Abstract
We characterized 102 kb of chromosome 19 containing the apolipoprotein (APO) E/C1/C4/C2 cluster and two flanking genes for common DNA variants associated with plasma low-density lipoprotein cholesterol (LDL-C) level. DNA variants were identified by comparing sequences of 48 haploid hybrid cell lines. We genotyped participants (1943 Whites and 2046 African-Americans) of the Coronary Artery Risk Development in Young Adults study for 115 variants. After controlling for the effects of the APOE ε2/3/4 polymorphism, a single nucleotide polymorphism, rs35136575, in the downstream hepatic control region 2 (HCR-2) was associated with LDL-C in Caucasians (P = 0.0004), accounting for 1% of variation. We genotyped rs35136575 in the Atherosclerosis Risk in Communities (ARIC) cohort (3679 African-Americans and 10 427 Whites) and in the Genetic Epidemiology Network of Arteriopathy (GENOA) sibships (1381 African-Americans in 592 sibships, 1116 Caucasians in 503 sibships and 1378 Mexican-Americans in 416 sibships), finding association with LDL-C level in ARIC Caucasians (P = 0.0064). Lower plasma LDL-C was observed with the rare allele. Plasma apoE level was strongly associated with HCR-2 variant genotype in all three GENOA samples (P ≤ 0.002), indicating an effect on apoE concentration. Patterns of association for plasma apo A-I, apoB, LDL-C, high-density lipoprotein cholesterol, total cholesterol and triglyceride levels with rs35136575 in the population-based samples evaluated in this study suggest a pleiotropic effect that may be context-dependent.
INTRODUCTION
Apolipoprotein (apo) E is a component of the triglyceride (TG)-rich chylomicron, chylomicron remnant, very low-density lipoprotein (VLDL) particles and high-density lipoprotein (HDL) subspecies in human plasma. ApoE participates in receptor-mediated uptake of these particles by liver. Variation in the APOE gene contributes to variation in the levels of plasma apoB (the major apolipoprotein component of chylomicron, low-density lipoprotein (LDL) and VLDL particles), high-density lipoprotein cholesterol (HDL-C), low-density lipoprotein cholesterol (LDL-C), TG and total cholesterol (TC) (reviewed in 1–3).
In many populations, the major ε2/3/4 protein isoform polymorphism of the APOE gene accounts for a significant proportion of lipid and apolipoprotein variability. For example, the ε2/3/4 genotype accounts for 3–6% of LDL-C variation in Dutch, Nigerian, Javanese and Mexican-American samples (4–7). The ε2/3/4 genotype accounted for 9–20% of plasma apoE variation among Caucasian samples from North Karelia, Finland and Rochester, MN, USA, and 15–16% among African-Americans of Jackson, MS, USA (8). In that study, statistically, significantly more plasma apoE variation was accounted for by the addition to the model of genotype information from one or more single nucleotide polymorphism (SNP) within 1 kb of the APOE gene (8).
This laboratory previously evaluated a sparse set of SNPs spanning the APOE/C1/C4/C2 gene cluster (16 SNPs across 48 Kb) in the Coronary Artery Risk Development in Young Adults study (CARDIA) and found strong association with plasma apoB, LDL-C and TC (9). However, SNPs in that study were selected for even spacing, rather than for the ability to tag common variation, precluding conclusions as to the number of common functional variants and their location relative to genes in the region.
The purpose of this study was (i) to characterize a large region (102 kb) of chromosome 19 around the APOE gene for the number, location and type of DNA variants; (ii) to evaluate the association between plasma LDL-C level and common (relative minor allele frequency ≥0.04) SNPs in the CARDIA cohort; and (iii) to evaluate SNPs significant in CARDIA for association with LDL-C and related traits in samples from two additional studies, the Atherosclerosis Risk in Communities (ARIC) cohort and the Genetic Epidemiology Network of Arteriopathy (GENOA) sibships.
RESULTS
In order to comprehensively catalogue DNA variation across a 102 kb region spanning the APOE/C1/C4/C2 gene cluster and the two genes flanking the cluster, the sequences in this region of 48 haploid hybrid cell lines (from 12 African-Americans and 12 Caucasians) were evaluated. Three hundred and five DNA variants were identified (Supplementary Material, Table A). Of these, 292 were SNPs, of which 95 appeared in only a single sequence. Thirteen variations were insertion/deletion polymorphisms. Polymorphisms that appeared in only a single haploid sequence were not considered for genotyping in the population-based samples because of the possibility that: (i) they represent sequencing artefacts or (ii) they represent very low-frequency variants for which there would be insufficient statistical power to detect association.
Table 1 summarizes the characteristics of the CARDIA, GENOA and ARIC participants. Mean levels of quantitative variables differed among races within study except ApoB in CARDIA and TC and LDL-C in ARIC. Mean levels of quantitative variables differed among studies within race (P ≤ 0.05) except for HDL-C in African-Americans.
Table 1.
Sample characteristics of the CARDIA, GENOA and ARIC participants used in these analyses
| CARDIA | GENOA | ARIC | |||||
|---|---|---|---|---|---|---|---|
| African-American | Caucasian | African-American | Caucasian | Mexican-American | African-American | Caucasian | |
| N | 1939 | 2054 | 1616 | 1334 | 1481 | 3679 | 10427 |
| Females (%) | 58.4 | 53.6 | 68.9 | 54.6 | 60.4 | 61.8 | 52.7 |
| Age (years) | 24.54 ± 3.78 | 25.62 ± 3.35a | 57.94 ± 10.27 | 55.92 ± 10.96 | 55.70 ± 12.09a | 53.55 ± 5.83b | 54.39 ± 5.72a,c |
| BMI (kg/m2) | 25.43 ± 5.67 | 23.71 ± 3.96a | 30.90 ± 6.59 | 30.38 ± 6.31 | 30.83 ± 6.09a | 29.60 ± 6.16b | 27.02 ± 4.87a,c |
| ApoA-I (mg/dl) | 141.62 ± 21.83 | 135.93 ± 19.34a | 158.77 ± 32.27 | 157.47 ± 31.36 | 145.02 ± 25.73a | 139.00 ± 32.27b | 130.81 ± 30.73a,c |
| ApoB (mg/dl) | 90.86 ± 24.02 | 90.73 ± 23.56 | 102.53 ± 25.97 | 106.65 ± 23.12 | 108.68 ± 26.39a | 92.46 ± 30.99b | 93.99 ± 28.33a,c |
| ApoE (mg/dl) | NA | NA | 5.31 ± 2.20 | 5.15 ± 1.65 | 5.47 ± 2.47 | NA | NA |
| HDL-C (mg/dl) | 54.60 ± 13.01 | 51.73 ± 12.42a | 55.49 ± 17.98 | 51.53 ± 16.35 | 44.95 ± 12.72a | 54.95 ± 17.53 | 50.40 ± 16.79a,c |
| LDL-C (mg/dl) | 110.95 ± 31.65 | 108.79 ± 29.10a | 121.62 ± 41.11 | 123.90 ± 34.67 | 120.00 ± 37.52a | 137.71 ± 43.17b | 137.65 ± 37.90c |
| TC (mg/dl) | 178.73 ± 33.61 | 176.21 ± 31.91a | 204.99 ± 46.92 | 210.49 ± 38.79 | 205.64 ± 47.11a | 214.55 ± 45.31b | 214.67 ± 40.76c |
| TG (mg/dl) | 65.91 ± 36.15 | 78.49 ± 52.01a | 145.40 ± 90.80 | 193.59 ± 103.58 | 236.54 ± 194.13a | 113.93 ± 80.14b | 137.99 ± 92.94a,c |
| Use of lipid-lowering meds (%) | NA | NA | 6.3 | 18.0 | 14.0 | 1.5 | 3.4 |
| Use of hypertension meds (%) | NA | NA | 94.9 | 94.9 | 96.4 | 32.8 | 22.5 |
CARDIA, Coronary Artery Risk Development in Young Adults; GENOA, the Genetic Epidemiology Network of Arteriopathy; ARIC, the Atherosclerosis Risk in Communities.
aA difference in mean levels (P ≤ 0.05) among races within the study.
bA difference in mean levels (P ≤ 0.05) among studies within African-Americans.
cA difference in mean levels (P ≤ 0.05) among studies within Caucasians.
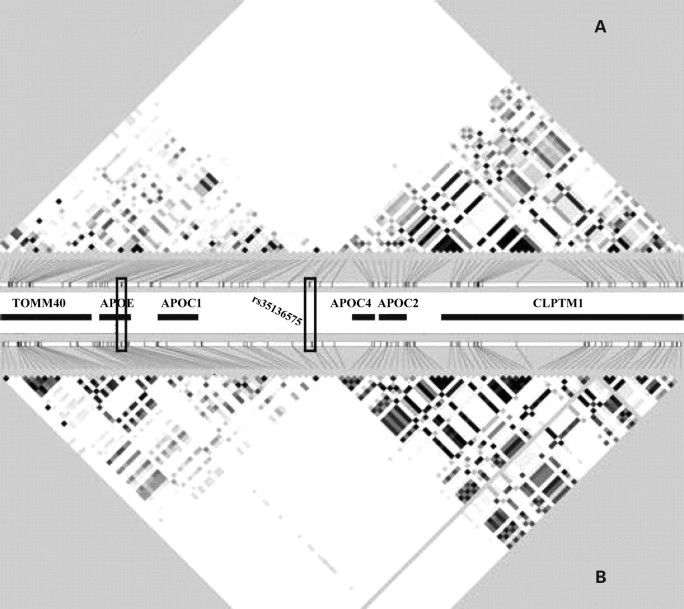
The high density of Alu repeats in this region (see Supplementary Material, Table A) presented some difficulties for the genotyping assay development process. Following successful assay development, CARDIA individuals were genotyped for 128 SNPs and one insertion/deletion polymorphism. Genotypes for 108 were deemed of sufficient quality (based on tests of missing genotype data) and informativeness (a relative minor allele frequency ≥0.001 in CARDIA) for inclusion in genotype–phenotype association analyses. In addition, six SNPs not observed in the haploid hybrid cell lines and one SNP observed in only a single sequence were genotyped in CARDIA based on a special interest in the location or on prior reports of statistical association. Genotype–phenotype association was evaluated for a total of 115 DNA variants in CARDIA (Supplementary Material, Table A). The pairwise linkage disequilibrium (LD) structure (r2) among SNPs in Hardy–Weinberg equilibrium (HWE) with a relative frequency of ≥0.001 in CARDIA African-Americans (109 SNPs) and Caucasians (107 SNPs) is shown in Figure 1. Average pairwise LD across the region was r2 = 0.06 in African-Americans and r2 = 0.08 in Caucasians. However, pairwise LD was unevenly distributed, with a clear break in LD occurring between the APOC1 and APOC4 genes. Such breaks in LD may correspond to hotspots of recombination (10).
Figure 1.
Pairwise linkage disequilibrium (LD) reported as r2 for 115 SNPs on chromosome 19 in (A) African-Americans and (B) Caucasians of CARDIA. The colour gradient indicates relative level of LD from black = complete LD (r2 = 1) to white = no LD (r2 = 0). Black boxes indicate the locations of rs429358 and rs7412, in the APOE gene, and of rs35136575.
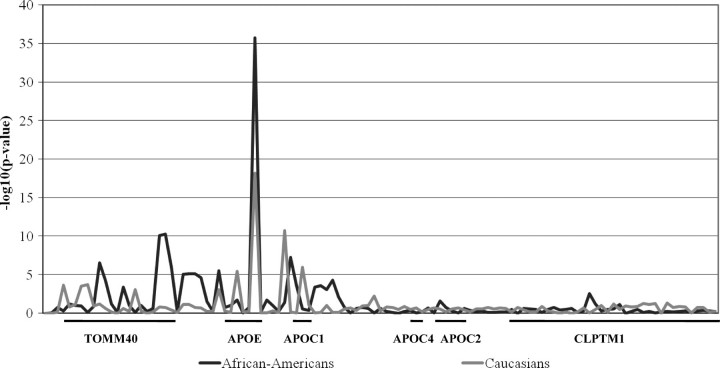
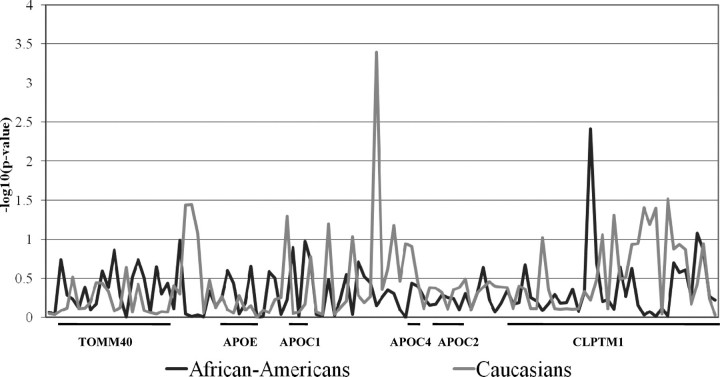
Figure 2 summarizes the results of association analyses with LDL-C in the African-American and Caucasian individuals of CARDIA. Plasma LDL-C level was most strongly associated with the APOE ε2/3/4 isoform polymorphism in both races (P < 0.0001). This polymorphism accounted for 10.5% of inter-individual LDL-C variation in African-Americans and 4.9% in Caucasians, a slightly higher proportion of variation than that reported for other population-based samples that we are aware of (3–6%) (4–7). In addition to APOE ε2/3/4, 17 SNPs in African-Americans and 9 SNPs in Caucasians were associated with LDL-C (P < 0.001). SNP association with LDL-C may be because of LD with APOE ε2/3/4. To evaluate APOE region SNPs for effects on LDL-C independent of APOE ε2/3/4, all other polymorphisms were evaluated for association within ε3 homozygotes and in the full sample after linear regression adjustment for ε2/3/4 genotype effect (Fig. 3). Weak association was suggested between ε2/3/4-adjusted LDL-C and APOS_087909 in the CLPTM1 intron 4 (P = 0.0038) in African-Americans. A single SNP, rs35136575, was strongly predictive of LDL-C level in Caucasians, independent of APOE ε2/3/4 genotypic state (P = 0.0025 prior to adjustment for ε2/3/4 effects, P = 0.0004 in the full sample after ε2/3/4 adjustment and P = 0.0050 in ε3 homozygotes). APOE ε2/3/4 and rs35136575 were not in LD (r2 = 0.001 in African-Americans and r2 = 0.015 in Caucasians). Plasma LDL-C was associated with rs35136575 in both genders of CARDIA Caucasians (P = 0.003 in females and P = 0.037 in males) without evidence for a gender-by-genotype interaction effect (P > 0.05). On the basis of sequence alignment with the APOE/C1/C4/C2 gene cluster regulatory regions (11), this SNP is located in the hepatic control region 2 (HCR-2) that regulates hepatic expression of all four apolipoproteins in the cluster, ∼27 kb downstream of the APOE gene. In addition, the location of this SNP corresponds to the distinct break in LD, already noted, between the APOC1 and APOC4 genes.
Figure 2.
Results of the tests of association between plasma LDL-C level and 115 SNPs [reported as –log10(false discovery rate (FDR)-adjusted P-value)] in CARDIA African-Americans and Whites. A P-value = 0.05 corresponds to a –log10(P-value) = 1.30. Consecutive SNPs within the genes of this region are noted below the X-axis.
Figure 3.
Results of the tests of association between plasma LDL-C level adjusted for APOE ε2/3/4 genotype and 115 SNPs [reported as –log10(FDR-adjusted P-value] in CARDIA African-Americans and Whites. P = 0.05 corresponds to a –log10(P-value) = 1.30. Consecutive SNPs within the genes of this region are noted below the X-axis.
To evaluate this association in other independent cohorts, rs35136575 was genotyped in GENOA and ARIC samples and found to have a similar allele frequency distribution in all samples (Table 2). Table 2 shows the plasma LDL-C means for rs35136575 genotype classes in CARDIA, GENOA and ARIC samples. In addition to CARDIA Caucasians, rs35136575 genotype was statistically significantly associated with APOE ε2/3/4-adjusted LDL-C level in ARIC Caucasians (P = 0.0065) and accounted for 1% of (adjusted) LDL-C variation. Results were similar for association analyses within ε3 homozygotes (not shown).
Table 2.
Relative frequency of the rs35136575 G allele (MAF) and genotype mean levels (±SD) of unadjusted plasma LDL-C and Apolipoprotein (Apo) E in the CARDIA, GENOA and ARIC samples
| CARDIA | GENOA | ARIC | |||||
|---|---|---|---|---|---|---|---|
| African-American | Caucasian | African-American | Caucasian | Mexican-American | African-American | Caucasian | |
| MAF | 0.18 | 0.26 | 0.19 | 0.25 | 0.17 | 0.18 | 0.25 |
| LDL-C (mg/dl) | |||||||
| CC | 111.93 ± 31.19 | 110.38 ± 30.15 | 122.66 ± 41.84 | 123.14 ± 35.69 | 120.47 ± 37.14 | 137.66 ± 42.63 | 137.06 ± 37.24 |
| CG | 110.44 ± 32.17 | 106.78 ± 27.41 | 120.08 ± 40.23 | 124.47 ± 33.26 | 119.36 ± 38.77 | 135.23 ± 42.28 | 136.77 ± 37.91 |
| GG | 108.75 ± 30.82 | 101.72 ± 26.95** | 115.43 ± 38.07 | 125.71 ± 33.71 | 108.51 ± 33.65 | 138.46 ± 47.30 | 134.28 ± 36.04* |
| ApoE (mg/dl) | |||||||
| CC | NA | NA | 5.41 ± 2.27 | 5.44 ± 1.76 | 5.61 ± 2.37 | NA | NA |
| CG | NA | NA | 5.15 ± 2.01 | 4.88 ± 1.42 | 5.14 ± 1.89 | NA | NA |
| GG | NA | NA | 4.74 ± 2.01** | 4.41 ± 1.40** | 4.36 ± 1.70** | NA | NA |
CARDIA, Coronary Artery Risk Development in Young Adults; GENOA, the Genetic Epidemiology Network of Arteriopathy; ARIC, the Atherosclerosis Risk in Communities.
*P ≤ 0.01 from the test of genotype–phenotype association.
**P ≤ 0.001.
Plasma apoE levels were available for GENOA participants and found to be strongly associated with rs35136575 genotype in all three races (P = 0.0017 in African-Americans and P < 0.0001 in Caucasians and Mexican-Americans) after accounting for variation because of APOE ε2/3/4. The rs35136575 variation accounted for 1% of variation in adjusted LDL-C in African-Americans, 3% in Caucasians and 2% in Mexican-Americans. In all cases, plasma apoE level decreased with increasing number of G alleles (Table 2).
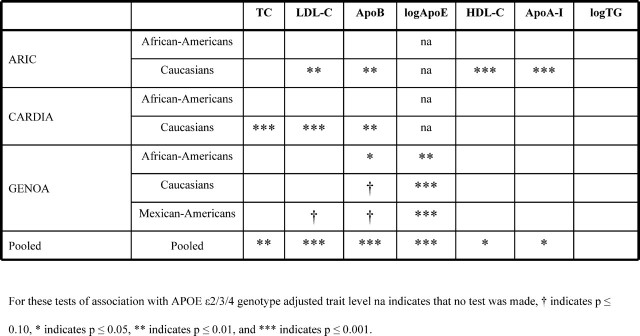
To further investigate the association between measures of plasma lipid metabolism and the HCR-2 polymorphism (independent of APOE ε2/3/4 genotype), we evaluated APOE ε2/3/4-adjusted apoA-I, apoB, HDL-C, TC and logTG for association with rs35136575 genotype in all samples. The strengths of association are summarized in Figure 4 and genotype class means are presented in Supplementary Material, Table B. A strong association (P ≤ 0.001) of rs35136575 with one or more plasma lipid or apolipoprotein measure was observed in all samples except those consisting of African-Americans, although in GENOA African-Americans a statistically significant association with plasma apoB was observed (P < 0.01). In analyses of data pooled across study and race, rs35136575 was associated with all plasma lipid and apolipoproteins (P < 0.05) except TGs after adjustment for APOE ε2/3/4 genotype effect.
Figure 4.
The strength of association between rs35136575 genotype and plasma levels of total cholesterol (TC), LDL cholesterol (LDL-C), apolipoprotein (Apo) B, log-transformed ApoE, HDL cholesterol (HDL-C), ApoA-I and log-transformed triglycerides (TG) after linear adjustment for age, age2, age3, BMI, gender and APOE genotype.
DISCUSSION
We report that rs35136575, downstream of the APOE gene, is associated with plasma apoE levels as well as levels of other plasma lipid and apolipoprotein traits. This SNP is within a known HCR that regulates transcription of multiple apolipoprotein genes, and within the break in LD observed in CARDIA for this region. This study represents a fairly comprehensive survey of common (relative minor allele frequency approximately ≥0.04) DNA variation in a 102 kb region of chromosome 19 containing the TOMM40, APOE, APOC1, APOC4, APOC2 and CLPTM1 genes. Some common DNA variants may not have been genotyped in CARDIA because they were rare or missing in the haploid hybrid cell lines used in the resequencing phase. Sampling error is inherent in any selection of 12 individuals each from two ethnic groups. Of the three DNA variants within the HCR-2 region observed in the haploid hybrid cell lines, two were common and genotyped in CARDIA, whereas the third was observed in only a single sequence and not genotyped. The pairwise LD between rs35136575 and other SNPs in the region was very low, averaging r2 = 0.004 in African-Americans (range = 0.00–0.047) and r2 < 0.001 in Whites (range = 0.00–0.13). This break in LD has also been noted as a putative recombination hotspot in HapMap (www.hapmap.org). This pattern of LD across the region makes it unlikely that rs35136575 is in LD with unmeasured variation, suggesting that the allelic state has direct functional consequences on APOE expression.
Association with LDL-C was observed in ARIC and CARDIA Caucasian samples, but not in GENOA Caucasians (Fig. 4). In post hoc analyses of GENOA individuals we further adjusted plasma lipid and apolipoproteins for plasma apoE level. LDL-C and TC levels were associated with rs35136575 genotype in GENOA Caucasians (P = 0.045 and P = 0.025) and TG was associated with all three GENOA groups (P = 0.0029 in African-Americans, P = 0.0001 in Caucasians and P < 0.0001 in Mexican-Americans). This suggests that the rs35136575 polymorphism influence on lipid metabolism is not restricted to ApoE level, and that it may also influence expression of other apolipoproteins in the APOE/C1/C4/C2 cluster.
Apolipoprotein synthesis occurs mostly in the liver and small intestine. Hepatic expression of the APOE, APOC1, APOC2 and APOC4 genes is primarily controlled by the far-downstream HCRs designated HCR-1 and HCR-2 (11). These regions are each approximately 600 bp long and are located approximately 15 and 27 Kb 3′ of the APOE gene (12). On the basis of alignment with the 535 bp sequence containing HCR-2 that was presented by Allan et al. (13), rs35136575 is located in a region of HCR-2 that is conserved with HCR1, and it contains protein-binding sequences as determined by in vivo footprint assays (footprint region 1b) (11). Although the binding proteins have not been characterized, rs35136575 could influence HCR-2 enhancer function by alteration of key sequence elements in this footprinted region. DNA expression experiments in transgenic mice have suggested that both HCR-1 and HCR-2 are capable of independently regulating liver-specific expression of the APOE, APOC1, APOC2 and APOC4 genes (12–14). In transgenic mice, constructs containing only the HCR-1 or HCR-2 elements were both able to direct gene expression at levels reflective of their expression in vivo, suggesting that either region is able to interact with proximal promoters of these genes to coordinate liver-specific transcription. It has been suggested, based on relative location, that the HCR-1 region may have a dominant effect on APOE and APOC1, and HCR-2 may have a dominant effect on APOC2 and APOC4 expression (reviewed in 15). Although not available in this study, it would be quite interesting to observe the effect of rs35136575 genotype on plasma concentrations of apoC-I, apoC-II and apoC-IV in human population-based samples.
In this study, one or more plasma lipid or apolipoprotein measure was associated with rs35136575 genotype in all samples except ARIC and CARDIA African-Americans. However, the overall pattern of genotype–phenotype association across the traits differed among samples. After pooling the seven study- and race-specific samples evaluated here, APOE ε2/3/4-adjusted levels of apoA-I, apoB, apoE, HDL-C, LDL-C and TC were associated with rs35136575 genotype (P < 0.05). Taken together, these patterns of association suggest that rs35136575 (or a functional variant marked by this SNP) has a pleiotropic effect, but that a comprehensive evaluation of context-dependent genotype–phenotype relationships might prove fruitful.
MATERIALS AND METHODS
DNA variation discovery in haploid hybrid cell lines from three samples
We constructed monosomic mouse/human hybrid cell lines that carry separated homologues for chromosome 19 from 24 individuals unselected for health (12 African-Americans from Jackson, MS, USA, and 12 Caucasians from Rochester, MN, USA) (16,17). We resequenced a 102 kb contiguous region on chromosome 19 that contains the APOE/C1/C4/C2 gene cluster, CLPTM1 and PEREC1/TOMM40 by amplification of overlapping mid-sized PCR fragments (2–4 kb). The PCR fragments were sequenced with internal primers on both strands using fluorescent methods (ABI 3730xl DNA Sequencer). DNA variants were identified by sequence alignment from the 48 monosomic hybrid cell lines (one cell line for each chromosome 19 homologue from each individual).
SNP-genotyping methods
Genotyping in CARDIA, ARIC and GENOA samples was performed using PCR amplification of genomic DNA, a short extension reaction across the polymorphic site, and mass spectrometry to detect allele-specific mass differences. Allele detection and genotype calling were performed using a MassARRAY System from Sequenom® (San Diego, CA, USA). The sequences of the PCR and extension primers are available from the authors upon request.
Characteristics of samples
Details of the CARDIA study have been described elsewhere (18,19). In brief, young adults aged 18–30 years were randomly sampled from the community in Birmingham, AL, USA; from selected census tracts in Chicago, IL, USA, and Minneapolis, MN, USA; and from the Kaiser-Permanente health plan membership in Oakland, CA, USA. Participants were recruited to be roughly equal in racial, gender, age (≤24 and >24 years) and education (high school or less versus higher than high school) groups. Study participants were given six examinations from the time of the study initiation (1985–1986). Results shown here pertain to data collected at the first exam. All participants gave written informed consent, including consent for genetic studies. The CARDIA study was approved by the Institutional Review Boards of the four participating field centres, and this ancillary study was approved by additional institutional review boards. TC and TG were measured enzymatically. HDL-C was determined by precipitation with dextran sulphate/magnesium chloride. LDL-C was calculated using the Friedewald equation, after excluding individuals whose plasma TGs exceeded 400 mg/dl (20). BMI was calculated as weight (kg) divided by height squared (m2). ApoA-I and ApoB were determined using radioimmunoassay. In CARDIA, genotypes of 115 SNPs were obtained on 3993 individuals (1939 African-Americans and 2054 Caucasians).
As part of the GENOA study, sibships from Jackson, MS, USA, and Rochester, MN, USA, were recruited if they contained at least two full siblings with essential hypertension, clinically diagnosed before the age of 60 years. Sibships from Starr County, TX, USA, were recruited if they contained at least 2 full siblings with type II diabetes. Sampling details, the clinical and laboratory protocols and baseline characteristics have been described by Daniels et al. (2004) (21). Results shown here pertain to the baseline data. All participants gave written informed consent, including consent for genetic studies. The GENOA study was approved by the Institutional Review Boards of the participating field centres, and by additional institutional review boards. Plasma TC and TG levels were measured by standard enzymatic methods. LDL particles were precipitated with polyethylene glycol 6000, and an aliquot of the supernatant was used for the determination of HDL-C by an enzymatic method. LDL-C was calculated using the Friedewald equation. Plasma apoA-I, apoB and apoE levels were measured by radioimmunoassay. Genotype and phenotype data were obtained for 1616 non-Hispanic African-Americans in 592 sibships, for 1481 non-Hispanic Whites in 505 sibships and for 1378 Mexican-American individuals in 416 sibships.
The ARIC study is a prospective investigation of atherosclerosis and its clinical sequelae involving 15 792 individuals aged 45–64 years at recruitment (1987–1989), with examinations every 3 years after baseline. Results shown here pertain to data collected at the first exam. Institutional review boards approved the ARIC study, and all participants provided their written informed consent. A detailed description of the ARIC study design and methods have been published elsewhere (22–24). Briefly, subjects were selected by probability sampling from four communities: Forsyth County, NC, USA; Jackson, MS, USA; northwestern suburbs of Minneapolis, MN, USA; and Washington County, MD, USA. Incidence of coronary heart disease (CHD) was determined by contacting participants annually to identify hospitalizations during the previous year and by surveying discharge lists from local hospitals and death certificates from state vital statistics offices for potential cardiovascular events. Plasma TC and TG was measured by an enzymatic method, and LDL-C was calculated using the Friedewald equation. HDL-C cholesterol was measured after dextran–magnesium precipitation of non-HDL lipoproteins. ApoA-I and apoB were determined by radioimmunoassay. The 14 106 participants (3679 African-Americans and 10 427 Caucasians) used in these analyses had both genotype and phenotype data, had no previous history of prevalent stroke, transient ischaemic attack/stroke symptoms or CHD at the initial clinical visit and had not prohibited use of their DNA for research purposes.
Statistical analysis
Allele frequencies were obtained by direct counting. HWE was evaluated using a χ2 goodness-of-fit test. Individuals were removed from the analyses if they had not fasted 4 h or more prior to the examination. Plasma phenotype values were adjusted prior to analysis by fitting a race-, and gender-specific linear regression model containing age, age2, age3 and BMI. The residuals from the regression model were added back to the race- and gender-specific grand mean to produce an adjusted phenotype value for each individual. Quantitative traits were further adjusted for gender using the same method. ApoE and TG levels were log transformed to reduce the non-normality of the distributions. The rs429358 and rs7412 genotypes were recoded as the APOE ε2/3/4 isoform polymorphism. Individuals were removed prior to trait adjustment and analysis if they reported use of lipid-lowing medication (448 ARIC individuals and 596 GENOA individuals). Reported use of antihypertension medication was evaluated for inclusion as a covariate in analyses of ARIC and GENOA. Because inclusion did not alter conclusions, reported P-values are for statistical models that did not incorporate information on use of antihypertension medication.
Differences in quantitative trait means among genotypes were evaluated by a one-way ANOVA in the samples of unrelated individuals (i.e. CARDIA and ARIC). r2 was used to estimate the proportion of phenotypic variation because of measured genetic variation. In GENOA, genotype–phenotype association was evaluated using a generalized estimating equation approach to account for phenotypic correlation among related individuals. In CARDIA, a P = 0.001 was taken as the threshold for statistical significance. In ARIC and GENOA, a P-value of 0.05 was taken as the threshold of statistical significance. Association between quantitative plasma lipid and apolipoprotein traits and rs35136575 genotype in a sample pooled across studies was evaluated by a general linear model that included terms for age, age2, age3, BMI, gender, study, race(study) and APOE ε2/3/4.
SUPPLEMENTARY MATERIAL
Supplementary material is available at HMG online.
Conflict of Interest statement. None declared.
FUNDING
This work was supported by the following NIH grants: CARDIA: HL072905 HL072810, HL072904 and GM065509. GENOA: HL039107, HL054457 and HL051021. ARIC: HL072810.
REFERENCES
- 1.Eichner J.E., Dunn S.T., Perveen G., Thompson D.M., Stewart K.E., Stroehla B.C. Apolipoprotein E polymorphism and cardiovascular disease: a HuGE review. Am. J. Epidemiol. 2002;155:487–495. doi: 10.1093/aje/155.6.487. [DOI] [PubMed] [Google Scholar]
- 2.Frikke-Schmidt R., Sing C.F., Nordestgaard B.G., Tybjaerg-Hansen A. Gender- and age-specific contributions of additional DNA sequence variation in the 5′ regulatory region of the APOE gene to prediction of measures of lipid metabolism. Hum. Genet. 2004;115:331–345. doi: 10.1007/s00439-004-1165-z. [DOI] [PubMed] [Google Scholar]
- 3.Song Y., Stampfer M.J., Liu S. Meta-analysis, Apolipoprotein E genotypes and risk for coronary heart disease. Ann. Intern. Med. 2004;141:137–147. doi: 10.7326/0003-4819-141-2-200407200-00013. [DOI] [PubMed] [Google Scholar]
- 4.Isaacs A., Sayed-Tabatabaei F.A., Aulchenko Y.S., Zillikens M.C., Sijbrands E.J.G., Schut A.F.C., Rutten W.P.F., Pols H.A.P., Witteman J.C.M., Oostra B.A., van Duijn C.M. Heritabilities, apolipoprotein E, and effects of inbreeding on plasma lipids in a genetically isolated population: the Erasmus Rucphen Family Study. Eur. J. Epidemiol. 2007;22:99–105. doi: 10.1007/s10654-006-9103-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kamboh M.I., Bunker C.H., Aston C.E., Nestlerode C.S., McAllister A.E., Ukoli F.A. Genetic association of five apolipoprotein polymorphisms with serum lipoprotein-lipid levels in African Blacks. Genet. Epidemiol. 1999;16:205–222. doi: 10.1002/(SICI)1098-2272(1999)16:2<205::AID-GEPI7>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 6.Gajra B., Candlish J.K., Saha N., Heng C.K., Soemantri A.G., Tay J.S. Influence of polymorphisms for apolipoprotein B (ins/del, Xbal, EcoRI) and apolipoprotein E on serum lipids and apolipoproteins in a Javanese population. Genet. Epidemiol. 1994;11:19–27. doi: 10.1002/gepi.1370110103. [DOI] [PubMed] [Google Scholar]
- 7.Hanis C.L., Hewett-Emmett D., Douglas T.C., Bertin T.K., Schull W.J. Effects of the Apolipoprotein E polymorphism on levels of lipids, lipoproteins, and apolipoproteins among Mexican-Americans in Starr County, Texas. Arterioscler. Thromb. 1991;11:362–370. doi: 10.1161/01.atv.11.2.362. [DOI] [PubMed] [Google Scholar]
- 8.Stengård J.H., Clark A.G., Weiss K.M., Kardia S., Nickerson D.A., Salomaa V., Ehnholm C., Boerwinkle E., Sing C.F. Contributions of 18 additional DNA sequence variations in the gene encoding apolipoprotein E to explaining variation in quantitative measures of lipid metabolism. Am. J. Hum. Genet. 2002;71:501–517. doi: 10.1086/342217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klos K.L.E., Sing C.F., Boerwinkle E., Hamon S.C., Rea T., Clark A.G., Fornage M., Liu K., Hixson J. Consistent effects of genes involved in reverse cholesterol transport on plasma lipid and apolipoprotein levels in CARDIA participants. Atheroscler. Thromb. Vasc. Biol. 2006;26:1828–1836. doi: 10.1161/01.ATV.0000231523.19199.45. [DOI] [PubMed] [Google Scholar]
- 10.Templeton A.R., Clark A.G., Weiss K.M., Nickerson D.A., Boerwinkle E., Sing C.F. Recombinational and mutational hotspots within the human lipoprotein lipase gene. Am. J. Hum. Genet. 2000;66:69–83. doi: 10.1086/302699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dang Q., Taylor J. In vivo footprinting analysis of the hepatic control region of the human apolipoprotein E/C-I/C-IV/C-II gene locus. J. Biol. Chem. 1996;271:28667–28676. doi: 10.1074/jbc.271.45.28667. [DOI] [PubMed] [Google Scholar]
- 12.Allen C.M., Taylor S., Taylor J.M. Two hepatic enhancers, HCR.1 and HCR.2, coordinate the liver expression of the entire human apolipoprotein E/C-I/C-IV/C-II gene cluster. J. Biol. Chem. 1997;272:29113–29119. doi: 10.1074/jbc.272.46.29113. [DOI] [PubMed] [Google Scholar]
- 13.Allan C.M., Walker D., Taylor J.M. Evolutionary duplication of a hepatic control region in the human apolipoprotein E gene locus. J. Biol. Chem. 1995;270:26278–26281. doi: 10.1074/jbc.270.44.26278. [DOI] [PubMed] [Google Scholar]
- 14.Simonet W.S., Bucay N., Lauer S.J., Taylor J.M. A far-downstream hepatocyte-specific control region directs expression of the linked human apolipoprotein E and C-I genes in transgenic mice. J. Biol. Chem. 1993;268:8221–8229. [PubMed] [Google Scholar]
- 15.Zannis V.I., Kan H.-Y., Kritis A., Zanni E., Kardassis D. Transcriptional regulation of the human apolipoprotein genes. Frontiers Biosci. 2001;6:456–504. doi: 10.2741/zannis. [DOI] [PubMed] [Google Scholar]
- 16.Andrés A.M., Clark A.G., Shimmin L., Boerwinkle E., Sing C.F., Hixson J.E. Understanding the accuracy of statistical haplotype inference with sequence data of known phase. Genet. Epidemiol. 2007;31:659–671. doi: 10.1002/gepi.20185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shimmin L.C., Natarajan S., Ibarguen H., Montasser M., Kim D.-K., Hanis C.L., Boerwinkle E., Wadhwa P.D., Hixson J.E. Coricotropin Releasing Hormone (CRH) gene variation: comprehensive resequencing for variant and molecular haplotype discovery in monosomic hybrid cell lines. DNA Seq. 2007;18:434–444. doi: 10.1080/10425170701388719. [DOI] [PubMed] [Google Scholar]
- 18.Hughes G.H., Cutter G., Donahue R., Friedman G.D., Hulley S., Hunkeler E., Jacobs D.R., Jr, Liu K., Orden S., Pirie P., et al. Recruitment in the Coronary Artery Disease Risk Development in Young Adults (Cardia) Study. Control. Clin. Trials. 1987;8:68S–73S. doi: 10.1016/0197-2456(87)90008-0. [DOI] [PubMed] [Google Scholar]
- 19.Friedman G.D., Cutter G.R., Donahue R.P., Hughes G.H., Hulley S.B., Jacobs D.R., Jr, Liu K., Savage P.J. CARDIA: study design, recruitment, and some characteristics of the examined subjects. J. Clin. Epidemiol. 1988;41:1105–1116. doi: 10.1016/0895-4356(88)90080-7. [DOI] [PubMed] [Google Scholar]
- 20.Friedewald W.T., Levy R.I., Frederickson D.S. Estimation of the concentration of low density lipoprotein cholesterol in plasma. Clin. Chem. 1972;18:499–501. [PubMed] [Google Scholar]
- 21.Daniels P.R., Kardia S.L.R., Hanis C.L., Brown C.A., Hutchinson R., Boerwinkle E., Turner S.T. Genetic Epidemiology Network of Arteriopath study. The familial aggregation of hypertension treatment and control in the GENOA study. Am. J. Med. 2004;116:676–681. doi: 10.1016/j.amjmed.2003.12.032. [DOI] [PubMed] [Google Scholar]
- 22.ARIC Investigators. The Atherosclerosis Risk in Communities (ARIC) Study: design and objectives. Am. J. Epidemiol. 1989;129:687–702. [PubMed] [Google Scholar]
- 23.White A.D., Folsom A.R., Chambless L.E., Sharret A.R., Yang K., Conwill D., Higgins M., Williams O.D., Tyroler H.A. Community surveillance of coronary heart disease in the Atherosclerosis Risk in Communities (ARIC) Study: methods and initial 2 years' experience. J. Clin. Epidemiol. 1996;49:223–233. doi: 10.1016/0895-4356(95)00041-0. [DOI] [PubMed] [Google Scholar]
- 24.Rosamond W.D., Folsom A.R., Chambless L.E., Wang C.H., McGovern P.G., Howard G., Copper L.S., Shahar E. Stroke incidence and survival among middle-aged adults: 9-year follow-up of the Atherosclerosis Risk in Communities (ARIC) cohort. Stroke. 1999;30:736–743. doi: 10.1161/01.str.30.4.736. [DOI] [PubMed] [Google Scholar]