E139D change was found to perturb the specificity of the C-SH2 domain considerably, making it more similar to that of the N-SH2 domain as indicated by the preference for hydrophobic residues at positions –2, +1, +3 and +5 (Fig. 4). Consistent with this finding, the C-SH2D139 domain shared more than half of its binders with the wild-type N-SH2 domain, having an overall correlation coefficient of 0.62, which was significantly higher than that observed for the wild-type C-SH2 domain (Table 1 and Supplementary Material, Table S3). Among the high-affinity binders recognized by the wild-type N-SH2 domain, a number of established SHP2 interactors, including PDGFRB (21,22), FRS2 (23), IRS1 (24), GAB2 (25,26) and SSTR2 (27), were shared with the C-SH2D139 mutant but not with the wild-type counterpart.
Table 1.
Number of interacting phosphopeptides, and specific and shared binders of SHP2's wild-type N-SH2 and C-SH2 domains and related NS-causing mutants
| Interacting peptides | N-SH2 | C-SH2 | ||
|---|---|---|---|---|
| Wild-type | T42A | Wild-type | E139D | |
| Total number of bindersa | 73 | 156 | 78 | 90 |
| Specific bindersb | 6 | 89 | 31 | 43 |
| Shared peptidesc | ||||
| Wild-type N-SH2 | — | 67 (0.72) | 24 (0.27) | 50 (0.62) |
| Wild-type C-SH2 | 24 (0.27) | 24 (0.33) | — | 47 (0.76) |
aInteracting peptides were defined as binders with a signal higher than the average signal plus 1 standard deviation.
bSpecific binders are phosphopeptides that were found to interact exclusively with individual SH2 domains.
cNumber of recognized peptides shared by SH2 domains. The numbers in brackets refer to the Pearson's correlation coefficient resulting from binding experiments performed with the same phosphopeptide array challenged with the two indicated domains. Each experiment was performed in duplicate and showed a correlation coefficient higher than 0.9.
Figure 4.
WebLogo representation of binding recognition specificity of SHP2’s SH2 domains. Sequence logo representation generated for the wild-type N-SH2 and C-SH2 domains (above) and derived mutants bearing the T42A or E139D substitution (below). Each logo abridges the residue enrichment at the positions flanking the phosphorylated tyrosine (pY) in the best ligand peptides and was obtained by comparing the amino acid recurrence of bound peptides with a signal higher than average plus 1 standard deviation. For each position, the overall stack height indicates the sequence conservation, while the height of symbols within each stack indicates the relative frequency of the indicated residue. Residue positions are numbered starting from the pY (indicated as 0).
Overall, these data indicate that the T42A and E139D have different consequences on SH2 domain's function. Whereas the former promotes a substantial increase in phosphopeptide binding affinity without altering appreciably the binding specificity of the N-SH2 domain, the latter perturbs the specificity of the C-SH2 domain to a large extent, making it more similar to that of the N-SH2 domain.
Y279C, I282V and T468M amino acid substitutions in the PTP domain
The 836A>G (Y279C) and 1403C>T (T468M) nucleotide transitions specifically occur in subjects with LS, where they account for more than 80% of total germline events (28). According to the published records, a largely invariant occurrence for the LS-causing Y279C mutation has been established (P = 1.5 × 10−26), while no other amino acid substitution affecting codon 468 has been reported so far (P = 2.7 × 10−23) (Supplementary Material, Table S1). Residue I282, which is always mutated into valine (844A>G) in NS (P = 7.2 × 10−11), was included for direct comparison with residue Y279, since the I282V and Y279C amino acid changes have opposite effects on SHP2's catalytic activity (12). These residues are completely conserved among SHP2's orthologs and paralogs (Supplementary Material, Fig. S1).
Biochemical analysis performed on mutants at codon 282 demonstrated that, with the exception of the SHP2L282 and SHP2M282 proteins that showed activities comparable to that observed for wild-type SHP2 (Fig. 1), mutants exhibited variably increased phosphatase activity basally, indicating that most mutations at this codon perturb the stability of the enzyme in its catalytically inactive conformation. Of note, the activity of the SHP2V282 mutant was only 3-fold higher than that of the wild-type protein and considerably lower than that observed for the SHP2N282, SHP2T282 and SHP2S282mutants, suggesting less destabilizing effect of this amino acid substitution on the N-SH2/PTP interdomain interaction. Following BTAM peptide stimulation, however, the SHP2N282 and SHP2S282 recombinant proteins did not appear to be appreciably further activated and the other mutants exhibited an enhanced catalytic activity that was significantly lower than that observed for the NS-causing SHP2V282 mutant (Student's t-test, P < 0.001, in all comparisons). This finding strongly suggested that amino acid changes resulting from a missense point mutation at codon 282 adversely affect SHP2's catalytic activity with the disease-causing I282V substitution being the sole exception. To test this hypothesis, each single-base mutation was introduced into a construct encoding for the isolated PTP domain (SHP2PTP, hereafter), and in vitro catalytic activity of bacterially expressed proteins was assessed to compare the intrinsic phosphatase activity of mutants. Phosphatase assays demonstrated that the I282V change promoted a 2-fold increase in SHP2PTP's catalytic activation, while all other amino acid substitutions impaired SHP2PTP's function (Student's t-test, P < 0.001, in all comparisons) (Fig. 1). This demonstrated that the isoleucine-to-valine substitution at codon 282 is the only amino acid change perturbing the stability of the catalytically inactive conformation of the protein without weakening catalysis and suggested that upregulated SHP2's function in this mutant results from a combinatorial effect of a mild destabilization of the N-SH2/PTP interdomain interaction and enhanced intrinsic catalytic activity.
We and others recently showed that the LS-associated SHP2C279 and SHP2M468 mutants are characterized by impaired catalytic activity (12,14,15). In vitro phosphatase assays performed on the other 10 mutants resulting from single-base changes affecting codons 279 and 468 documented comparable biochemical behavior for the majority of proteins, including the rare LS-causing SHP2S279 mutant, both basally and following BTAM peptide stimulation (Fig. 1). This finding provided evidence that the impaired phosphatase activity per se does not explain the highly prevalent occurrence of the Y279C and T468M substitutions. The reduced intrinsic catalytic capability of these mutants was confirmed by in vitro biochemical assays performed on recombinant SHP2PTP proteins (Fig. 1), which ruled out the possibility that an increased stability of the N-SH2/PTP interdomain interaction promoted by the replacement of Y279 or T468 might exert any inhibitory structural role on SHP2's catalytic activation.
Aside from impairing catalysis, these mutations were hypothesized to have dominant negative effects resulting from disruption of the basal inhibitory interaction between the N-SH2 and PTP domains and enhanced/prolonged binding to signaling partners (12,14). To verify this model experimentally, SPR assays were performed. All mutants at codon 468 except SHP2S468 exhibited enhanced phosphopeptide binding affinity compared with the wild-type protein (Fig. 2), supporting the idea that these amino acid substitutions cause a significant disruption of the N-SH2/PTP interacting surface. Unexpectedly, the highest binding affinity was documented for the SHP2P468 and SHP2K468 recombinant proteins, which have not been identified in LS so far. To rule out the possibility that the increased phosphopeptide binding affinity of mutants at this codon could depend upon a direct interaction between the impaired catalytic site of mutants and the immobilized phosphopeptide, SPR analysis was performed on recombinant SHP2PTP proteins, and sensorgrams of proteins/peptide interaction demonstrated the inability of the active site to bind the BTAM motif (data not shown), excluding this possibility. Unexpectedly, SPR assays performed on mutants at codon 279 indicated that the wild-type enzyme had a binding affinity comparable or higher than those observed for all of the mutants, including the LS-causing SHP2C279 and SHP2S279 proteins, supporting the view that mutations at this codon do not disrupt the N-SH2/PTP interacting surface significantly (Fig. 2). These data suggest that the Y279C substitution could either lead to a milder dominant negative effect due to impairment of the mutant's catalytic activity or dysregulate SHP2's function through a different, as-yet-uncharacterized molecular mechanism. Of note, the present findings do not provide a clear explanation for the largely invariant occurrence of the Y279C and T468M changes among subjects with LS.
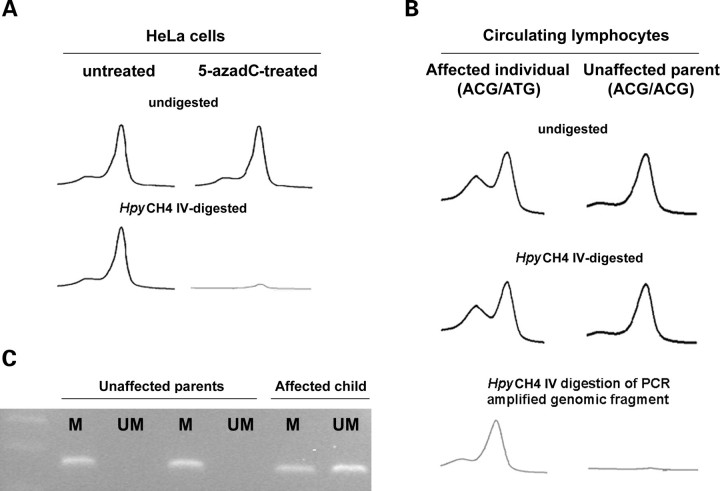
To elucidate the driving forces for the recurrent occurrence of the Y279C and T468M mutations, a genetic mechanism based on sequence-context mutagenesis was considered. Although we were not able to identify a molecular cause that could explain the high recurrence of the Y279C change, we noted that the triplet coding for the threonine at position 468 encompassed a CpG dinucleotide, making it more susceptible to mutagenesis when methylated as a consequence of deamination of the 5-methylcytosine to thymine (29). As a first attempt, the methylation status of this codon was analyzed in HeLa cells. Following treatment with the DNA demethylating agent 5-azadeoxycytidine (5-azadC), a PCR-based restriction analysis assay was performed to check for methylation of the cytosine at position 1403 utilizing the methylation-sensitive enzyme HpyCH4 IV, which was used to cut the unmethylated ACGT sequence specifically. The effect of 5-azadC treatment and endonuclease digestion was determined by PCR amplification of exon 12 of the PTPN11 gene, which encompasses the mutational hot spot. This assay showed loss of the specific PCR product in DNA obtained from 5-azadC-treated cells, compared to the untreated cells, indicating that the HpyCH4 IV site within exon 12 was methylated in both alleles (Fig. 5A). To confirm this finding, genomic DNA obtained from peripheral lymphocytes of four unrelated subjects with LS who carryed the heterozygous 1403C>T transition, as well as their unaffected parents, was HpyCH4 IV-digested, PTPN11 exon 12-amplified and PCR products were analyzed by denaturing high performance liquid chromatography (DHPLC) (Fig. 5B). Consistent with the methylation status of the CpG site, amplification products of HpyCH4 IV-digested or undigested templates resulted in two elution peaks in affected individuals (AmCGT/ATGT genotype) and one single peak in unaffected parents (AmCGT/AmCGT genotype). Efficiency of the HpyCH4 IV treatment was confirmed by digestion of a PCR-amplified genomic fragment encompassing exon 12 and subsequent exon 12 re-amplification of the digested product, which showed the presence of a single peak in the affected individuals and no product in unaffected parents. Finally, the methylation status of the CpG site was definitely proved by using a methylation-specific PCR approach (Fig. 5C). Genomic DNAs of subjects heterozygous for the 1403C>T mutation and unaffected parents were denatured by NaOH treatment and subjected to sodium bisulfite modification of unmethylated cytosines to uracils. PCR amplifications performed using primers designed to amplify either the methylated or the unmethylated alleles confirmed the methylated status of the cytosine at position 1403 in all cases. Taken as a whole, these data provide strong evidence that the CpG dinucleotide within codon 468 is methylated, at least in HeLa cells and human circulating lymphocytes, and strongly suggest that the invariant occurrence of the T468M change in LS is due to the enhanced mutability of this codon as a result of methylation-induced deamination of 5-methylcytosine.
Figure 5.
CpG methylation status at codon 468. (A) DHPLC profiles showing missing of PTPN11 exon 12 PCR product in HeLa cells treated with the DNA demethylating agent 5-azadC and digested with the methylation-sensitive enzyme HpyCH4 IV. (B) DHPLC profiles showing amplification of PTPN11 exon 12 from genomic DNA obtained from peripheral lymphocites of a subject heterozygous for the LS-causing C1403T mutation and unaffected parents, digested or not with the HpyCH4 IV endonuclease. Efficiency of the treatment was confirmed by digestion of an amplified genomic fragment encompassing exon 12 and subsequent re-amplification of the digested product. (C) Methylation-specific PCR assay performed on genomic DNA obtained from the same members of the family. Amplification was carried out using primers opportunely designed to amplify either the methylated (M) or the unmethylated (UM) alleles.
DISCUSSION
In this study, we investigated the driving forces underlying the largely invariant occurrence of the NS- or LS-causing T42A, E139D, Y279C, I282V and T468M amino acid substitutions and explored the molecular mechanisms involved in SHP2's functional dysregulation. Our findings demonstrate that the NS-causing T42A, E139D and I282V are the only changes promoting upregulation of SHP2's function among all possible substitutions resulting from a single-base mutation affecting each codon, while a higher mutation rate due to deamination of methylated cytosine at position 1403 is predicted to represent the driving factor leading to the invariant prevalence of the T468M mutation in LS.
Available crystallographic and biochemical data are consistent with a model of SHP2's functional regulation in which the N-SH2 domain acts as an allosteric switch with two spatially non-overlapping sites that are involved in an intramolecular interaction with the PTP domain and in binding to a pY-containing signaling partner (11,30). These sites function with negative cooperativity and mediate the conformational transition controlling the inactive and active states of the phosphatase (11,17,31). In the absence of a phosphorylated ligand, SHP2 assumes a closed conformation as evidenced by the low catalytic activity of the unstimulated protein, whereas the N-SH2 domain loses surface complementarity for its binding site on the PTP domain (11) and overall binding affinity with it (30) in the phosphopeptide-bound state. From these observations, it is predicted that dysregulation of SHP2's function can result from structural perturbations involving each of the two interactions. Indeed, previous work by our group and others demonstrated that different mechanisms are operating for disease-causing PTPN11 mutations. In most cases, mutations affect the N-SH2/PTP interdomain binding network, which destabilize the closed, inactive conformation. Alternatively, they can act by increasing SHP2's binding affinity for pY-containing signaling partners. Although these mechanisms are distinct molecularly, they are likely to be equivalent functionally since both are predicted to act by enhancing SHP2 targeting to activated receptors or docking proteins and prolonging this interaction. Consistent with previously published data, this study provided evidence that the invariant NS-causing I282V mutation as well as most other amino acid substitutions resulting from a single-base change involving this codon, perturb the stability of the N-SH2/PTP interaction required to maintain SHP2 in its catalytically inactive conformation. Unexpectedly, the isoleucine-to-valine change was found to cause only a slight destabilization of the closed conformation compared with other mutations at that residue. However, this substitution enhanced SHP2's intrinsic phosphatase activity while all other changes impaired catalysis, suggesting the requirement of an uncompromised catalytic function for SHP2 mutants at that codon. Of note, the I282V mutation has not been observed in the context of malignancy. Most PTPN11 mutations associated with cancer appear to have greater gain-of-function than NS-associated defects. Since the vast majority of cancer-associated defects alter the N-SH2 domain, their mutant SHP2 proteins have normal phosphatase catalytic activity but highly dysregulated N-SH2/PTP binding. This leads us to speculate that the extent of destabilization of the N-SH2/PTP binding network plays a major role in the degree of SHP2's functional upregulation. We cannot, however, exclude that an increased catalytic activity in a protein with otherwise normal biochemical behavior might have a distinct effect on intracellular signaling. Biochemical characterization of a larger set of mutants at the PTP domain, including those involving residue M504, which is invariably substituted by valine in NS, is required to address this issue.
The present data also demonstrated that the recurrent and invariant T42A mutation, but not other amino acid changes affecting this codon, promotes an increased phosphopeptide binding affinity of the phosphatase while retaining binding specificities similar to those of its wild-type counterpart. While an increased binding affinity for the BTAM peptide was also demonstrated for the SHP2D139 mutant, phosphopeptide array binding analyses showed that the C-SH2 domain bearing the E139D substitution acquired binding properties that differed substantially from those of its wild-type counterpart. These new binding affinities were more similar to those observed for the N-SH2 domain, revealing a previously unrecognized behavior for a SHP2 mutant and a novel mechanism involved in SHP2's functional dysregulation. As originally proposed by Hof et al. (11), the C-SH2 domain is predicted to increase the binding affinity of the N-SH2 domain to the pY-containing ligand by compensating for the energetic costs of disrupting its intramolecular interaction with the PTP domain. This model is supported by the evidence that a biphosphorylated peptide promoting simultaneous engagement of both SH2 domains more potently stimulates SHP2's activation than does the binding of singly phosphorylated ligands (32). Experimental data also indicate that the C-SH2 domain greatly enhances binding specificity by virtue of its tandem arrangement with the N-SH2 domain, since efficient binding would be strictly dependent upon the intrinsic specificity of the individual SH2 domains plus structural constrains from both domains with pY-containing ligands (31,33). On the basis of these observations, the altered binding specificity of the C-SH2 domain of SHP2D139 can be expected to perturb binding of that mutant protein to activated receptors and docking proteins qualitatively and quantitatively. SHP2 binds to BTAM motifs with widely spaced phosphotyrosines, a notable difference from other signal transducers with tandemly arranged SH2 domains. This feature makes it possible for SHP2's SH2 domains to bind simultaneously to phosphorylated sites within two adjacent signaling partners. The increased similitude in binding specificity between the wild-type N-SH2 and C-SH2D139 domains could potentially permit the SHP2D139 mutant to bind activated homodimeric receptors that have a single recognition sites in each monomer. Such interactions would be unlikely for the wild-type protein due to the different binding specificities of the N-SH2 and C-SH2 domains. As elegantly discussed by Barford and Neel (34), it is also possible that the engagement of the C-SH2 domain alone could recruit SHP2 to the membrane without concomitant catalytic activation. This could allow SHP2 to function as an adaptor molecule, independently from its phosphatase activity. Again, SHP2D139 would be predicted to behave differently in this context. These possibilities are currently under study.
In contrast to the NS- and leukemia-associated amino acid changes, biochemical data indicate that the recurrent LS-causing Y279C and T468M substitutions impair catalysis and presumably cause a diametrically opposite effect on SHP2's function (12,14,15). The findings that the expression of LS-associated mutants impaired ERK activation in response to multiple growth factors and that these mutants displayed enhanced binding to GAB1 would be consistent with a dominant negative effect (14). SPR assays performed in the present study support the view that most of mutants at codon 468, including the LS-causing SHP2M468 protein, affect the stability of the closed conformation of the enzyme. None of the mutants at codon 279, however, showed a BTAM binding affinity higher then that observed for the wild-type protein, suggesting that these mutations do not perturb significantly the N-SH2/PTP interacting surface. Although this finding does not exclude a milder dominant negative effect of the SHP2C279 and SHP2S279 mutants, at least in limiting the concentration of binding partners, our new data suggest that the LS-causing mutations affecting codons 279 and 468 are unlikely to be equivalent functionally. The findings that overexpression of the Y258C (Y279C) or T592M (T468M) csw allele in fruit flies results in a gain-of-function phenotype and does not impair signal flow through the EGFR-RAS/MAPK axis (Oishi and Gelb, unpublished data), and overexpression of the same Shp2 mutants in zebrafish has consequences on RhoA signaling but does not affect the Ras/MAPK pathway (35), indicate that the perturbing consequences of LS-causing mutants on intracellular signaling remain to be determined.
A major finding of the present study was the relatively homogeneous behavior of mutants at codons 279 and 468, which excluded impaired catalysis as the sole driving factor responsible for the invariant or prevalent occurrence of LS-associated Y279C and T468M substitutions. By providing evidence that the CpG dinucleotide at codon 468 is methylated in HeLa cells and primary human circulating lymphocytes, we suggest that the frequent occurrence of the T468M change is due to the enhanced mutability of this codon as a result of methylation-induced deamination of 5-methylcytosine. On the basis of this evidence, we expect that the catalytically impaired SHP2P468, SHP2K468 and SHP2R468 mutants might rarely occur among subjects with LS. DNA methylation is a major contributor to point mutations leading to human genetic disease as indicated by germline lesions within a CpG site recurring among patients affected by achondroplasia (36,37), Li–Fraumeni syndrome (38) and hemophilia (39). Although we failed in identifying a similar sequence context-specific mechanism for the high recurrence of the 836A>G transition producing the Y279C substitution, it is of interest that adenine is mutated into guanine in approximately two-thirds of total germline mutational events [see The Human Gene Mutation Database, statistics for missense mutations (http://www.hgmd.cf.ac.uk/ac/hoho1.php)]. Similarly, the A>C transversion (Y279S) at the second position of that codon recurs with a higher frequency than the A>T change (Y279F). Of note, the specificity of single-base changes is also known to be subject to neighboring-nucleotide effects (40). Indeed, the A>G transition is favored by the presence of two thymines immediately 5′ and 3′ to the adenine, as is the case of 836A>G. However, on the basis of these observations, a similar prevalence of the 835T>C mutation (Y279H) would be expected, which has not been documented in LS so far. While it is possible to speculate that the higher intrinsic catalytic activity of the SHP2H279 mutant might result in a subclinical or distinct phenotype, further studies are required to address this issue.
In conclusion, our findings document that either site-specific increased mutability or selection-by-function can act as primary driving forces for the invariant occurrence of disease-causing mutations in the PTPN11 gene, and uncovered novel mechanisms for SHP2's functional dysregulation. Our data also suggest that the LS-causing Y279C and Y468M mutations are unlikely to be equivalent functionally, although studies are required to understand the consequences of these mutations on intracellular signaling in more detail.
MATERIALS AND METHODS
SHP2 constructs
The full-length human His-tagged PTPN11 cDNA was cloned in a pET-26b vector (Novagen) using the HindIII and XhoI restriction sites. The 31 single-base changes resulting in amino acid substitutions at codons 42, 139, 279, 282 and 468 (Supplementary Material, Table S1) were introduced by site-directed mutagenesis [QuikChange Site-Directed Mutagenesis Kit (Stratagene)]. The isolated PTP domain (residues 246–527) constructs were generated by PCR amplification of the full-length constructs (forward primer: 5′-CACACAAAGCTTCTCAAGCAGCCCCTTAACACG-3′, reverse primer: 5′-CACACACTCGAGTTCGTGCCCTTTCCTCTTGC-3′) and subcloned in the pET-26b vector. The isolated GST-tagged N-SH2 and C-SH2 domain constructs (residues 6–103 and 112–215, respectively) were also PCR generated (N-SH2, forward primer: 5′-AGACTGGATCCTGGTTTCATGGACATCTCTCTG-3′, reverse primer: 5′-AGACTGAATTCGTACTACAACTCAAGCAGCCCC-3′; C-SH2, forward primer: 5′-AGACTGGATCCTGGTTTCACCCAAATATCACTG-3′, reverse primer: 5′-AGACTGAATTCAGTTCAGAGGATATTTAAGCTC-3′) and cloned in a pGEX-2TK vector (Amersham Bioscences), using the BamHI and EcoRI restriction sites.
SHP2 expression and purification
Recombinant proteins were expressed in E. coli (DE3) Rosetta2 competent cells (Novagen). After IPTG-induction at 30°C, harvesting and cell lysis, full-length SHP2 and SHP2PTP proteins were purified by chromatography with the use of Ni-NTA magnetic agarose beads (Qiagen) and stored at −20°C in the presence of 5 mm DTT, whereas the isolated SH2 proteins were purified by chromatography using the glutathione–Sepharose beads (Amersham Biosciences) and stored at −80°C in 20% glycerol.
Biochemical studies
In vitro phosphatase assays were performed using 20 pmol of purified recombinant proteins in 200 µl of PTP buffer (25 mm Hepes, pH 7.4; 50 mm NaCl; 2.5 mm EDTA; 60 mg/ml BSA and 5 mm DTT) supplemented with 20 mm of pNPP (Sigma) as substrate, either in basal condition or with the PTPNS1 BTAM peptide (DITpYADLNLPKGKKPAPQAAEPNNHTEpYASIQTS-NH2) (Primm) (10 µm) and incubated for 30 min (full-length proteins) or 15 min (SHP2PTP proteins) at 30°C. Reactions were stopped by the addition of 800 µl of 0.1 N NaOH. pNPP dephosphorylation was evaluated by measuring absorbance at 410 nm. Amount, purity and integrity of recombinant proteins were evaluated using the Protein Assay Kit (Bio-Rad) and Coomassie staining.
SPR analysis
SPR measurements were performed using a BIAcoreX instrument (BIAcore Intl. AB) equipped with two flow-cell sensor chips. Biotinylated PTPNS1 BTAM peptide (80 µg/ml) was resuspended in a buffer containing 10 mm HEPES (pH 7.4), 150 mm NaCl, 3 mm EDTA, 1 mm DTT and 0.005% (v/v) surfactant P20 and injected over a streptavidin-coated sensor chip (SA) at a flow rate of 10 µl/min for 2 min, to obtain an immobilization level of 800 resonance units. The biotin was linked to the N-terminus of the peptide by a stretch of poly-glycine. Wild-type SHP2 and mutants were resuspended in the same buffer and applied at a concentration of 125 nm, with a flow rate of 30 µl/min. The amount of protein bound to the sensor chip was monitored by the change in refractive index [given in arbitrary response units (RU)] at 25°C, as a function of time. At the end of the sample plug, the buffer was passed over a sensor surface to allow dissociation. The sensor surface was regenerated for the next sample using a 2 µl pulse of 10 mm NaOH. To minimize the possible dephosphorylation of bound PTPNS1, mutants were injected following the inverse order of their phosphatase activity. Integrity of bound PTPNS1 was tested by using an anti-phosphotyrosine antibody (Clone 4G10, Upstate) applied following each injection. Finally, no binding was observed when SHP2 proteins were run on the flow-cell sensor chip with immobilized biotin.
Phosphopeptide library chip design and array binding analysis
Chip slides (JPT Peptide Technologies GmbH) consisted of 6057 pY-containing peptides. Between them, 1604 were chosen since they were found to be phosphorylated in high or low throughput experiments aimed at the characterization of the human phosphoproteome (41). The remaining 4453 peptides were considered since they received a high score according to the NetPhos Neural Network predictive algorithm (42). Each chip included three identical replicated subarrays, which contained phosphorylated peptides and control spots arranged in a grid of 6400 dots. Control spots included PBS (used as negative control), IgG or IgM antibodies (to check for secondary antibody binding), GST protein (used as positive control for the primary antibody) and triple FLAG-epitope peptide (used as negative control). Finally, TAMRA and Cy3 dyes were spotted to facilitate grid orientation. To verify within-array reproducibility, 60 peptides were spotted in triplicate. Following 1 h blocking in 5% BSA-containing PBS 1×, chip slides were probed with 1 mg/ml of GST-SH2 protein (1 h at room temperature). After several washes in PBS 1×, chips were incubated with a 1:1000 diluition of an anti-GST Cy-5 conjugated antibody (Amersham-Pharmacia) (1 h at room temperature). Finally, chips were extensively washed in PBS 1× and fluorescence intensity was revealed with ScanArray Gx Plus (Perkin Elmer). In order to determine a positive data set of interacting peptides, the intensity of each spot was measured, and the mean intensity of the three replicated spots was computed for each of the 6400 positions. Binders (positive data set) were defined as those spots with a signal above the ‘total average’ (average of the spots with a signal greater than the background) plus 1 standard deviation (positive threshold). The enrichment of specific residues in the positive data set was visualized by WebLogo (43). Binomial testing and P-value <0.05 were applied.
MD simulations
Starting coordinates of the N-SH2 domain of human SHP2 bound to the PDGFRB's SVLpYTAVQP phosphopeptide were taken from the available X-ray crystal structure (Protein Data Bank, http://www.rcsb.org/pdb/home/home.do, entry [1AYA, chains A and P]) (17).
The program DeepView 3.7 (44) was used to determine the position of missing atoms. Residues 6 and 7 of the phosphopeptide, whose coordinates were missing in the PDB file, were not simulated. The phosphopeptide N- and C-termini were assumed to be neutral. MD simulations were performed with GROMACS 3.2.1, using the GROMOS96 ffG43a1 force field (45). Simulations were performed as described previously (46), except for some details. Briefly, after energy minimization in vacuo, the protein was centered in a triclinic box (5.1×5.4×5.8 nm) and hydrated with 5269 water molecules. Following initial energy minimization and a 100 ps MD run during which the protein atoms were position restrained, the temperature of solute and solvent was raised to 300 K in a stepwise manner, performing four MD runs, 50 ps each, at different temperatures (50, 100, 200 and 250 K). Pressure was kept constant (1 bar) using a Rahman–Parrinello isotropic barostat (1 ps time constant). Electrostatic interactions were calculated by the use of the particle mesh Ewald method, and a 1.2 nm cut-off radius for the real part calculation. Van der Waals interactions were calculated using a cut-off radius (1.2 nm). All simulated trajectories were 10 ns long. Calculations were performed on the HPC facilities at the CASPUR consortium. Molecular graphics were produced with the MOLMOL software (47). Translational and rotational motions of the protein were removed by least-square fitting the positions of the Cα carbon atoms of the most rigid residues in the protein (residues: 5–8, 26–34, 41–47, 51–56, 63, 74–79, 96–102) to their starting coordinates.
PCR-based methylation assays
HeLa cells were grown in DMEM (Invitrogen) supplemented with 10% FCS, 100 mg/l streptomycin and 60 mg/l penicillin. Cells were counted and seeded at the concentration of 1×106 ml−1. Treatment with 1 µm 5-azadC (Sigma) was started the day after seeding. Twenty-four hours later, fresh 5-azadC was added every day to keep its concentration constant at 0.5 µm. A control flask was also grown without 5-azadC addition. After 3 weeks, both treated and untreated cultures were recovered. Genomic DNA was extracted using the PUREGENE DNA purification kit (Gentra Systems), according to manufacturer's instructions. HpyCH4 IV (New England Biolabs) digestion of genomic DNA was performed at 37°C for 2 h. Primers were utilized to amplify exon 12 (forward primer: 5′-GCTCCAAAGAGTAGACATTGTTTC-3′, reverse primer: 5′-GACTGTTTTCGTGAGCACTTTC-3′) and the genomic portion encompassing exons 11 to 13 (forward primer: 5′-CAAAAGGAGACGAGTTCTGGGAAC-3′, reverse primer: 5′-CGTATCCAAGAGGCCTAGCAAG-3′) of the PTPN11 gene, and PCR amplifications were optimized using the following conditions: 94°C, 12 min (first denaturing step); 94°C, 45 s, 56°C (exon 12) or 60°C (exons 11–13), 30 s, 72°C, 45 s (exon 12) or 4 min (exons 11–13), 33 cycles; 72°C, 12 min (final extension). The amplimers were analyzed by DHPLC with the use of the Wave 2100 System (Transgenomics) at column temperatures recommended by the Navigator version 1.5.4.23 software. Methylation-specific PCR was performed as previously described (48), using the CpGenome DNA modification kit (CHEMICON International). Briefly, 1 µg of genomic DNA was denatured by 200 mm NaOH and modified by 3.3 m sodium bisulfite. Primers were designed to amplify the methylated (M-MSP: forward primer: 5′-TGGAATTGGTCGGATAGGGAC-3′, reverse primer: 5′-TCTCTTCCCCAAACTATTTTCG-3′) and unmethylated (UM-MSP: forward primer: 5′-TGGAATTGGTTGGATAGGGAT-3′, reverse primer:5′-TCTCTTCCCCAAACTATTTTCA-3′) alleles, and amplifications were optimized using the following conditions: 94°C, 12 min (first denaturing step); 94°C, 45 s, 60°C (M-MSP) or 58°C (UM-MSP), 30 s, 72°C, 45 s, 33 cycles; 72°C, 12 min (final extension). PCR products were separated by electrophoresis on 2% agarose gel.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by Telethon-Italy grants (GGP04172, GGP07115 to M.T.), Programma di Collaborazione Italia-USA/malattie rare (to M.T.), National Institutes of Health (HL074728, HL71207 to B.D.G.) and Sixth European Union Framework Programme (Interaction Proteome Integrated project to G.C.).
ACKNOWLEDGEMENTS
We are indebted to the CASPUR consortium (Rome) for providing computational resources and supporting A.G.
Conflict of Interest statement. None of the authors has a financial interest related to this work.
REFERENCES
- 1.Tartaglia M., Mehler E.L., Goldberg R., Zampino G., Brunner H.G., Kremer H., van der Burgt I., Crosby A.H., Ion A., Jeffery S., et al. Mutations in PTPN11, encoding the protein tyrosine phosphatase SHP2, cause Noonan syndrome. Nat. Genet. 2001;29:465–468. doi: 10.1038/ng772. [DOI] [PubMed] [Google Scholar]
- 2.Tartaglia M., Kalidas K., Shaw A., Song X., Musat D.L., van der Burgt I., Brunner H.G., Bertola D.R., Crosby A., Ion A., et al. PTPN11 mutations in Noonan syndrome: molecular spectrum, genotype-phenotype correlation, and phenotypic heterogeneity. Am. J. Hum. Genet. 2002;70:1555–1563. doi: 10.1086/340847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Digilio M.C., Conti E., Sarkozy A., Mingarelli R., Dottorini T., Marino B., Pizzuti A., Dallapiccola B. Grouping of multiple-lentigines/ LEOPARD and Noonan syndromes on the PTPN11 gene. Am. J. Hum. Genet. 2002;71:389–394. doi: 10.1086/341528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Legius E., Schrander-Stumpel C., Schollen E., Pulles-Heintzberger C., Gewillig M., Fryns J.P. PTPN11 mutations in LEOPARD syndrome. J. Med. Genet. 2002;39:571–574. doi: 10.1136/jmg.39.8.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Noonan J. Hypertelorism with Turner phenotype. A new syndrome with associated congenital heart disease. Am. J. Dis. Child. 1968;116:373–380. doi: 10.1001/archpedi.1968.02100020377005. [DOI] [PubMed] [Google Scholar]
- 6.Gorlin J.R., Anderson R.C., Blaw M. Multiple lentigines syndrome. Am. J. Dis. Child. 1969;117:652–662. doi: 10.1001/archpedi.1969.02100030654006. [DOI] [PubMed] [Google Scholar]
- 7.Tartaglia M., Gelb B.D. Germ-line and somatic PTPN11 mutations in human disease. Eur. J. Med. Genet. 2005;48:81–96. doi: 10.1016/j.ejmg.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 8.Neel B.G., Gu H., Pao L. The ‘Shp'ing news: SH2 domain-containing tyrosine phosphatases in cell signaling. Trends Biochem. Sci. 2003;28:284–293. doi: 10.1016/S0968-0004(03)00091-4. [DOI] [PubMed] [Google Scholar]
- 9.Tartaglia M., Niemeyer C.M., Shannon K.M., Loh M.L. SHP-2 and myeloid malignancies. Curr. Opin. Hematol. 2004;11:44–50. doi: 10.1097/00062752-200401000-00007. [DOI] [PubMed] [Google Scholar]
- 10.Chan R.J., Feng G.S. PTPN11 is the first identified proto-oncogene that encodes a tyrosine phosphatase. Blood. 2007;109:862–867. doi: 10.1182/blood-2006-07-028829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hof P., Pluskey S., Dhe-Paganon S., Eck M.J., Shoelson S.E. Crystal structure of the tyrosine phosphatase SHP2. Cell. 1998;92:441–450. doi: 10.1016/s0092-8674(00)80938-1. [DOI] [PubMed] [Google Scholar]
- 12.Tartaglia M., Martinelli S., Stella L., Bocchinfuso G., Flex E., Cordeddu V., Zampino G., Burgt I., Palleschi A., Petrucci T.C., et al. Diversity and functional consequences of germline and somatic PTPN11 mutations in human disease. Am. J. Hum. Genet. 2006;78:279–290. doi: 10.1086/499925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keilhack H., David F.S., McGregor M., Cantley L.C., Neel B.G. Diverse biochemical properties of Shp2 mutants: implications for disease phenotypes. J. Biol. Chem. 2005;280:30984–30993. doi: 10.1074/jbc.M504699200. [DOI] [PubMed] [Google Scholar]
- 14.Kontaridis M.I., Swanson K.D., David F.S., Barford D., Neel B.G. PTPN11 (SHP2) mutations in LEOPARD syndrome have dominant negative, not activating, effects. J. Biol. Chem. 2006;281:6785–6792. doi: 10.1074/jbc.M513068200. [DOI] [PubMed] [Google Scholar]
- 15.Hanna N., Montagner A., Lee W.H., Miteva M., Vidal M., Vidaud M., Parfait B., Raynal P. Reduced phosphatase activity of SHP-2 in LEOPARD syndrome: consequences for PI3K binding on Gab1. FEBS Lett. 2006;580:2477–2482. doi: 10.1016/j.febslet.2006.03.088. [DOI] [PubMed] [Google Scholar]
- 16.O'Reilly A.M., Pluskey S., Shoelson S.E., Neel B.G. Activated mutants of SHP-2 preferentially induce elongation of Xenopus animal caps. Mol. Cell. Biol. 2000;20:299–311. doi: 10.1128/mcb.20.1.299-311.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee C.H., Kominos D., Jacques S., Margolis B., Schlessinger J., Shoelson S.E., Kuriyan J. Crystal structures of peptide complexes of the amino-terminal SH2 domain of the Syp tyrosine phosphatase. Structure. 1994;2:423–438. doi: 10.1016/s0969-2126(00)00044-7. [DOI] [PubMed] [Google Scholar]
- 18.Songyang Z., Shoelson S.E., Chaudhuri M., Gish G., Pawson T., Haser W.G., King F., Roberts T., Ratnofsky S., Lechleider R.J., et al. SH2 domains recognize specific phosphopeptide sequences. Cell. 1993;72:767–778. doi: 10.1016/0092-8674(93)90404-e. [DOI] [PubMed] [Google Scholar]
- 19.Qin C., Wavreille A.S., Pei D. Alternative mode of binding to phosphotyrosyl peptides by Src homology-2 domains. Biochemistry. 2005;44:12196–12202. doi: 10.1021/bi050669o. [DOI] [PubMed] [Google Scholar]
- 20.Sweeney M.C., Wavreille A.S., Park J., Butchar J.P., Tridandapani S., Pei D. Decoding protein–protein interactions through combinatorial chemistry: sequence specificity of SHP-1, SHP2, and SHIP SH2 domains. Biochemistry. 2005;44:14932–14947. doi: 10.1021/bi051408h. [DOI] [PubMed] [Google Scholar]
- 21.Ekman S., Kallin A., Engström U., Heldin C.H., Rönnstrand L. SHP2 is involved in heterodimer specific loss of phosphorylation of Tyr771 in the PDGF beta-receptor. Oncogene. 2002;21:1870–1875. doi: 10.1038/sj.onc.1205210. [DOI] [PubMed] [Google Scholar]
- 22.Rönnstrand L., Arvidsson A.K., Kallin A., Rorsman C., Hellman U., Engström U., Wernstedt C., Heldin C.H. SHP-2 binds to Tyr763 and Tyr1009 in the PDGF beta-receptor and mediates PDGF-induced activation of the Ras/MAP kinase pathway and chemotaxis. Oncogene. 1999;18:3696–3702. doi: 10.1038/sj.onc.1202705. [DOI] [PubMed] [Google Scholar]
- 23.Hadari Y.R., Kouhara H., Lax I., Schlessinger J. Binding of Shp2 tyrosine phosphatase to FRS2 is essential for fibroblast growth factor-induced PC12 cell differentiation. Mol. Cell. Biol. 1998;18:3966–3973. doi: 10.1128/mcb.18.7.3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuhné M.R., Pawson T., Lienhard G.E., Feng G.S. The insulin receptor substrate 1 associates with the SH2-containing phosphotyrosine phosphatase Syp. J. Biol. Chem. 1993;268:11479–11481. [PubMed] [Google Scholar]
- 25.Arnaud M., Mzali R., Gesbert F., Crouin C., Guenzi C., Vermot-Desroches C., Wijdenes J., Courtois G., Bernard O., Bertoglio J. Interaction of the tyrosine phosphatase SHP2 with Gab2 regulates Rho-dependent activation of the c-fos serum response element by interleukin-2. Biochem. J. 2004;382:545–556. doi: 10.1042/BJ20040103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Crouin C., Arnaud M., Gesbert F., Camonis J., Bertoglio J. A yeast two-hybrid study of human p97/Gab2 interactions with its SH2 domain-containing binding partners. FEBS Lett. 2001;495:148–153. doi: 10.1016/s0014-5793(01)02373-0. [DOI] [PubMed] [Google Scholar]
- 27.Ferjoux G., Lopez F., Esteve J.P., Ferrand A., Vivier E., Vely F., Saint-Laurent N., Pradayrol L., Buscail L., Susini C. Critical role of Src and SHP2 in sst2 somatostatin receptor-mediated activation of SHP-1 and inhibition of cell proliferation. Mol. Biol. Cell. 2003;14:3911–3928. doi: 10.1091/mbc.E03-02-0069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sarkozy A., Digilio M.C., Zampino G., Dallapiccola B., Tartaglia M., Gelb B.D. LEOPARD syndrome: clinical aspects and molecular pathogenesis. In: Zenker M., editor. Monographs in Human Genetics—Noonan Syndrome and Related Disorders: A Matter of Deregulated RAS Signalling. Vol. 17. Basel, Switzerland: Karger Press; 2008. in press. [Google Scholar]
- 29.Cooper D.N., Youssoufian M. The CpG dinucleotide and human genetic disease. Hum. Genet. 1988;78:151–155. doi: 10.1007/BF00278187. [DOI] [PubMed] [Google Scholar]
- 30.Dechert U., Adam M., Harder K.W., Clark-Lewis I., Jirik F. Characterization of protein tyrosine phosphatase SH-PTP2. Study of phosphopeptide substrates and possible regulatory role of SH2 domains. J. Biol. Chem. 1994;269:5602–5611. [PubMed] [Google Scholar]
- 31.Eck M.J., Pluskey S., Trüb T., Harrison S.C., Shoelson S.E. Spatial constraints on the recognition of phosphoproteins by the tandem SH2 domains of the phosphatase SH-PTP2. Nature. 1996;379:277–280. doi: 10.1038/379277a0. [DOI] [PubMed] [Google Scholar]
- 32.Pluskey S., Wandless T.J., Walsh C.T., Shoelson S.E. Potent stimulation of SH-PTP2 phosphatase activity by simultaneous occupancy of both SH2 domains. J. Biol. Chem. 1995;270:2897–2900. doi: 10.1074/jbc.270.7.2897. [DOI] [PubMed] [Google Scholar]
- 33.Ottinger E.A., Botfield M.C., Shoelson S.E. Tandem SH2 domains confer high specificity in tyrosine kinase signaling. J. Biol. Chem. 1998;273:729–735. doi: 10.1074/jbc.273.2.729. [DOI] [PubMed] [Google Scholar]
- 34.Barford D., Neel B.G. Revealing mechanisms for SH2 domain mediated regulation of the protein tyrosine phosphatase SHP-2. Structure. 1998;6:249–254. doi: 10.1016/s0969-2126(98)00027-6. [DOI] [PubMed] [Google Scholar]
- 35.Jopling C., van Geemen D., den Hertog J. Shp2 knockdown and Noonan/LEOPARD mutant Shp2-induced gastrulation defects. PLoS Genet. 2007;3:e225. doi: 10.1371/journal.pgen.0030225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rousseau F., Bonaventure J., Legeai-Mallet L., Pelet A., Rozet J.M., Maroteaux P., Le Merrer M., Munnich A. Mutations in the gene encoding fibroblast growth factor receptor-3 in achondroplasia. Nature. 1994;371:252–254. doi: 10.1038/371252a0. [DOI] [PubMed] [Google Scholar]
- 37.Bellus G.A., Hefferon T.W., Ortiz de Luna R.I., Hecht J.T., Horton W.A., Machado M., Kaitila I., McIntosh I., Francomano C.A. Achondroplasia is defined by recurrent G380R mutations of FGFR3. Am. J. Hum. Genet. 1995;56:368–373. [PMC free article] [PubMed] [Google Scholar]
- 38.Rideout W.M., III, Coetzee G.A., Olumi A.F., Jones P.A. 5-Methylcytosine as an endogenous mutagen in the human LDL receptor and p53 genes. Science. 1990;249:1288–1290. doi: 10.1126/science.1697983. [DOI] [PubMed] [Google Scholar]
- 39.Pattinson J.K., Millar D.S., McVey J.H., Grundy C.B., Wieland K., Mibashan R.S., Martinowitz U., Tan-Un K., Vidaud M., Goossens M., et al. The molecular genetic analysis of hemophilia A: a directed search strategy for the detection of point mutations in the human factor VIII gene. Blood. 1990;76:2242–2248. [PubMed] [Google Scholar]
- 40.Krawczak M., Ball E.V., Cooper D.N. Neighboring-nucleotide effects on the rates of germ-line single-base-pair substitution in human genes. Am. J. Hum. Genet. 1998;63:474–488. doi: 10.1086/301965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Diella F., Cameron S., Gemünd C., Linding R., Via A., Kuster B., Sicheritz-Pontén T., Blom N., Gibson T.J. Phospho.ELM: a database of experimentally verified phosphorylation sites in eukaryotic proteins. BMC Bioinformatics. 2004;5:79. doi: 10.1186/1471-2105-5-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blom N., Gammeltoft S., Brunak S. Sequence- and structure-based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol. 1999;294:1351–1362. doi: 10.1006/jmbi.1999.3310. [DOI] [PubMed] [Google Scholar]
- 43.Crooks G.E., Hon G., Chandonia J.M., Brenner S.E. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guex N., Peitsch M.C. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 45.van Gunsteren W.F., Billeter S.R., Eising A.A., Hünenberger P.H., Krüger P., Mark A.E., Scott W.R.P., Tironi I.G. Biomolecular Simulation: The GROMOS96 Manual and User Guide. Zürich, Switzerland: Hochschulverlag AG an der ETH Zurich Press; 1996. [Google Scholar]
- 46.Bocchinfuso G., Stella L., Martinelli S., Flex E., Carta C., Pantaleoni F., Pispisa B., Venanzi M., Tartaglia M., Palleschi A. Structural and functional effects of disease-causing amino acid substitutions affecting residues Ala72 and Glu76 of the protein tyrosine phosphatase SHP2. Proteins. 2007;66:963–974. doi: 10.1002/prot.21050. [DOI] [PubMed] [Google Scholar]
- 47.Koradi R., Billeter M., Wuthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graph. 1996;14:29–32. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 48.Herman J.G., Graff J.R., Myöhänen S., Nelkin B.D., Baylin S.B. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc. Natl Acad. Sci. USA. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]