Abstract
microRNAs (miRNAs) are ∼22-nucleotide small RNAs that act as endogenous regulators of gene expression by base-pairing with target mRNAs. Here we analyze the function of the six members of the Caenorhabditis elegans miR-51 family of miRNAs (miR-51, miR-52, miR-53, miR-54, miR-55, miR-56). miR-51 family miRNAs are broadly expressed from mid-embryogenesis onward. The miR-51 family is redundantly required for embryonic development. mir-51 family mutants display a highly penetrant pharynx unattached (Pun) phenotype, where the pharyngeal muscle, the food pump of C. elegans, is not attached to the mouth. Unusually, the Pun phenotype in mir-51 family mutants is not due to a failure to attach, but instead a failure to maintain attachment during late embryogenesis. Expression of the miR-51 family in the mouth is sufficient to maintain attachment. The Fat cadherin ortholog CDH-3 is expressed in the mouth and is a direct target of the miR-51 family miRNAs. Genetic analysis reveals that miR-51 family miRNAs might act in part through CDH-3 to regulate pharynx attachment. This study is the first to assign a function to the miR-51/miR-100 miRNA family in any organism.
MicroRNAs (miRNAs) are a widespread class of noncoding ∼22-nucleotide (nt) endogenous RNAs found in animals, plants. and algae (Lagos-Quintana et al. 2001; Lau et al. 2001; Lee and Ambros 2001; Arazi et al. 2005; Grimson et al. 2008). These RNAs modulate gene expression by blocking translation and/or destabilizing target mRNAs (Bartel 2004, 2009). Individual miRNAs are complexed with proteins of the Argonaute family to carry out their function (Bartel 2004). The first miRNAs described, lin-4 and let-7, are required for developmental timing in the nematode Caenorhabditis elegans (Wightman et al. 1993; Reinhart et al. 2000; Slack et al. 2000; Lee and Ambros 2001). Since then, a number of approaches, including reverse and forward genetics, have identified functions for miRNAs in animal and plant development, physiology, and disease. For example, in C. elegans the lin-4 miRNA and the let-7 family of miRNAs control the timing of aspects of larval development (Chalfie et al. 1981; Ambros and Horvitz 1984; Lee et al. 1993; Wightman et al. 1993; Reinhart et al. 2000; Abbott et al. 2005); the C. elegans lsy-6 miRNA acts in the asymmetric differentiation of the left and right ASE chemosensory neurons (Johnston and Hobert 2003), and the miRNA miR-61 is a target of Notch signaling during vulval patterning (Yoo and Greenwald 2005).
Although new sequencing technologies have resulted in a dramatic increase in the number of known miRNAs (Griffiths-Jones 2004; Griffiths-Jones et al. 2006), the functions of the majority of miRNAs remain unknown. One approach to determine miRNA function is to identify direct targets. Animal 3′ untranslated regions (UTRs) often contain short sequence motifs that are partially complementary to miRNA sequences. These sequence motifs have been conserved during evolution at higher rates than expected by chance (Brennecke et al. 2005; Krek et al. 2005; Lewis et al. 2005; Xie et al. 2005). Such putative miRNA binding sites match the 5′ region of the miRNA, termed the “seed” sequence (nucleotides 2–8), which is thought to be the main determinant of miRNA target specificity. While many mRNAs may be under positive selection to maintain miRNA target sites, the “seed” sequence tends to be the most highly conserved region of the miRNA. Consequently, miRNAs are often grouped into families on the basis of “seed” sequence identity (Lewis et al. 2005).
Previously, we showed that deletion of most miRNA genes in C. elegans individually resulted in no obvious abnormal phenotypes (Miska et al. 2007). Here we analyze in detail multiply mutant animals that lack the function of a whole family of miRNAs in C. elegans.
MATERIALS AND METHODS
Nematode culture, strains, and alleles:
C. elegans were grown under standard conditions at 20° (Wood 1988). The food source used was Escherichia coli strain HB101 (Caenorhabditis Genetics Center, University of Minnesota, Twin Cities, MN). The wild-type strain was var. Bristol N2 (Brenner 1974). nDf67 is a 4433-bp deletion covering mir-51 and mir-53 (Alvarez-Saavedra and Horvitz 2010). Breakpoints are AGGAGATCAAGTTCAATACTGGAGC/TGAGCTTGAATCAGGACAAGTGAGCT…TTTCAATAATTATAATTGGAGTTGAACAGA/GTATGTATGTCTTAGTGACATTACTAGTTACATGAC. Other miRNA deletion alleles were described previously (Miska et al. 2007). All strains and DNA constructs used are listed in the supporting information, Table S1 and Table S2, respectively.
Molecular biology:
All PCRs used Phusion Taq polymerase (Finnzymes, Espoo, Finland). Restriction enzymes were purchased from New England Biolabs (Ipswich, MA). Ligations were calculated with a 3:1 molar ratio and carried out with a Rapid DNA ligation kit (Roche, Basel, Switzerland). TA cloning into TOPO pCR2.1 was done according to the manufacturer's instructions (Invitrogen, Carlsbad, CA). Gateway cloning, including use of the Multisite Gateway Three-Fragment vector construction kit, was according to the manufacturer's instructions (Invitrogen). All constructs were confirmed by sequencing.
Transgenes:
Transgenes were created by microinjection of 100 ng/μl total DNA (Mello et al. 1991) or by using transposon-mediated homologous recombination (Frøkjaer-Jensen et al. 2008). A 1-kb ladder (Invitrogen) was added to a final concentration of 100 ng/μl in all microinjection experiments. miRNA rescue arrays were injected at 20 ng/μl along with dlg-1∷dlg-1∷mcherry (a gift from Andrea Hutterer) at 5 ng/μl into SX123 and SX173 to generate homozygotes rescued by transgenes. The pha-4∷mir-52∷unc-54 construct was injected at 10 ng/μl with pGFP-N (Portereiko and Mango 2001) (a gift from Susan Mango) at 10 ng/μl into SX356. col-10∷gfp∷cdh-3 constructs (WT or MUT) were injected at 20 ng/μl along with myo-2∷mcherry∷unc-54 (5 ng/μl) (Lehrbach et al. 2009) into SX356. bath-15∷gfp∷cdh-3 constructs (WT, MUT, or DEL) were created by transposon-mediated homologous recombination in EG4322. A cdh-3-containing fosmid, WRM066aH05 (Geneservice, Cambridge, UK), was injected at 20 ng/μl into SX947, and pGFP-N was used as a marker at 20 ng/μl. miRNA GFP reporter constructs were injected at 20 ng/μl together with lin-15 genomic sequence at 80 ng/μl into MT1642 and integrated using X rays. No integrant was recovered for mir-54–56∷gfp. All integrants were outcrossed twice prior to analysis.
Phenotypic assays:
For the pharynx unattached (Pun) assay, adult animals were allowed to lay embryos for 24 hr and transferred to a fresh plate. L1 progeny were scored for a Pun phenotype 24 hr later. RNA interference was performed as described previously (Piano et al. 2002). For the growth assay, animals were scored as slow growing if embryonic and larval development were retarded by at least 25% under standard conditions. Proportions of life stages present were not quantified. For the mating assay, mating was assayed as described previously (Wood 1988). Successful mating was never observed for some mutant strains, as indicated.
Immunofluorescence imaging:
Progeny of nDf67n4114; nDf58; mjEx123 animals were released from gravid adults by alkaline hypochlorite treatment, aged in complete S-basal for ∼5 hr, and fixed on slides as reported previously (Le Bot et al. 2003). Rabbit α-DsRed antibody (recognizing mCHERRY, Living Colors, Clontech, Mountainview, CA) and monoclonal α-MH27 antibody (Developmental Studies Hybridoma Bank, Iowa City, IA) were used as primary antibodies for 16 hr at 4° in PBS, 0.1% Triton-X, 0.1% BSA. Primary antibodies were removed with three washes in PBS, 0.1% Triton-X. α-Mouse and rabbit Alexa Fluor-488 and -568, respectively, were used as fluorescent secondary antibodies. Secondary antibodies were washed off and DAPI was added in the final PBS wash. Embryos were mounted in Vectashield (Vector Laboratories, Burlingame, CA). Animals were imaged on an Olympus Upright FV1000 confocal microscope (Olympus, Southend-on-Sea, UK).
Live imaging:
Animals were mounted on 2% agarose pads and anesthetized in 2 mm tetramisol (Sigma-Aldrich, Gillingham, UK). Animals were imaged on a Olympus Upright FV1000 confocal or Perkin Elmer (Fremont, CA) spinning disc confocal microscope. Mean pixel intensity of arcade cells was determined using ImageJ (http://rsb.info.nih.gov/ij/index.html) by drawing around two or three arcade cells each in 30 animals. Average background fluorescence was subtracted.
Mammalian tissue culture and luciferase assays:
HeLa cells were grown in DMEM (Invitrogen) with 10% FBS in 5% CO2 at 37° (Invitrogen) (Miska et al. 1999) and seeded at a density of 1 × 104 cells/well into 96-well plates. Cells were transfected 24 hr later with Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol with a total of 150 ng of 3′ UTR luciferase reporter vectors (see Table S2 for details) and 50 nmol of miR-52 mimic (OnTarget siRNAs, Dharmacon, Lafayette, CO) or the standard control short interfering RNAs provided by the manufacturer (Dharmacon) in triplicate. Cells were incubated for 48 hr after transfection. Dual-Glo (Promega, Madison, WI) luciferase assay kit was used according to manufacturer's protocol to detect firefly and Renilla luciferase activity. Illuminescence was detected with a Glomax luminometer (Turner BioSystems, Sunnyvale, CA). Firefly luciferase activity was normalized using Renilla luciferase activity.
RESULTS AND DISCUSSION
The miR-51 family is essential for embryonic development in C. elegans:
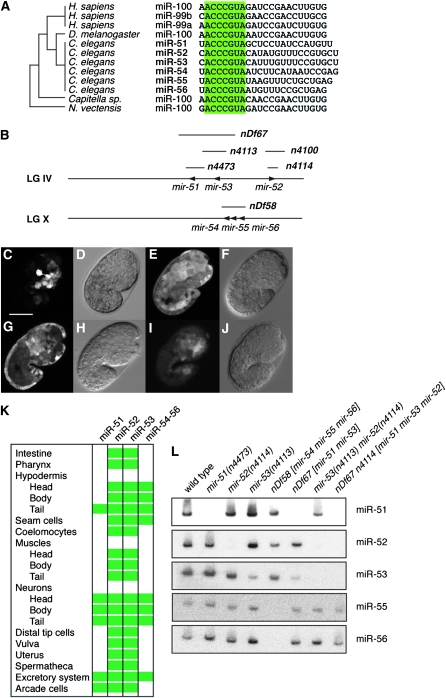
The miRNAs of the miR-51 family in C. elegans are members of the oldest family of animal miRNAs described to date, the miR-100 family. miR-100 family miRNAs emerged prior to the bilaterian split and can be found in Cnidaria and many Bilateria, including annelids, nematodes, flies, and humans (Figure 1A) (Grimson et al. 2008). There are six miR-51 family miRNAs in C. elegans, miR-51, miR-52, miR-53, miR-54, miR-55, and miR-56 (Figure 1B). miR-51/miR-53 and miR-54–56 are clustered in the genome on chromosomes IV and X, respectively. miR-54–56 are likely derived from the same transcript (data not shown). We generated promoter∷gfp fusion constucts for mir-51, mir-52, mir-53, and mir-54–56 by cloning upstream sequences of 1.3, 2.9, 0.7, and 2.9 kb, respectively, in front of gfp in pPD95.79 (see Table S2 for details). miR-51 family miRNAs are expressed in many tissues from mid-embryogenesis onward (Figure 1, C–J). miR-52 and miR-53 GFP reporters were expressed most widely (Figure 1K) in hypodermal, muscle, neural, and interstitial cell types in both embryos and larvae. mir-52 was most strongly expressed in the pharynx and anterior embryo (Figure 1E) but was detectable in most other tissues. mir-53 was more strongly expressed in the hypodermis and neurons and more weakly expressed in the gut and anterior cells around the pharynx (Figure 1G). miR-51 and miR-54–56 GFP reporters were comparatively restricted in expression to anterior and ventral cells identified as neurons, tail hypodermal cells, cells of the excretory system, and in the case of miR-51 GFP, the arcade cells. These expression patterns are in agreement with previous Northern data (Lim et al. 2003). These data are also largely in agreement with a previous study of miRNA expression in C. elegans using similar GFP transgenes (Martinez et al. 2008). One exception is miR-52, which is one of the most highly expressed miRNAs in C. elegans by Northern (Lim et al. 2003) and high-throughput sequencing (Kato et al. 2009), but was not found to be expressed strongly by promoter fusion experiments in the earlier study (Martinez et al. 2008). Another study found that mir-54–56 drives expression ubiquitously in the soma (Zhang and Emmons 2009).
Figure 1.—
The conserved miR-51 family is widely expressed throughout development in C. elegans. (A) The miR-51 is highly conserved. Its origin predates the origin of Bilateria. Bases 2–8 (“seed region”) are highlighted. (B) Genomic location of miR-51 family genes in the C. elegans genome. Mutant alleles shown in boldface type. mir-54, mir-55, and mir-56 are likely expressed from the same transcript. Not drawn to scale. (C–J) GFP fluorescence and DIC/Nomarski imaging of animals carrying promoter∷gfp fusion transgenes. mir-51∷gfp is expressed in anterior ventral cells (C and D, Z-projection; 10 μm), and mir-52∷gfp is ubiquitously expressed in the soma (E and F). mir-53∷gfp is also ubiquitously expressed, but expression is weak in the gut and in anterior pharyngeal cells (G and H), and mir-54–56∷gfp is very weakly expressed in anterior ventral cells (I and J, gain adjusted, Z-projection; 10 μm). Germline expression cannot be assessed using these reporters due to possible transgene silencing. Coelomocyte expression may represent uptake of GFP exported from other cells. Developmental stage: embryos at 1.5-fold stage. Scale bar: 20 μm. (K) Summary of miR-51 family expression patterns as assessed by GFP expression in transgenes in embryos and/or larvae. (L) Northern blotting confirms the absence of miR-51 family miRNAs in mir-51 family mutants. A total of 20 μg total RNA extracted from mixed-stage animals was used for each lane. Two blots were probed and stripped three times in the following order: (i) miR-51, miR-52, miR-54; (ii) miR-53, miR-56, miR-55. The probe for miR-54 was not specific and is not shown, but probes for miR-55 and miR-56 confirmed the absence of the miR-54–56 cluster in nDf58 mutants. miR-52 is detected by the miR-53 probe (compare lanes 4 and 7) because these two miRNAs differ by only one nucleotide. However, miR-53 is not detected by the miR-52 probe at the same exposure, likely because miR-53 is expressed at a lower level than miR-52.
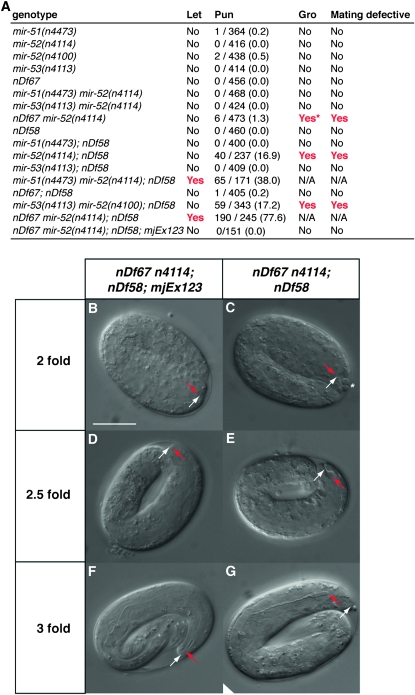
The function of the miR-51/miR-100 family of miRNAs is unknown in any organism. Previously, we found that C. elegans mutants of individual mir-51 family miRNAs were superficially wild type (Miska et al. 2007). However, given the overlap in miR-51 family expression patterns, we were interested in testing whether miR-51 family miRNAs might act redundantly. We therefore used a number of deletion alleles that we previously described (Miska et al. 2007) and a new deletion allele, nDf67, covering mir-51 and mir-53, to generate a panel of mir-51 family mutants that lack more than one mir-51 family member (Figure 1, B and L). We find that combining mir-51 family mutants reveals a number of synthetic phenotypes (Figure 2A). Deletion of all members of the mir-51 family is lethal. We refer to this mutant as the mir-51 family mutant. Most of these animals die during embryogenesis or shortly after hatching. The lethality of the mir-51 family mutant can be rescued using transgenes carrying genomic constructs for any mir-51 family miRNA (Figure 2A and data not shown). A wild-type copy of any miRNA of the mir-51 family, except for mir-53, is able to suppress this lethality. Several of the viable mutant strains display a variety of abnormalities including slow growth (Gro) and an inability of males to mate (Figure 2A and data not shown). Generally, mutants lacking mir-52 display the most severe phenotypes. This is consistent with miR-52 being highly abundant and widely expressed (Figure 1K). The earliest defect that we observed in mutants of the mir-51 family is a Pun phenotype in late embryos. Deletion of all mir-51 family miRNAs results in 77.6% Pun animals (n = 245). Mutant strains lacking mir-52 and mir-54–56 are at least 15% Pun (n > 200). The Pun phenotype of mir-51 family mutants can be rescued using transgenes containing genomic fragments corresponding to any mir-51 family miRNA (Figure 2A and data not shown). Redundant rescue is intriguing, given that the GFP reporters do not entirely overlap in their expression patterns; mir-53 is able to rescue the Pun phenotype as a rescuing transgene but not in mir-51 mir-52 mir-54 mir-55 mir-56 mutants; mir-54–56 is able to rescue the Pun phenotype despite not being expressed in the pharynx or arcade cells (Figure 1K). The miR-53 GFP reporter confirms weaker expression in the anterior pharynx, and endogenous levels alone may be insufficient to maintain pharyngeal attachment. Another study has demonstrated a broader expression pattern for mir-54–56 (Zhang and Emmons 2009) using a 6.0-kb promoter, suggesting a lack of necessary upstream sequences in the reporter used in this study. Alternatively, GFP expression levels may be too weak to be detectable by this miR-54–56 reporter.
Figure 2.—
The miR-51 family is redundantly required for embryogenesis, growth, male mating, and pharyngeal attachment. (A) Summary of abnormal phenotypes observed in mir-51 family mutants. The nDf58 allele is a deletion covering mir-54, mir-55, and mir-56. The nDf67 allele is a deletion covering mir-51 and mir-53. mjEx123 is an extrachromosomal transgene that includes a mir-52 genomic fragment. Single mutants of the mir-51 family show no obvious abnormal defects, but animals multiply mutant for mir-51 family genes show synthetic abnormalities. Let, lethal. “Yes” in the “Let” column indicates that a strain was not viable under normal laboratory conditions. Pun, pharynx unattached. Number of animals scored as Pun and total number of animals scored are given (percentage is in parentheses). Gro, slow growth. The asterisk in A indicates that only ∼5% of progeny show a slow growth phenotype. Mating defective column: “Yes” indicates that males fail to mate successfully under standard conditions. For nonviable genotypes, phenotypes were assessed in offspring of rescued homozygotes. (B–G) In embryos lacking all members of the miR-51 family, the pharynx detaches from the anterior hypodermis. mir-51 family mutant animals carrying (B, D, and F) or not carrying a (C, E, and G) a miR-52 expressing extrachromosomal array (mjEx123) were observed. Six different animals are shown. Red arrow: anterior pharynx. White arrow: anterior hypodermis. Scale bar: 20 μm.
The miR-51 family is required to maintain pharyngeal attachment to the hypodermis:
As deletion of all members of the mir-51 family miRNAs is lethal, we generated a strain in which this lethality was rescued by an extrachromosomal array, mjEx123, that contains a genomic fragment spanning the mir-52 locus and a visible marker, dlg-1∷mcherry. This strain segregates both viable rescued embryos expressing DLG-1∷mCHERRY and nonviable mir-51 family mutant embryos that have lost the extrachromosomal array (Figure 2, B–G). In animals carrying mjEx123, the pharynx (red arrow in Figure 2B) has attached to the anterior hypodermis (white arrow in Figure 2B), as in wild type. In mir-51 family mutants, the pharynx is also attached at the twofold stage (Figure 2C). In some animals, a few unidentified cells that have lost contact with the rest of the embryo are visible in the anterior sensory depression (asterisk in Figure 2C), which is enlarged. At the 2.5-fold stage, the pharynx is elongating and the posterior and anterior bulbs and isthmus begin to form (Figure 2D). At this stage in mir-51 family mutants, the pharynx remains attached, but pharynx extension is variable (Figure 2E). At the threefold stage, the pharynx is normally well extended, and the bulbs and isthmus become recognizable. However, in mir-51 family mutants, the pharynx has detached and there is little separation between the bulbs and isthmus (Figure 2G). Nevertheless, the basement membrane around the pharynx is intact. These defects are not observed in rescued animals. We conclude that miR-51 family miRNAs are required for maintenance but not for establishment of pharyngeal attachment.
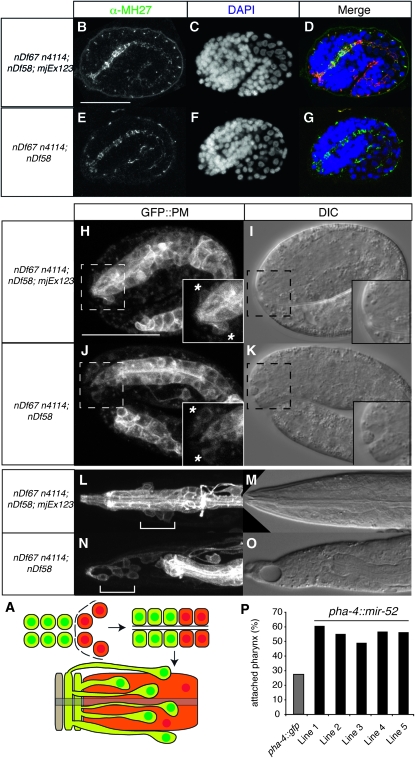
Early morphogenesis of the pharynx occurs in several phases (Sulston et al. 1983; Portereiko and Mango 2001; Mango 2009). In the first phase, the lumen of the pharynx elongates through polarization of the anterior pharyngeal cells (Figure 3A). In the second phase, a cylindrical epithelium is formed by six pharyngeal cells and nine arcade cells that in phase 3 connects to the anterior hypodermis to form the mouth. Later on, the buccal cavity consists of a ring of anterior hypodermis, two arcade rings, and the anterior pharynx. At this point, the nuclei of the arcade cells have been displaced posteriorly but remain attached to the arcade ring through cytoplasmic bridges (Figure 3A, bottom). Genetic analyses have identified Pun mutants that fail at different times during buccal morphogenesis (Portereiko and Mango 2001; Mango 2009). To check for polarization of the anterior pharyngeal cells in our mutants, we visualized adherens junctions through staining of embryos with an α-MH27 antibody, which recognizes AJM-1 (Figure 3, B–G). In both rescued (Figure 3, B–D) and mir-51 family mutant embryos (Figure 3, E–G), AJM-1 expression is continuous from the developing mouth to the intestine, indicating that the arcade cells, pharyngeal epithelial cells, and pharyngeal muscle cells can polarize and form a continuous epithelium in the absence of miR-51 family miRNAs. However, while the arcade cell extensions contribute to mouth formation in mir-51 family mutant embryos, their cell bodies are often incorrectly positioned (Figure 3, H–K). We visualized the arcade and pharyngeal cell bodies using a plasma membrane localized GFP (GFP∷PM) transgene pxIs10 (Portereiko and Mango 2001) in mir-51 family mutant and mjEx123 rescued embryos. In rescued embryos, the arcade cell bodies are positioned posterior to the pharyngeal attachment site and are closely associated with the basement membrane of the pharynx, and the anterior depression is small (Figure 3H), as in wild type. In mir-51 family mutant embryos, the arcade cell bodies are not associated with the pharyngeal basement membrane (Figure 3J). The anterior depression is large, and in some cases the arcade cell bodies have moved anteriorly (asterisk in Figure 3, H and J). We conclude that in mir-51 family mutants the arcade cells commit to their mesenchymal-to-epithelial transition, polarize, and form the arcade ring, but their cell bodies migrate abnormally. Later, the arcade is detached from the pharynx in most animals (Figure 3, L–O). In mir-52 rescued animals, arcade cell bodies (white bracket, Figure 3L) are positioned adjacent to the metacorpus just anterior to the anterior bulb and extend projections to the arcade ring at the tip of the buccal cavity. In mir-51 family mutants (Figure 3N), the pharynx becomes detached from the arcade ring. This detachment occurs between the posterior arcade ring and the pharynx or, more rarely, between the anterior and posterior arcade rings, but not between the anterior arcade ring and the hypodermis. These data suggest that mir-51 mutants fail to maintain the arcade–pharyngeal connection during elongation when mechanical stress requires a tight adhesion. Next, we examined where the miR-51 family miRNAs are required for normal pharyngeal morphogenesis. On the basis of their expression patterns (Figure 1K), miR-51 family miRNAs might act in hypodermal or pharyngeal cells. We generated animals expressing pha-4∷mir-52 transgenes in the mir-51 family mutant background. The pha-4 promoter drives expression in the arcade cells, pharynx, and gut (Portereiko and Mango 2001). Five independent transgenic lines increased pharyngeal attachments two- to threefold compared to mir-51 family mutants (Figure 3P). We conclude that the miR-51 family is sufficient in either the arcade or the pharynx to control pharyngeal attachment.
Figure 3.—
The miR-51 family is sufficient in the pharynx to maintain attachment of the pharynx to the mouth. (A) Diagrammatic representation of morphogenesis of the buccal cavity (mouth). Arcade cells and arcade (green), pharyngeal epithelium and pharynx (orange), and the hypodermal cell Hyp1 (beige) are shown. (B–G) The anterior pharynx is polarized in wild-type and mir-51 family mutant embryos. Embryos were fixed and stained using an antibody against mCHERRY to identify those carrying a mir-52 rescuing transgene marked by DLG-1∷mCHERRY (mjEx123) (data not shown) and with α-MH27 antibody (B and E), which is specific for AJM-1, to visualize adherens junctions. (C and F) DAPI stain. (D and G) Merge. (H–O) Arcade cells are often incorrectly positioned in mir-51 family mutant embryos. Live imaging. (H and J) plasma membrane localized GFP in arcade and pharynx. Insets show the region bounded by the dotted lines in more detail. (L–O) Arcade cells become detached from the developing pharynx. Live imaging as in H–K. (L and N) White brackets: arcade cell bodies. (P) Tissue-specific rescue of the Pun phenotype. Animals carrying pha-4∷mir-52∷unc-54 transgenic arrays in the mir-51 family mutant background. Five independent transgenic lines showed an increased number of animals with an attached pharynx as compared to a control pha-4∷gfp∷unc-54 transgenic array.
miR-51 family miRNAs directly regulate the Fat cadherin ortholog CDH-3:
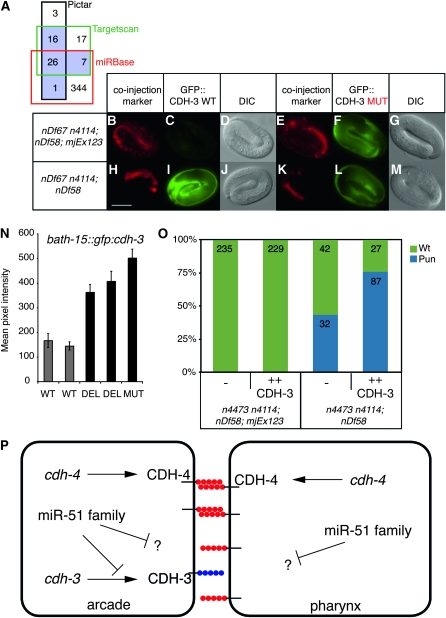
To better understand the function of the miR-51 family, we aimed to identify direct in vivo targets. Three target prediction algorithms, miRBase, PicTar, and TargetScan, identified many potential targets with an overlap of 26 genes (Figure 4A) (Lewis et al. 2005; Griffiths-Jones et al. 2006, 2008; Lall et al. 2006). We selected 10 targets for experimental validation using a heterologous luciferase reporter assay system (see Table S2). Using this assay, we found that the 3′ UTR of cdh-3 can confer a significant miR-52-dependent inhibition of translation (Figure S1). We then tested whether the miR-51 family can regulate the cdh-3 3′ UTR in the hypodermis, in which the miR-51 family is expressed and GFP expression can be assayed easily (Figure 1K). Using a similar mir-51 family mutant strain carrying mjEx123 as described above (Figures 2 and 3), we find that in mir-52 rescued embryos, marked by DLG-1∷mCHERRY (Figure 4B), and in otherwise wild-type embryos (Figure S2), a col-10∷gfp∷cdh-3 transgene under the control of the wild-type cdh-3 3′ UTR (WT, Figure 4C) is downregulated. Importantly, a similar transgene but with a 4-bp mutation in the seed region of the predicted miR-52 binding site (Figure S3, MUT) does not show this effect (Figure 4F). In contrast, both WT and MUT transgenes are expressed in mir-51 family mutant embryos (Figure 4, I and L). A second transgene with an unrelated 3′ UTR (myo-2∷mcherry∷unc-54) is not regulated in a miR-52-dependent manner (Figure 4, B, E, H, and K). Taken together, these data show that miR-52 directly regulates the CDH-3 3′ UTR in vivo. We also tested if the cdh-3 3′ UTR is regulated by the miR-51 family in the arcade cells, where regulation of CDH-3 by miR-51 family miRNAs may be required for pharyngeal attachment. As the bath-15 promoter drives transgene expression in the arcade cells during late embryogenesis and larval development (Hunt-Newbury et al. 2007), we generated animals expressing bath-15∷gfp∷cdh-3 transgenes. We generated constructs corresponding to wild-type (WT) and two different mutant cdh-3 3′ UTRs (DEL, MUT, Figure S3). Expression of single-copy transgenes carrying a mutant cdh-3 3′ UTR is greater than that of transgenes carrying the wild-type cdh-3 3′ UTR (Figure 4N, Figure S4). In addition, we found that GFP expression from the bath-15∷gfp∷cdh-3 transgene is upregulated in mir-51 family mutants as compared to rescued animals (Figure S5). These data suggest that the miR-51 family directly regulates cdh-3 expression in the arcade cells.
Figure 4.—
The miR-51 family acts in part through CDH-3 to regulate pharyngeal attachment. (A) Venn diagram showing the number of unique genes predicted to be targeted by all six members of the miR-51 family by three different target prediction algorithms. Those predicted by two or more programs are outlined in black. (B–M) miR-52 can regulate the cdh-3 3′ UTR in the hypodermis. Transgenes: dlg-1∷mcherry, col-10∷gfp∷cdh-3 (WT, MUT), and myo-2∷mcherry∷unc-54. (N) cdh-3 3′ UTR regulation through miR-52 was assessed in the arcade cells using bath-15∷gfp∷cdh-3 transgenes. Wild-type (WT) and two different mutant cdh-3 3′ UTRs were used (DEL, MUT; see Figure S3). Larvae were imaged at the L4 stage. The mean pixel intensity for arcade cells was quantified. Error bars indicate the standard error of the mean (n > 30, P > 0.0001, Student's t-test). (O) Overexpression of CDH-3 (mjEx288 or mjEx289) enhances the unattached pharynx phenotype of mir-51 mir-52 mir-54 mir-55 mir-56 mutants (n4473 n4114; nDf58). The Pun phenotype was quantified in these mutants and in mir-52 rescued embryos (mjEx123) in the presence or absence of a transgene carrying cdh-3 genomic sequence (P < 0.0001, χ2 test). (P) Hypothetical model of miR-51 family activity during pharyngeal adhesion. CDH-3 is in blue; CDH-4 is in red.
In some instances, abnormal phenotypes of miRNA mutants can be suppressed by mutations in direct targets, e.g., the case of the lin-4 miRNA and lin-14 (Lee et al. 1993). We therefore tested whether the Pun phenotype of mir-51 family mutants could be rescued by reduction of cdh-3 dosage. However, neither injection of double-stranded RNA against cdh-3 nor a putative loss-of-function mutant of cdh-3 (Pettitt et al. 1996) failed to suppress the Pun phenotype of mir-51 family mutants (Figure S6 and data not shown). With the caveat that CDH-3 function may not be completely removed in either experiment, we conclude that the miR-51 family targets at least one additional gene to regulate pharyngeal attachment. Therefore, we tested the role of CDH-3 in pharyngeal attachment using gain of function. Overexpression of CDH-3 through a transgene containing a cdh-3 genomic fragment did not induce a Pun phenotype in wild-type or mir-52 rescued animals (Figure 4O and data not shown). However, in a mir-51mir-52mir-54mir-55mir-56 mutant background, CDH-3 overexpression results in an increase of the percentage of animals displaying a Pun phenotype from 43% to 76% (Figure 4O). These data suggest that at least one function of the miR-51 family during pharyngeal morphogenesis is to downregulate CDH-3.
Previously, we reported that many miRNAs are individually not required for embryogenesis (Miska et al. 2007). In addition, most miRNA families are also not required for embryogenesis (Alvarez-Saavedra and Horvitz 2010). Here we demonstrate that the miR-51 family is redundantly required for embryonic development in C. elegans and analyze the mir-51 family mutant phenotype. While expression patterns and mutant phenotypes vary among family members, all miR-51 family miRNAs share an essential role during development. Although early embryogenesis in mir-51 family mutants appears to be normal, the pharynx fails to maintain its attachment to the mouth late in embryogenesis. Interestingly, this phenotype is shared by loss-of-function mutants in the Fat cadherin ortholog cdh-4 (Schmitz et al. 2008), which is expressed in the hypodermis, the arcade, and the pharynx. We have shown that another Fat cadherin ortholog, cdh-3, is a direct target of the miR-51 miRNA family. While cdh-3 does not appear to be required for pharyngeal attachment, its promoter is active in arcade cells (Pettitt et al. 1996). We find that cdh-3 overexpression enhances the Pun phenotype of a mir-51 family mutant. We propose a model in which the miR-51 family downregulates target genes in the arcade cells to control the adhesive properties of the cells forming the buccal cavity of C. elegans (Figure 4P). Failure to downregulate CDH-3 in these cells might interrupt putative homophilic interactions of CDH-4. It is likely that additional miR-51 family miRNA targets are also involved in pharyngeal attachment. Mutations in cdh-3 failed to suppress the mir-51 family mutant Pun phenotype. Indeed, many miRNAs are thought to function through a number of redundant targets not unlike transcription factors (Bartel 2009). Interestingly, another cadherin, CDH-12, is also a predicted target of the mir-51 family (Lewis et al. 2005). Fat cadherins have been implicated in adhesion, tube formation, and signaling during planar cell polarity in other systems (Sopko and Mcneill 2009). It would be interesting to investigate whether the miR-51/100 family has a conserved role in regulating any or all of these processes.
Acknowledgments
We thank the Caenorhabditis Genetics Center funded by the National Institute of Health for providing strains, Susan Mango and Andrea Hutterer for constructs, and Cherie Blenkiron for advice on luciferase assays. J.A. contributed the luciferase data, N.J.L. carried out some of the microinjections, and W.R.S. was supported by a Ph.D. studentship from the Wellcome Trust (United Kingdom). This work was supported by a Cancer Research UK Programme Grant to E.A.M. and by core funding to the Wellcome Trust/Cancer Research UK Gurdon Institute provided by the Wellcome Trust and Cancer Research UK.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.110.117515/DC1.
References
- Abbott, A. L., E. Alvarez-Saavedra, E. A. Miska, N. C. Lau, D. P. Bartel et al., 2005. The let-7 microRNA family members mir-48, mir-84, and mir-241 function together to regulate developmental timing in Caenorhabditis elegans. Dev. Cell 9 403–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Saavedra, E., and H. R. Horvitz, 2010. Many families of C. elegans microRNAs are not essential for development or viability. Curr. Biol. 20 367–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros, V., and H. R. Horvitz, 1984. Heterochronic mutants of the nematode Caenorhabditis elegans. Science 226 409–416. [DOI] [PubMed] [Google Scholar]
- Arazi, T., M. Talmor-Neiman, R. Stav, M. Riese, P. Huijser et al., 2005. Cloning and characterization of micro-RNAs from moss. Plant J. 43 837–848. [DOI] [PubMed] [Google Scholar]
- Bartel, D. P., 2004. microRNAs: genomics, biogenesis, mechanism, and function. Cell 116 281–297. [DOI] [PubMed] [Google Scholar]
- Bartel, D. P., 2009. microRNAs: target recognition and regulatory functions. Cell 136 215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke, J., A. Stark, R. B. Russell and S. M. Cohen, 2005. Principles of microRNA-target recognition. PLoS Biol. 3 e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner, S., 1974. The genetics of Caenorhabditis elegans. Genetics 77 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalfie, M., H. R. Horvitz and J. E. Sulston, 1981. Mutations that lead to reiterations in the cell lineages of C. elegans. Cell 24 59–69. [DOI] [PubMed] [Google Scholar]
- Frøkjaer-Jensen, C., M. W. Davis, C. E. Hopkins, B. J. Newman, J. M. Thummel et al., 2008. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat. Genet. 40 1375–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones, S., 2004. The microRNA Registry. Nucleic Acids Res. 32 D109–D111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones, S., R. J. Grocock, S. van Dongen, A. Bateman and A. J. Enright, 2006. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 34 D140–D144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones, S., H. K. Saini, S. van Dongen and A. J. Enright, 2008. miRBase: tools for microRNA genomics. Nucleic Acids Res. 36 D154–D158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson, A., M. Srivastava, B. Fahey, B. J. Woodcroft, H. R. Chiang et al., 2008. Early origins and evolution of microRNAs and Piwi-interacting RNAs in animals. Nature 455 1193–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt-Newbury, R., R. Viveiros, R. Johnsen, A. Mah, D. Anastas et al., 2007. High-throughput in vivo analysis of gene expression in Caenorhabditis elegans. PLoS Biol. 5 e237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston, R. J., and O. Hobert, 2003. A microRNA controlling left/right neuronal asymmetry in Caenorhabditis elegans. Nature 426 845–849. [DOI] [PubMed] [Google Scholar]
- Kato, M., A. de Lencastre, Z. Pincus and F. Slack, 2009. Dynamic expression of small non-coding RNAs, including novel microRNAs and piRNAs/21U-RNAs, during Caenorhabditis elegans development. Genome Biol. 10 R54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krek, A., D. Grün, M. N. Poy, R. Wolf, L. Rosenberg et al., 2005. Combinatorial microRNA target predictions. Nat. Genet. 37 495–500. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana, M., R. Rauhut, W. Lendeckel and T. Tuschl, 2001. Identification of novel genes coding for small expressed RNAs. Science 294 853–858. [DOI] [PubMed] [Google Scholar]
- Lall, S., D. Grün, A. Krek, K. Chen, Y.-L. Wang et al., 2006. A genome-wide map of conserved microRNA targets in C. elegans. Curr. Biol. 16 460–471. [DOI] [PubMed] [Google Scholar]
- Lau, N. C., L. P. Lim, E. G. Weinstein and D. P. Bartel, 2001. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294 858–862. [DOI] [PubMed] [Google Scholar]
- Le Bot, N., M. C. Tsai, R. K. Andrews and J. Ahringer, 2003. TAC-1, a regulator of microtubule length in the C. elegans embryo. Curr. Biol. 13 1499–1505. [DOI] [PubMed] [Google Scholar]
- Lee, R. C., and V. Ambros, 2001. An extensive class of small RNAs in Caenorhabditis elegans. Science 294 862–864. [DOI] [PubMed] [Google Scholar]
- Lee, R. C., R. L. Feinbaum and V. Ambros, 1993. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75 843–854. [DOI] [PubMed] [Google Scholar]
- Lehrbach, N., J. Armisen, H. Lightfoot, K. Murfitt, A. Bugaut et al., 2009. LIN-28 and the poly(U) polymerase PUP-2 regulate let-7 microRNA processing in Caenorhabditis elegans. Nat. Struct. Mol. Biol. 16 1016–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, B. P., C. B. Burge and D. P. Bartel, 2005. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120 15–20. [DOI] [PubMed] [Google Scholar]
- Lim, L. P., N. C. Lau, E. G. Weinstein, A. Abdelhakim, S. Yekta et al., 2003. The microRNAs of Caenorhabditis elegans. Genes Dev. 17 991–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mango, S., 2009. The molecular basis of organ formation: insights from the C. elegans foregut. Annu. Rev. Cell Dev. Biol. 25 597–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez, N. J., M. C. Ow, J. S. Reece-Hoyes, M. I. Barrasa, V. R. Ambros et al., 2008. Genome-scale spatiotemporal analysis of Caenorhabditis elegans microRNA promoter activity. Genome Res. 18 2005–2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello, C. C., J. M. Kramer, D. Stinchcomb and V. Ambros, 1991. Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10 3959–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miska, E. A., C. Karlsson, E. Langley, S. J. Nielsen, J. Pines et al., 1999. HDAC4 deacetylase associates with and represses the MEF2 transcription factor. EMBO J. 18 5099–5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miska, E. A., E. Alvarez-Saavedra, A. L. Abbott, N. C. Lau, A. B. Hellman et al., 2007. Most Caenorhabditis elegans microRNAs are individually not essential for development or viability. PLoS Genet. 3 e215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettitt, J., W. B. Wood and R. H. Plasterk, 1996. cdh-3, a gene encoding a member of the cadherin superfamily, functions in epithelial cell morphogenesis in Caenorhabditis elegans. Development 122 4149–4157. [DOI] [PubMed] [Google Scholar]
- Piano, F., A. J. Schetter, D. G. Morton, K. C. Gunsalus, V. Reinke et al., 2002. Gene clustering based on RNAi phenotypes of ovary-enriched genes in C. elegans. Curr. Biol. 12 1959–1964. [DOI] [PubMed] [Google Scholar]
- Portereiko, M. F., and S. E. Mango, 2001. Early morphogenesis of the Caenorhabditis elegans pharynx. Dev. Biol. 233 482–494. [DOI] [PubMed] [Google Scholar]
- Reinhart, B. J., F. J. Slack, M. Basson, A. E. Pasquinelli, J. C. Bettinger et al., 2000. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403 901–906. [DOI] [PubMed] [Google Scholar]
- Schmitz, C., I. Wacker and H. Hutter, 2008. The Fat-like cadherin CDH-4 controls axon fasciculation, cell migration and hypodermis and pharynx development in Caenorhabditis elegans. Dev. Biol. 316 249–259. [DOI] [PubMed] [Google Scholar]
- Slack, F. J., M. Basson, Z. Liu, V. Ambros, H. R. Horvitz et al., 2000. The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol. Cell 5 659–669. [DOI] [PubMed] [Google Scholar]
- Sopko, R., and H. McNeill, 2009. The skinny on Fat: an enormous cadherin that regulates cell adhesion, tissue growth, and planar cell polarity. Curr. Opin. Cell Biol. 21 717–723. [DOI] [PubMed] [Google Scholar]
- Sulston, J. E., E. Schierenberg, J. G. White and J. N. Thomson, 1983. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 100 64–119. [DOI] [PubMed] [Google Scholar]
- Wightman, B., I. Ha and G. Ruvkun, 1993. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75 855–862. [DOI] [PubMed] [Google Scholar]
- Wood, W., 1988. The Nematode Caenorhabditis elegans. Cold Spring Harbor Laboratory Press, New York.
- Xie, X., J. Lu, E. J. Kulbokas, T. R. Golub, V. Mootha et al., 2005. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature 434 338–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo, A. S., and I. Greenwald, 2005. LIN-12/Notch activation leads to microRNA-mediated down-regulation of Vav in C. elegans. Science 310 1330–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H., and S. W. Emmons, 2009. Regulation of the Caenorhabditis elegans posterior hox gene egl-5 by microRNA and the polycomb-like gene sop-2. Dev. Dyn. 238 595–603. [DOI] [PMC free article] [PubMed] [Google Scholar]