Abstract
In comparison to human leukocyte antigen (HLA) polymorphism, the impact of allelic sequence variation within T cell receptor (TCR) loci is much less understood. Particular TCR loci have been associated with autoimmunity, but the molecular basis for this phenomenon is undefined. We examined the T cell response to an HLA-B*3501–restricted epitope (HPVGEADYFEY) from Epstein-Barr virus (EBV), which is frequently dominated by a TRBV9*01+ public TCR (TK3). However, the common allelic variant TRBV9*02, which differs by a single amino acid near the CDR2β loop (Gln55→His55), was never used in this response. The structure of the TK3 TCR, its allelic variant, and a nonnaturally occurring mutant (Gln55→Ala55) in complex with HLA-B*3501HPVGEADYFEY revealed that the Gln55→His55 polymorphism affected the charge complementarity at the TCR–peptide-MHC interface, resulting in reduced functional recognition of the cognate and naturally occurring variants of this EBV peptide. Thus, polymorphism in the TCR loci may contribute toward variability in immune responses and the outcome of infection.
MHC molecules play a critical role in protective immunity by presenting antigenic peptide fragments for T cell recognition (Davis and Bjorkman, 1988). MHC polymorphism enhances immune defense across the population by ensuring wide variation in the T cell response to infecting pathogens through presentation of a broad array of target epitopes (Lawlor et al., 1990; Germain and Margulies, 1993). There are ∼4,000 different variants of HLA (Robinson et al., 2003), with polymorphism generally concentrated in the antigen-binding cleft, controlling the size and diversity of the peptide repertoire presented by each HLA molecule. Although HLA molecules can differ from each other by >30 amino acids, differences of only a few amino acids (micropolymorphism) can have a major impact on immune responses (Archbold et al., 2009). Namely, HLA micropolymorphism can influence the repertoire of peptides presented on the surface of APCs (Macdonald et al., 2003; Burrows et al., 2007), the conformation of HLA-bound peptide (Hülsmeyer et al., 2004; Tynan et al., 2005c), the dependence on chaperones for antigen loading (Zernich et al., 2004), αβ TCR recognition (Tynan et al., 2005a,b, 2007), and susceptibility to infectious disease (Limou et al., 2009).
To engage the vast repertoire of MHC-bound antigenic peptides, TCRs are diversified through the random rearrangement of V and J genes at the TCRα locus, and V, D, and J genes at the TCRβ locus of developing thymic T cells. Further potential diversity is created through untemplated addition or deletion of a variable number of nucleotides at the V-(D)-J junctional sites, called N regions. The residual repertoire of unique TCRs after thymic selection is between 10 and 100 million in humans (Arstila et al., 1999). Despite this vast potential repertoire, immune responses often show strong unexplained biases in TCR selection, resulting in immunodominance of certain “public” TCRs that are widely used in individuals with shared MHC types (Acha-Orbea et al., 1988; Argaet et al., 1994; Burrows et al., 1995; Turner et al., 2006; Gras et al., 2008). The first and second complementarity-determining regions (CDRs) of the TCR are germline encoded within the TRAV and TRBV gene segments, whereas the CDR3 regions are derived from the V-(D)-J and N regions. From the growing number of unique TCR–peptide-MHC (pMHC)–I structures determined, it is apparent that a rough docking mode is preserved in which the Vα domain is positioned over the MHC-I α2 helix and the N-terminal end of the peptide, whereas the Vβ domain is more often positioned over the MHC-I α1 helix and the C-terminal end of the peptide, although the precise interatomic interactions vary considerably between TCR–pMHC complexes (Rudolph et al., 2006; Godfrey et al., 2008).
As with the MHC genes, allelic sequence variation is also a feature of the TCR loci; however, the full extent of TCR polymorphism and its functional significance in influencing protective immunity is unknown. Nevertheless, several single nucleotide polymorphism (SNP) studies have revealed considerable polymorphism within the TRAV and TRBV gene segments (Subrahmanyan et al., 2001; Mackelprang et al., 2006). In one study, the TCR loci from 40 individuals across four ethnic groups were fully sequenced, and >550 SNPs were found, with many being situated in coding/regulatory regions of functional TCR genes and several causing null and nonfunctional mutations. On average, the coding region of each TCR variable gene contained two SNPs, with many more found in the 5′, 3′, and intronic sequences of these segments. Furthermore, a total of 51 SNPs in the TRA locus and 72 SNPs in the TRB locus resulted in amino acid changes (Subrahmanyan et al., 2001; Mackelprang et al., 2006), although the structural and functional consequences of this sequence variation have not been investigated. Importantly, particular TCR loci have been associated with increased susceptibility to common immune diseases such as multiple sclerosis (Seboun et al., 1989; Hibberd et al., 1992; Hockertz et al., 1998), asthma (Moffatt et al., 1994, 1997; Cho et al., 2001), and narcolepsy (Hallmayer et al., 2009).
In this study, we have investigated the functional and structural impact of natural micropolymorphism within genes encoding a public TCR that recognizes an 11–amino acid epitope, 407HPVGEADYFEY417 (referred to as HPVG), derived from the EBNA-1 protein of EBV. This epitope is highly immunogenic in EBV-exposed healthy individuals expressing HLA-B*3501 (Blake et al., 1997; Lee et al., 2004; Tellam et al., 2004; Miles et al., 2006). Although EBV is a genetically stable DNA virus, sequence variation within the HPVG epitope has been previously described (Snudden et al., 1995; Wang et al., 2002; Zhang et al., 2004; Dolan et al., 2006). Unrelated EBV+, HLA-B*3501+ individuals frequently generate CTLs against the HPVG epitope that express immunodominant public TCR α and β chains characterized by TRAV20, TRAJ58, TRBV9, and TRBJ2-2 usage (Miles et al., 2006). We now show that allelic variation within this TRBV9 gene, which led to a Gln (TRBV9*01) to His (TRBV9*02) substitution at position 55 (ImMunoGeneTics unique numbering; Lefranc, 2003), resulted in a reduction in TCR binding affinity and diminished functional recognition of the cognate viral epitope as well as the naturally occurring variants of this epitope. These factors dictate the preferential selection of the TRBV9*01 TCR β chain gene and the exclusion of TRBV9*02 in this antiviral immune response. Our data therefore illustrate both the sensitivity and significance of allelic polymorphism within the TCR loci in protective immunity.
RESULTS
Allelic variation in the TRBV9 gene
The HLA-B*3501–restricted CTL response to the HPVGEADYFEY epitope from EBV is characterized by type III–biased TCR usage (Miles et al., 2006; Turner et al., 2006), with biases in the TRAV and TRBV genes as well as conserved length and sequence motifs in the CDR3 loops. Specifically, in multiple clones from unrelated individuals, this biased response was defined by closely related TCRs, which comprised TRAV20/TRAJ58 combined with TRBV9/TRBJ2-2 (Miles et al., 2006). Notably, the CDR3α and CDR3β loops were largely germline encoded, although the CDR3α loops displayed a strongly biased selection of the random N-nucleotide–encoded Leu, whereas the CDR3β loops were characterized by a 3–amino acid “ARS/ART/VRT/APT” motif (Miles et al., 2006). Sequence data for three HPVG-specific CTL clones, isolated from three unrelated HLA-B*3501+ individuals, are shown in Fig. 1 a. These strong gene biases and selection of recurrent motifs suggested that they play a crucial role in determining the specificity toward the HLA-B*3501HPVG complex.
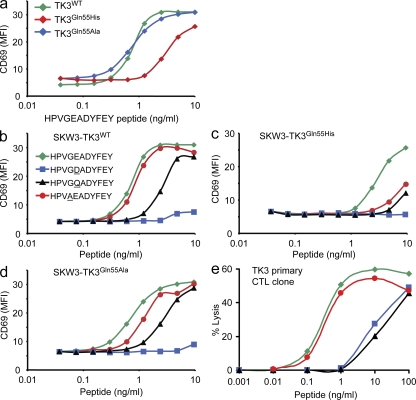
Figure 1.
TRBV9*01 is required for optimal recognition by a public TCR that dominates the response to the HPVG viral epitope. (a) Highly conserved V-(D)-J junctional region sequences of TCR α and β chains from CTL clones that recognize HLA-B*3501HPVG, isolated from three unrelated individuals. The nucleotide sequences are presented, and the one-letter code designating the translated amino acid is shown above the first nucleotide in each codon. Colored areas designate nucleotides of germline origin. (b) HLA-B*3501HPVG multimer staining, and CD3 and CD8 cell-surface expression for JurkatCD8 cells transduced with the TK3WT, TK3Gln55His (two clones), or TK3Gln55Ala TCR. Cells were stained with OKT3 (anti-CD3), anti-CD8a, or an HLA-B*3501HPVG multimer at 4°C. Data for the untransfected parental line was left out of the histogram for simplicity. (c) The TCR-transduced JurkatCD8 cells were also stained at 37°C for 4 h with the HLA-B*3501HPVG pentamer using a range of multimer concentrations (final concentration indicated on the x axis). Mean fluorescence intensity (MFI) of HLA-B*3501HPVG pentamer staining is shown on the y axis. (d) Activation of the TCR-transduced JurkatCD8 cells with various concentrations of the HLA-B*3501HPVG pentamer. CD69 up-regulation was used as a marker for cell activation. The binding of HLA-B*3501HPVG pentamer was performed at 37°C for 4 h. The cells were then washed and costained with APC-labeled anti-CD69 antibody for 30 min on ice. The experiments were conducted at least twice with similar results.
Polymorphism within the TRBV9 gene is common across various ethnic groups, with the TRBV9*01 allele occurring more frequently than the TRBV9*02 allele (TRBV9*01: 82, 79, and 97%; and TRBV9*02: 18, 21, and 3% in Caucasians, African Americans, and Chinese, respectively; Brzezinski et al., 2005). A single amino acid difference distinguishes these two alleles, with TRBV9*01 encoding a Gln and TRBV9*02 encoding a His at position 55, which is a framework residue lying directly adjacent to the CDR2 loop of the β chain. Accordingly, we investigated the possibility that this TCR micropolymorphism could influence recognition of HLA-B*3501HPVG.
PBMCs were collected from 27 healthy, HLA-B35+, EBV-exposed individuals, and genomic TRBV sequencing revealed that 21 donors were homozygous for the TRBV9*01 allele, 6 donors were heterozygous for TRBV9*01 and TRBV9*02, and 0 donors were homozygous for the TRBV9*02 allele. To determine if both of these alleles could be used in the response to the HPVG epitope, an HLA-B*3501HPVG multimer was used to sort T cells specific for the HPVG epitope from five of the heterozygous donors who were confirmed to be HLA-B*3501+. cDNA from the expressed TRBV9 gene products was cloned into Escherichia coli, and sequence analysis of at least 31 clones from each donor revealed that all were from the TRBV9*01 allele, indicating that TRBV9*02 was not used in the response to this EBV epitope (Table I). To confirm that these five heterozygous donors carried T cells expressing both TRBV9*01 and TRBV9*02 in their peripheral repertoire, the same cDNA sequencing procedure was used on unsorted PBMCs. Although T cells expressing TRBV9*01 were more common in the peripheral repertoire than those expressing TRBV9*02, these data confirmed that TRBV9*02-expressing T cells were available in all five donors for potential use in antiviral T cell immunity (Table I).
Table I.
Expression of TRBV9 alleles in T cells from HLA-B*3501+, EBV-sero+, TRBV9*01–TRBV9*02 heterozygous individuals
| Donor | HLA-B*3501HPVG multimer–sorted cells | Unsorted PBMCs | ||
| TRBV9*01 | TRBV9*02 | TRBV9*01 | TRBV9*02 | |
| Donor 7 | 31/31 | 0/31 | 16/26 | 10/26 |
| Donor 18 | 31/31 | 0/31 | 24/27 | 3/27 |
| Donor 23 | 31/31 | 0/31 | 20/29 | 9/29 |
| Donor 26 | 32/32 | 0/32 | 25/31 | 6/31 |
| Donor 27 | 36/36 | 0/36 | 19/29 | 10/29 |
Differential functional recognition
Next, we aimed to establish why the TRBV9*01 allele was repeatedly selected in the response to HLA-B*3501HPVG, whereas the TRBV9*02 allele was not. Further TCR sequence analysis for the TK3 CTL clone (Fig. 1 a) confirmed that its β chain was encoded by the TRBV9*01 allele, and it was therefore studied further as a prototype of this response. The TCR-negative human T cell line Jurkat, which had been engineered to stably express CD8 (Beddoe et al., 2009), was transduced with retroviral expression constructs encoding (a) the TK3 TCR cDNA (JurkatCD8-TK3WT), (b) the TK3 TCR with the naturally occurring Gln→His β-chain substitution corresponding to position 55 of TRBV9 (JurkatCD8-TK3Gln55His), or (c) a corresponding alanine mutant (JurkatCD8-TK3Gln55Ala). After cloning, these CD8+ Jurkat T cells expressing the wild-type and mutant TK3 TCRs were closely matched for levels of both CD3 and CD8 (Fig. 1 b). An HLA-B*3501HPVG multimer was used to assess antigen recognition by the TK3 TCR and its variants. The multimer incubation was performed at 4°C, and strong staining was observed for all cell lines (Fig. 1 b), confirming equivalent levels of TCR expression by the different transduced JurkatCD8 cell lines. However, when the multimer staining was repeated using a 37°C incubation instead of 4°C and a wide range of multimer concentrations, minor differences in staining intensity were observed between the cell lines, with two clones transduced with the TK3Gln55His TCR staining with the lowest intensity (Fig. 1 c). This temperature sensitivity in multimer association suggested that the TK3Gln55His TCR may bind with a lower avidity to the MHC–viral peptide complex in comparison to the TK3WT TCR.
Antigen-specific activation of the Jurkat transformants was then assayed by measuring up-regulation of cell-surface CD69 after incubation with varying concentrations of the HLA-B*3501HPVG multimer. Although the JurkatCD8-TK3WT and JurkatCD8-TK3Gln55Ala cell lines were activated by the multimer, the JurkatCD8-TK3Gln55His cells failed to show significant CD69 up-regulation after exposure to the pMHC complex (Fig. 1 d).
The JurkatCD8-TK3WT, JurkatCD8-TK3Gln55His, and JurkatCD8-TK3Gln55Ala cell lines were compared in dose–response CD69 up-regulation experiments using synthetic HPVGEADYFEY peptide presented on the HLA-deficient C1R cell line that had been transfected to express HLA-B*3501. In contrast to data shown in Fig. 1 d for the HLA-B*3501HPVG multimer, no significant differences in CD69 up-regulation were observed between the different cell lines after peptide stimulation (not depicted). We reasoned that the high levels of CD8 expression on the JurkatCD8 cells could mask TCR–pMHC affinity differences, and therefore the SKW3 thymoma cell line, which is TCR α and β deficient and lacks high levels of CD8αβ expression (Hundhausen et al., 1992), was also transduced to express either the TK3WT, TK3Gln55His, or TK3Gln55Ala TCRs. When these cell lines were tested in CD69 up-regulation experiments using various concentrations of the HPVGEADYFEY peptide presented on C1R–HLA-B*3501 APCs, the SKW3-TK3Gln55His cell line was found to require much higher peptide concentrations for equivalent CD69 up-regulation in comparison to the SKW3-TK3WT and SKW3-TK3Gln55Ala cell lines (Fig. 2 a). Collectively, these results establish that the TK3WT TCR can recognize the HLA-B*3501HPVG complex more efficiently than the allelic variant TK3Gln55His.
Figure 2.
T cell recognition of naturally occurring variants of the HPVGEADYFEY epitope. (a) Activation of SKW3-TK3WT, SKW3-TK3Gln55His, and SKW3-TK3Gln55Ala cell lines with various concentrations of the HPVGEADYFEY peptide, presented on C1R–HLA-B*3501 APCs. (b–d) Activation of SKW3-TK3WT (b), SKW3-TK3Gln55His (c), and SKW3-TK3Gln55Ala (d) cell lines with various concentrations of the four variants of the HPVGEADYFEY sequence, presented on C1R–HLA-B*3501 APCs. Mean fluorescence intensity (MFI) of CD69 was determined and plotted against the concentration of peptide as indicated. (e) The primary TK3 CTL clone was tested for recognition of peptides corresponding to the HPVGEADYFEY epitope or three variant sequences in dose–response cytotoxicity assays using HLA-B*3501+ PHA blasts as target cells. The experiments were conducted at least twice with similar results.
HPVG sequence variants
Given the sensitivity of the TK3 TCR to micropolymorphism within the TRBV9 gene, we next evaluated the impact of naturally occurring mutations in the HPVG epitope on recognition by the TK3WT, TK3Gln55His, and TK3Gln55Ala TCRs. The dominant EBV strains infecting Caucasians encode the HPVGEADYFEY sequence, as demonstrated by a recent study that found that 42 out of 43 Australian Caucasians carried strains encoding this epitope (Bell et al., 2008). In contrast, the dominant strain in Chinese individuals includes a conserved amino-acid substitution at position 5 (HPVGDADYFEY; Wang et al., 2002; Zhang et al., 2004), and this variant was also found in six out of six viral strains from Papua New Guinea (unpublished data). Furthermore, the Ag876 strain of EBV, which represents the prototype type-2 EBV strain, encodes another variant of this epitope, HPVAEADYFEY (Dolan et al., 2006). A third variant, HPVGQADYFEY, has also been described in type-1 EBV isolates (Snudden et al., 1995; Bell et al., 2008).
The SKW3-TK3WT cell line recognized the HPVGEADYFEY and HPVAEADYFEY peptides very efficiently, whereas suboptimal recognition was observed for the HPVGQADYFEY variant. The HPVGDADYFEY variant failed to stimulate significant CD69 up-regulation at any peptide concentration (Fig. 2 b). As mentioned in the previous section, the SKW3-TK3Gln55His cells were weakly responsive to the HPVGEADYFEY peptide, and the amino-acid substitutions at positions 4 or 5 resulted in further loss of stimulatory capacity (Fig. 2 c). The SKW3-TK3Gln55Ala cell line showed similar specificity for the peptide variants as the SKW3-TK3WT cell line (Fig. 2 d).
The TK3 CTL clone was raised from a healthy HLA-B*3501+, EBV-sero+ Caucasian individual (Miles et al., 2006). This primary CTL clone was next used in peptide dose–response cytotoxicity assays, using HLA-B*3501+ PHA blasts as target cells, to examine whether it could recognize the peptide variants. Although the HPVAEADYFEY peptide was recognized very well by the TK3 CTLs, the two mutations at position 5 in the EBV epitope led to a significant reduction in CTL lysis at limiting peptide concentrations (Fig. 2 e).
These data demonstrate that the public TCR expressed by the TK3 CTL clone was finely tuned for specific recognition of the HPVGEADYFEY epitope, although it is equally effective at recognizing the HPVAEADYFEY sequence encoded by type-2 strains of EBV. Furthermore, the data establish that selection of the TRBV9*01 allele over the TRBV9*02 allele for this public EBV-specific TCR ensures efficient recognition of the stimulating viral epitope and a greater capacity to tolerate mutations in the viral epitope.
Affinity measurements
To better understand the basis for the differential recognition of HLA-B*3501HPVG and the HPVG variants by the TK3WT, TK3Gln55His, and TK3Gln55Ala TCRs, surface plasmon resonance (SPR) studies were undertaken. The TK3WT TCR and its two variants were expressed, refolded, and purified, and the yields of refolded protein were equivalent for each TCR. Further, the TK3WT, TK3Gln55His, and TK3Gln55Ala TCRs appeared structurally intact in that they reacted with a conformationally specific anti-TCR mAb. By SPR analysis, the affinity (Kd) of the TK3WT TCR for HLA-B*3501HPVG was determined to be 2.2 µM, whereas the affinity of the TK3Gln55His and TK3Gln55Ala TCRs for HLA-B*3501HPVG was 34.7 and 5.8 µM, respectively (Table II and Fig. 3). Interestingly, the TK3WT TCR–HLA-B*3501HPVG interaction had a ninefold slower off rate (0.085 ± 0.003 s−1) when compared with that of the TK3Gln55His TCR–HLA-B*3501HPVG interaction (0.787 ± 0.137 s−1; Table II and Fig. 3).
Table II.
Affinity measurements of the TK3, TK3Gln55His, and TK3Gln55Ala TCRs with HLA-B*3501 in complex with the HPVG peptide and its variants
| Immobilized ligand | Analyte | Kd | Kon | Koff | Kd calc |
| μM | x104/Ms | 1/s | μM | ||
| TK3WT | B*3501-HPVGEADYFEY | 2.2 ± 0.2 | 4.01 ± 0.22 | 0.085 ± 0.003 | 2.21 ± 0.25 |
| TK3Gln55Ala | B*3501-HPVGEADYFEY | 5.76 ± 0.07 | 1.65 ± 0.39 | 0.115 ± 0.004 | 7.345 ± 1.505 |
| TK3Gln55His | B*3501-HPVGEADYFEY | 34.66 ± 1.66 | 3.08 ± 0.25 | 0.787 ± 0.137 | 25.6 ± 1.42 |
| TK3WT | B*3501-HPVAEADYFEY | 4.8 ± 0.53 | 8.33 ± 2.35 | 0.2 ± 0.001 | 3.195 ± 0.115 |
| TK3Gln55Ala | B*3501-HPVAEADYFEY | 12 ± 0.2 | ND | ND | ND |
| TK3Gln55His | B*3501-HPVAEADYFEY | 74.5 ± 1.4 | ND | ND | ND |
| TK3WT | B*3501-HPVGDAYGFEY | 189 ± 6 | ND | ND | ND |
| TK3Gln55Ala | B*3501-HPVGDAYGFEY | 179 ± 6 | ND | ND | ND |
| TK3Gln55His | B*3501-HPVGDAYGFEY | >200 | ND | ND | ND |
| TK3WT | B*3501-HPVGQAYGFEY | 52.03 ± 12.83 | 0.151 ± 0.042 | 0.076 ± 0.002 | 54.75 ± 14.45 |
| TK3Gln55Ala | B*3501-HPVGQAYGFEY | 99.6 ± 1.4 | 0.08 ± 0.001 | 0.131 ± 0.002 | 163.5 ± 5.5 |
| TK3Gln55His | B*3501-HPVGQAYGFEY | >200 | ND | ND | ND |
Underlining indicates the variant amino acid.
Figure 3.
SPR response sensograms for the TK3WT, TK3Gln55Ala, and TK3Gln55His TCRs. (a–c) A range of concentrations of the refolded HLA-B*3501HPVG complex was used for SPR response analysis with the TK3WT (a), TK3Gln55Ala (b), and TK3Gln55His (c) TCRs. (d) Binding curves for the three TCRs are shown, using graded concentrations of HLA-B*3501HPVG (only showing from 50 to 0 µM). The experiments have been conducted in triplicate and the error bars are shown for each data point (means ± SD).
We next used SPR to investigate the impact of the sequence variation within the HPVG epitope on recognition by the TK3, TK3Gln55His, and TK3Gln55Ala TCRs. Although the TK3WT TCR bound the HLA-B*3501HPVAEADYFEY variant with an affinity (4.8 µM) comparable to the cognate HLA-B*3501HPVGEADYFEY epitope, its interaction with the HLA-B*3501HPVGQADYFEY and HLA-B*3501HPVGDADYFEY variants was diminished (52 and 189 µM, respectively; Table II and Fig. S1). Although the TK3Gln55Ala TCR recognized the three peptide variants with comparable affinity to that of the TK3WT TCR, the TK3Gln55His TCR recognized the three peptide variants with much lower affinity (75 µM for the HPVAEADYFEY variant, and >200 µM for the HPVGDADYFEY and HPVGQADYFEY variants; Table II and Fig. S1).
Thus, consistent with the earlier functional data, the TK3WT TCR interacted with the HLA-B*3501–restricted HPVG epitope and its variants with a higher affinity when compared with the TK3Gln55His TCR. This difference in affinity was primarily related to a much slower off rate for the wild-type TCR.
Overview of TK3 TCR–HLA-B*3501HPVG structure
Given the different affinities of the TK3 and TK3Gln55His TCRs toward the HLA-B*3501HPVG complex, we hypothesized that these two TCRs would interact differently with the HLA-B*3501HPVG complex. To investigate this hypothesis and the structural basis for the observations described in the previous section, we purified the TK3 TCR–HLA-B*3501HPVG complex, and solved the structure at 2-Å resolution to an Rfac and Rfree of 22.5 and 28.6%, respectively (Table S1). The initial experimental phases clearly showed unbiased electron density for the HPVG peptide (unpublished data), and moreover, the electron density at the TK3 TCR–HLA-B*3501HPVG interface was unambiguous. The TK3 TCR docked at ∼66° across the long axis of the HLA-B*3501HPVG complex (Fig. 4, a and d) and thus falls within the range of roughly conserved docking orientations previously observed in TCR–pMHC complexes (Rudolph et al., 2006). Moreover, the TK3 TCR interacted with positions 65, 69, and 155 of HLA-B*3501, which is consistent with the observation that TCRs invariably interact with these three positions on pMHC-I (Tynan et al., 2005b; Rudolph et al., 2006). The TK3 TCR docked centrally on the HLA-B*3501HPVG complex, in which the total buried surface area (BSA) at the interface was ∼2,050 Å2, with 172 van der Waals (vdw) interactions, 18 H bonds, and 3 salt bridges (Tables S2 and S3).
Figure 4.
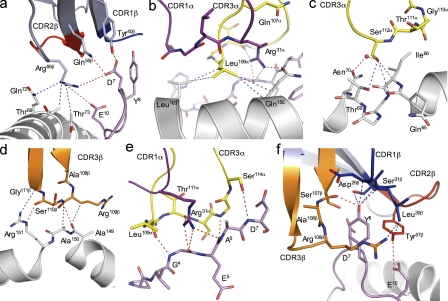
Overview and footprint of the TK3WT, TK3Gln55His, and TK3Gln55Ala TCRs in complex with HLA-B*3501HPVG. (a–c) Ribbon representation of the TK3WT (a), TK3Gln55His (b), and TK3Gln55Ala (c) TCRs in complex with HLA-B*3501HPVG. The TCR α chain is in pale pink; the TCR β chain is in pale blue, green, or gray in TK3WT, TK3Gln55His, or TK3Gln55Ala, respectively; HLA-B*3501 is in gray; and the HPVG peptide is represented as a black stick. (d–f) The footprints of the TK3WT (d), TK3Gln55His (e), and TK3Gln55Ala (f) TCRs on the HLA-B*3501HPVG complex. Residues contacted by the CDR loops are colored in purple (CDR1α), green (CDR2α), yellow (CDR3α), blue (CDR1β), red (CDR2β), or orange (CDR3β) in all complexes. The interaction with the framework of the TCR β chain is colored in pale blue. The black spheres in d, e, and f represent the orientation of the Vα and Vβ chains for each TCR on the HLA-B*3501HPVG complex, calculated by center of mass.
The Vα and Vβ domains contributed roughly equally to the BSA at the TK3 TCR–HLA-B*3501HPVG interface (55 and 45%, respectively; Fig. 4, a and d). All six CDR loops of the TK3 TCR contributed to interacting with HLA-B*3501HPVG, albeit to varying degrees, and notably, a large number of framework-derived residues (12% BSA) participated in this interaction. Namely, residues within the framework region of the TK3 TCR contacted the peptide (see following section), and Arg66β contacted residues Thr69, Gln72, and Thr73 on the α1 helix of HLA-B*3501 (Fig. 5 a and Table S2). Notably, Arg66β is unique to TRBV9 in that it is not found in any other TRBV genes. The CDR1α and CDR2α loops of the TK3 TCR contributed 17 and 5% to the BSA, respectively, and the CDR1β and CDR2β loops contributed 1 and 12% to the BSA, respectively. The CDR3 loops collectively played the most prominent role, with the CDR3α and CDR3β loops contributing 32 and 19% of the BSA at the interface, respectively, thereby providing clues toward the restricted length and repeated CDR3 usage.
Figure 5.
TK3WT contacts with HLA-B*3501 and with the HPVG epitope. (a–d) Contacts between the HLA-B*3501 molecule and the TK3WT FWβ (a), CDR1α and CDR3α (b), CDR3α (c), and CDR3β (d) are shown, with the HLA-B*3501 and selected side chains depicted in white. The CDR loops and selected side chains of the TCR are colored as in Fig. 4 for all of the panels, and the peptide is represented as a pink stick. The blue and red dashed lines represent the vdw and polar interactions, respectively, and the red sphere represents a water molecule. (e and f) Interactions between the HPVG peptide and the TCR. (e) The interaction between the CDR1α and CDR3α with residues P4–P7 of the HPVG peptide. (f) The interaction between the three CDRβ loops with the HPVG peptide.
The CDR2α loop was located peripheral to the α2 helix, and consequently its interactions were limited to the bulky aromatic ring of Tyr57α packing against the side chain of Arg151 (unpublished data). The CDR1α loop sat above and predominantly interacted with the α2 helix, in which Arg31α interdigitized between Gln107α and Leu109α of the CDR3α to plug a cavity between the HPVG peptide and the TCR α chain, and H bond to Gln155 (Fig. 5 b). The unusual interplay between the CDR1α and CDR3α loops underpinned the restricted length of the CDR3α loop and provided a basis for understanding the repeated usage of the N region–encoded Leu109α residue. Moreover, Leu109α formed several specificity-governing interactions with Gln155, the main chain of Tyr159 and Leu163. The small side chain of N region–encoded Gly110α assisted in enabling the Jα-encoded Ser112α to form several vdw interactions with the main chain of Gln65, Ile66, and Thr69 and a water-mediated H bond with Asn70 (Fig. 5 c). The CDR1β loop did not appreciably contact the MHC-I, whereas the CDR2β loop interacted with the α1 helix of HLA-B*3501: Glu61β and Tyr57β from the CDR2β loop contacted Gln72 and Glu76, respectively (Table S2). The 108ARS110 motif and Gly111β of the CDR3β loop contacted the tip of the α2 helix (residues 149–151). Namely, the Arg109β side chain interacted with Ala149, whereas its main chain interacted with Ala150. Ser110β H bonded the main chain of Ala149 and contacted Arg151, the latter of which also contacted Gly111β, and collectively, these interactions underpinned the biased selection of the “ARS” motif (Fig. 5 d).
Having determined the structure of HLA-B*3501HPVG in the nonliganded state (Miles et al., 2006), we were able to observe the conformational changes that took place in HLA-B*3501 upon complexation with the TK3 TCR (conformational changes within the peptide are discussed in the following section). Although the overall conformation of HLA-B*3501 remained relatively rigid upon ligation (root mean square deviation = 0.4 Å), several HLA-B*3501 residues changed conformation to avoid clashes with or to form specificity-governing contacts with the TK3 TCR. For example, the side chain of Arg151 flipped conformation to fit between the aromatic ring of Tyr57α from the CDR2α loop and Gln155 of HLA-B*3501 (Fig. S2 a). The latter residue also changed conformation as a result of Arg31α from the CDR1α loop, which inserted its guanadinium head group between the α2 helix of HLA-B*3501 and the HPVG peptide. Within the α1 helix there were five MHC residues (Asp61, Arg62, Gln65, Gln72, and Gln76) that changed conformation upon TK3 TCR ligation (Fig. S2 a). For example, the Arg62 was pushed outside the peptide-binding groove via the CDR3α loop, which caused Arg62 to break its H bond with Gln65, which then transmitted to Gln65 forming a new interaction with Asp61. Accordingly, the principle role of the CDR3 loops in interacting with HLA-B*3501 was consistent with the biased CDR3 usage exhibited in this response.
Peptide-mediated contacts
In the nonliganded state, the central region (residues 5–8) of the HPVG peptide was highly mobile within the antigen-binding cleft of HLA-B*3501 (Miles et al., 2006). Upon TK3 TCR ligation, this region of the peptide was stabilized, whereby the TK3 TCR contacted the peptide substantially and contributed 42% BSA at the pMHC interface, a value that is at the higher end when compared with the TCR–pMHC database (Rudolph et al., 2006). Collectively, five of the CDR loops interacted with the HPVG peptide, contacting residues P3-Val to P8-Tyr and P10-Glu, forming a total of 81 vdw contacts, 13 H bonds, and 2 salt bridges (Table S3). P3-Val to P7-Asp were contacted by the CDR1α and the CDR3α loops, in which the role of the CDR1α loop was restricted to Arg31α interacting with P4-Gly and P6-Ala. The CDR3α loop made extensive contact with the peptide (Fig. 5 e), with five residues of the HPVG peptide (P3-Val to P7-Asp) contacted by five CDR3α residues (Leu109α and from Thr111α to Ser114α; Fig. 5 e). Because of its small side chains, the 111TSGS114 motif (found exclusively in TRAJ58) was in very close proximity to the HPVG peptide, making a large number of contacts from P4-Gly to P7-Asp. The C-terminal region of the peptide, from P7-Asp to P10-Glu, was contacted by the three CDRβ loops (Fig. 5 f) and also by three residues of the β-chain framework (Tyr40β, Gln55β, and Arg66β; Fig. 6 a). The P8-Tyr side chain was enveloped by the three CDRβ loops, in which the CDR1β was located above the P8-Tyr, forming contacts with Asp29β, Leu30β, and Ser31β. This latter observation further explains TRBV9 usage, as none of the other 54 TRBV genes have the “DLS” motif in CDR1β. In addition, the P8-Tyr side chain was flanked by the Tyr57β of the CDR2β loop and the CDR3β (Ser107β, Ala108β, and Arg109β) in a manner that permitted the Arg109β side chain to wrap around the aromatic ring of P8-Tyr (Fig. 5 f). The CDR2β loop sat above the P10-Glu, which formed an H bond with Tyr57β as well as a salt bridge with the framework residue, Arg66β (Fig. 5 f and Fig. 6 a). The P7-Asp formed vdw interactions with the CDR1β and CDR3β loops, an H bond with Ser107β, and vdw contacts and an H bond with the framework residue Tyr40β (Fig. 5 f). Additionally, P7-AspOδ2-Oδ1 H bonded to the polymorphic residue Gln55βNε2 (Fig. 6 a and Table S3).
Figure 6.
Interactions with the polymorphic TCR residue. (a–c) Interaction between the TK3WT (a), TK3Gln55His (b), and TK3Gln55Ala (c) TCRs with the peptide, localized at the polymorphic site of the β chain. The different chains are colored as in Fig. 4, the residues involved in the interaction are represented as sticks, and all of the panels show the same orientation of the different structures. (b) Loss of a direct H bond between the Arg66β and D7, and also between the Arg115α and the Ser112α in the TK3Gln55His structure when compared with the TK3WT (a). The loss of contacts is greater in the TK3Gln55Ala structure (c) when compared with the TK3WT complex (a). The blue and red dashed lines represent the vdw and polar interactions, respectively, and the red sphere represents a water molecule.
As mentioned in the previous paragraph, an earlier study found that the central region (residues 5–8) of the HPVG peptide was highly mobile within the antigen-binding cleft of HLA-B*3501 in the nonliganded state and, therefore, the peptide conformation could not be modeled in its entirety (Miles et al., 2006). However, the bulged peptide was more stable when bound to HLA-B*3508, which differs from HLA-B*3501 at a single amino acid (position 156; Miles et al., 2006). We were therefore able to compare the conformation of the central bulged region of the peptide in the HLA-B*3508HPVG unliganded structure and in the TK3 TCR–HLA-B*3501HPVG complex. This comparison suggested that the conformation of the peptide bulge was altered in the TCR-ligated complex, allowing the TCR to maximize MHC-I contacts (Fig. S2 b). The TCR regions responsible for these changes in peptide conformation are the CDR3α loop and the framework residues Arg66β and Gln55β. Accordingly, the TK3 TCR substantially contacted the HPVG epitope and included an interaction with the polymorphic Gln55β residue.
The structural impact of micropolymorphism within the TRBV9 gene
To investigate the structural impact of the sequence micropolymorphism between the TRBV9*01 and TRBV9*02 alleles, we next determined the structure of the TK3Gln55His TCR–HLA-B*3501HPVG complex to 2.1-Å resolution with an Rfac and Rfree of 22.8 and 27.6%, respectively (Fig. 4, b and e). Additionally, we solved the structure of the TK3Gln55Ala TCR–HLA-B*3501HPVG complex to 2.7-Å resolution to establish why the Gln55Ala mutation did not affect the affinity of the interaction when compared with that of the cognate interaction (Fig. 4, c and f; and Tables S1–S3). Both ternary complexes crystallized in the same space group and unit cell dimensions as that of the cognate complex, and thus, any structural differences observed could be attributable to the mutation at TRBV9 position 55 in the TK3 TCR.
The TK3Gln55His and TK3Gln55Ala TCRs engaged HLA-B*3501HPVG similarly to that of the TK3 TCR–HLA-B*3501HPVG complex (overall root mean square deviation = 0.62 and 0.92 Å, respectively; Fig. 4, b and c), burying a similar surface area (2,025 and 1,920 Å2 for TK3Gln55His and TK3Gln55Ala, respectively). However, the Gln55His mutation caused a decrease in the number of contacts (155 vdw contacts, 15 H bonds, and 2 salt bridges) with HLA-B*3501HPVG when compared with the TK3 TCR–HLA-B*3501HPVG complex (172 vdw contacts, 18 H bonds, and 3 salt bridges), and the number of contacts with the TK3Gln55Ala TCR was decreased further (129 vdw, 13 H bonds, and 1 salt bridge; Tables S2 and S3). Compared with the interaction involving Gln55β in the cognate complex (Fig. 6 a), His55β H bonded to P7-Asp of the HPVG peptide in a similar manner (Fig. 6 b). In the TK3Gln55Ala TCR–HLA-B*3501HPVG complex, the interaction between position 55 and the HPVG epitope was lost (Fig. 6 c). Thus, the loss of contacts with HLA-B*3501HPVG at position 55 with P7-Asp was not the sole discriminator for the weaker affinity of the TK3Gln55His TCR when compared with the TK3 TCR, as the TK3Gln55Ala TCR displayed a loss of contacts without decreasing the affinity for HLA-B*3501HPVG.
Within the TK3 TCR, the neutral Gln55β residue sat within a basic pocket, flanked by Arg115α and Arg66β, and additionally surrounded by the main chain N atoms of R115α, S114α, and G113α. Upon ligation with HLA-B*3501HPVG, the negative charge of the P7-Asp residue was effectively neutralized by H bonding to Gln55β and salt bridging to Arg66β. However, in the TK3Gln55His TCR, position 55 was replaced by the positively charged His residue, thereby creating a more electropositively charged pocket within the TK3 TCR. To accommodate this additional positive charge, the conformations of these neighboring Arg residues were subtly altered, resulting in the loss of a direct salt bridge between P7-Asp and Arg66β, the loss of a direct H bond between Arg115α and Ser112α-O, and a reduction in the vdw contacts with HLA-B*3501 (Fig. 6 b and Fig. S2 c). Consequently, the TK3Gln55His TCR less readily accommodates the negative charge of the P7-Asp, which accounts for the 10 times faster dissociation of the TK3Gln55His–HLA B*3501HPVG complex when compared with both the TK3WT and TK3Gln55Ala TCRs in complex with the same pHLA (Table II). Accordingly, altered charge complementarity at the interface between the TK3 TCR and HLA-B*3501HPVG underpinned the weaker interaction and faster dissociation with the TK3Gln55His TCR, and the preferential selection of TRBV9*01 over TRBV9*02 in this response.
DISCUSSION
Given the central role that T cells play in immune responses and immune-related disorders, polymorphism in the TCR genes has long been suspected as a potential candidate for disease susceptibility, and recent studies identifying extensive allelic sequence variation in the TCR V genes support this contention (Subrahmanyan et al., 2001; Mackelprang et al., 2006). Indeed, genomic polymorphisms of TCR genes have been found in association with common immune diseases such as multiple sclerosis (Seboun et al., 1989; Hibberd et al., 1992; Hockertz et al., 1998), asthma (Moffatt et al., 1994, 1997; Cho et al., 2001), and narcolepsy (Hallmayer et al., 2009); however, the mechanisms controlling these effects are not understood. Although these associations imply that micropolymorphism within the TCR genes can influence T cell recognition and/or repertoire selection, our present study is the first to directly characterize the structural and functional impact of allelic sequence variation in the TCR loci. Our demonstration that a single amino-acid difference within a TRBV gene can reduce TCR binding affinity for a viral peptide–MHC complex and preclude its dominant selection in an antiviral T cell response sheds light on how allelic polymorphism in the TCR loci affects the adaptive immune response. Furthermore, the molecular basis for these functional differences was revealed by structural studies showing that micropolymorphism within the TRBV9 gene affects stabilizing interactions between the TCR β chain and the antigenic peptide.
By examining TCR usage in the response to the HPVG viral epitope in donors heterozygous for two common alleles of the TRBV9 gene segment, we showed that TRBV9*01-expressing T cells, but not TRBV9*02-expressing T cells, were selected in this highly biased T cell response. The basis for this observation became clear from experiments that showed reduced affinity and functional recognition of the HLA-B*3501HPVG complex when the TRBV9*02 TCR was used in place of the wild-type TRBV9*01 TCR. In contrast, an earlier study demonstrated that the TRBV9*02 allele was used by an HLA-A2–restricted human immunodeficiency virus pol-specific CTL clone, and that introducing a Gln (as in TRBV9*01) in place of the His residue at TRBV position 55 resulted in loss of recognition of the viral peptide (Vessey et al., 1996). These contrasting observations suggest a functional basis for the retention of these closely related TRBV9 alleles in diverse human populations. Thus, selection pressure from various pathogens may maintain both TRBV9*01 and TRBV9*02 in the population because each imparts distinct T cell specificity characteristics that may favor recognition of different peptide–MHC complexes. Further, a previous report demonstrated that allelic polymorphism in the CDR1 domain of a mouse Vβ10 gene segment affected recognition of a foreign peptide derived from an HLA molecule (Bour et al., 1999), supporting the conclusion that TCR gene polymorphism can influence specificity and TCR selection in CD8+ T cell responses.
Our previous TCR repertoire analysis of the response to the HPVG epitope showed that TRBV9+ T cells dominated in four out of five HLA-B*3501+ donors, but a fifth donor used both TRBV9+ and TRBV28+ T cells in roughly equal numbers (Miles et al., 2006), establishing that TRBV9 is not an absolute requirement for recognition of this HLA-B*3501HPVG complex. Although 0 out of 27 HLA-B35+, EBV-exposed individuals recruited for the present study was homozygous for the TRBV9*02 allele, it is likely that such donors would respond to the HPVG epitope with TCRs encoded by Vβ genes other than TRBV9. Thus, retention of the TRBV9*02 allele in the population may increase the overall TCR repertoire diversity against this epitope, thereby protecting the population against viral epitope variants that escape recognition by the dominant public TCR. The HPVGDADYFEY epitope variant, for example, which is found in EBV strains infecting Chinese individuals, was recognized suboptimally by the public TRBV9*01+ TK3 TCR and particularly poorly by the TK3Gln55His TCR, and therefore, alternative TCRs are likely to be recruited in the response to this peptide.
A study of polymorphism within the human TRBV gene segments showed that nonsynonymous variants are evenly distributed between the framework and CDR regions (50 in framework regions and 8 in CDR regions), and it was suggested that the SNPs mapped to the CDR regions may alter the avidity or specificity of the TCR (Subrahmanyan et al., 2001). Our data demonstrate that coding polymorphisms within framework residues are also functionally relevant, directly affecting TCR affinity toward HLA-B*3501HPVG, thereby raising the possibility that coding polymorphisms anywhere along the TRBV gene could potentially influence immune function.
Recently, structural studies have suggested that the germline-encoded regions of a TCR dictate MHC bias (Feng et al., 2007; Dai et al., 2008; Scott-Browne et al., 2009). However, we show in this study that the germline-encoded polymorphic residue within TRBV9 exclusively contacted the viral epitope, and this interaction effectively determined the functional outcome of this MHC-restricted response.
This study has highlighted the structural and functional relevance of nonsynonymous TCR V gene polymorphism, suggesting that it may have evolved to further diversify the immune repertoire. Given the structural and functional impact of the TRBV gene polymorphism documented in this paper, it will be important to examine the complementary role of TCR polymorphism and MHC associations in autoimmune disease involving well-defined self-antigens. Our data provide insights into the mechanisms controlling disease associations with allelic sequence variation in the TCR loci, and suggest its potentially important role in shaping the antiviral T cell repertoire, thereby contributing toward interindividual variability in T cell responses and the outcome of infection.
MATERIALS AND METHODS
TCR sequencing.
The CTL clones TK3, MW1, and CS1 have been characterized previously, and TCR α and β chain sequences were determined as previously described (Miles et al., 2006). For TRBV9 genotyping, genomic DNA samples were obtained from PBMCs from 27 healthy, HLA-B*35+ EBV-exposed Caucasian individuals. The polymorphic region of the TRBV9 gene segment was amplified by PCR using the primers 5′-TTCAGGCTCCTCTGCTGTGT-3′ and 5′-GTGGGGCAGGAATGTTATTG-3′. The PCR products were purified using the MinElute PCR Purification Kit (QIAGEN) and directly sequenced in both directions using the BigDye Terminator reaction kit (Applied Biosystems).
To examine TRBV9*01 versus TRBV9*02 expression in the T cell repertoire, total RNA was extracted using TRIzol reagent from PBMCs or HLA-B*3501HPVG–specific T cells isolated by FACS after staining with a pMHC pentamer (ProImmune). A one-step SuperScript II PCR reaction kit (Invitrogen) was used with a TRBV9 family–specific primer (5′-AGTCACACAAACCCCAAAGC-3′) and a constant region primer (5′-TTCTGATGGCTCAAACAC-3′). The PCR products were purified and cloned into the pGEM-T vector system (Promega) and sequenced using the BigDye Terminator reaction kit. Human protocols were approved by the Queensland Institute of Medical Research Human Research Ethics Committee.
TCR cloning, protein expression, and purification.
cDNA was reverse transcribed from TK3 T cell RNA using SuperScript II. PCR products encoding the TK3 TCR α and β chains were cloned into pGEM-T Easy before cotransferring into the pMIG vector (Holst et al., 2006). The primers were as follows (italicized nucleotides represent restriction sites and lowercased nucleotides represent TCR gene sequences): P1, 5′-CGGAATTCGCTAGCCACCatggagaaaatgttggagtgt-3′; P2, 5′-GGGCCCTGGGTTCTCTTCGACGTCGCCGGCCTGCTTAAGCAGCGAGAAATTGGTGGCGCCGGATCCgctggaccacagccgcagcgt-3′; P3, 5′-GGATCCGGCGCCACCAATTTCTCGCTGCTTAAGCAGGCCGGCGACGTCGAAGAGAACCCAGGGCCCatgggctgcaggctcctctgc-3′; and P4, 5′-CCGCTCGAGCTGCAGctagcctctggaatcctttct-3′.
The variant TCRs TK3βGln55His and TK3βGln55Ala were generated from the parental pGEM-TK3αWT-2A-TK3βWT plasmid DNA through site-directed mutagenesis. The corresponding inserts were transferred to the pMIG vector and the resulting constructs were denoted as pMIG-TK3WT (TK3αWT-2A-TK3βWT), pMIG-TK3Gln55His (TK3αWT-2A-TK3βGln55His), and pMIG-TK3Gln55Ala (TK3αWT-2A-TK3βGln55Ala). The mutagenesis primers were as follows (underlined nucleotides represent mutation codons): Gln55Ala P5, 5′-CTCCAGTTCCTCATTGCGTATTATAATGGAGAAG-3′; P6, 5′-CTTCTCCATTATAATACGCAATGAGGAACTGGAG-3′; Gln55His P7, 5′-CTCCAGTTCCTCATTCACTATTATAATGGAGAAG-3′; and P8, 5′-CTTCTCCATTATAATAGTGAATGAGGAACTGGAG-3′.
The TK3WT, TK3Gln55His, and TK3Gln55Ala TCRs were expressed, refolded, and purified using an engineered disulfide linkage in the constant domains between the TRAC and TRBC. Both the α and β chains of the TK3WT TCR and the mutant versions were expressed separately as inclusion bodies in the BL21 E. coli strain. Inclusion bodies were resuspended in 8 M urea, 20 mM Tris-HCl, pH 8, 0.5 mM Na-EDTA, and 1 mM dithiothreitol. The TCRs were refolded by flash dilution in a solution containing 3 M urea, 100 mM Tris, pH 8, 2 mM Na-EDTA, 400 mM l-arginine–HCl, 0.5 mM of oxidized glutathione, and 5 mM of reduced glutathione. The refolding solution was dialyzed to eliminate the urea. The resulting protein solution was purified by gel filtration and HiTrap-Q anion exchange chromatography.
Soluble class I heterodimers containing the HPVG peptide (or variant peptides) were prepared as described previously (Macdonald et al., 2002). In brief, a truncated form (amino-acid residues 1–276) of the HLA-B*3501 heavy chain and full-length β2-microglobulin (β2m) were expressed in E. coli as inclusion bodies. Each HLA-B*3501–peptide complex was refolded by diluting the heavy chain and β2m inclusion body preparations into refolding buffer containing a molar excess of peptide ligand. The refolded complexes were concentrated and purified by anion exchange chromatography. The complexes were further purified by gel filtration and HiTrap-Q anion exchange chromatography.
Retroviral transduction into JurkatCD8 and SKW3 cells.
JurkatCD8 cells were generated previously, and they lack a TCR β chain (TCRα+β−; Beddoe et al., 2009). SKW3 cells are TCRα−β− (double deficient), and they were purchased from the German Collection of Microorganisms and Cell Cultures.
JurkatCD8–TK3WT, JurkatCD8–TK3Gln55Ala, and JurkatCD8–TK3Gln55His cell lines were created via retroviral transduction as described previously (Holst et al., 2006; Beddoe et al., 2009). In brief, the plasmids pMIG-TK3WT, pMIG-TK3Gln55Ala, or pMIG-TK3Gln55His (4 µg) were individually combined with the packaging vectors pPAM-E (4 µg) and pVSV-g (2 µg) and cotransfected into 106 293T cells in a 10-cm dish with FuGENE 6 (Roche) as previously described (Holst et al., 2006). The transiently transfected 293T cells were further cultured for 5 d. During this time, the retrovirus-containing supernatant was collected twice daily and used to transduce JurkatCD8 cells along with 6 µg/ml polybrene. At the end of the transduction, JurkatCD8 cells were analyzed for expression of the TK3 αβ TCR with HLA-B*3501HPVG pentamer staining by FACS. JurkatCD8-TK3αβ–positive cells were enriched by FACS and were subsequently cloned by single-cell sorting using flow cytometry. SKW3-TK3WT, SKW3-TK3Gln55Ala, and SKW3-TK3Gln55His cell lines were generated similarly. In these transduced cell lines, GFP expression levels correlated very well with the surface expression levels of CD3.
FACS.
2 × 105 JurkatCD8-TK3 or SKW3-TK3 cells were stained with OKT3 antibody (anti-CD3) on ice for 30 min or with peptide-loaded HLA class I multimers (HLA-B*3501HPVG pentamer) on ice or at 37°C for 45 min. When antibody costaining was also performed, pentamer binding was performed first, followed by costaining antibodies. After washing twice with FACS buffer (PBS containing 2% fetal calf serum and 0.02% azide), the cells were run through a FACSCalibur or FACSAria with CellQuest software (all from BD) and analyzed with FlowJo software (Tree Star, Inc.). For cell sorting, cells were similarly stained and all steps were performed in a biohazardous hood and with filter-sterile reagents.
T cell activation assay.
105 JurkatCD8-TK3 cells were cultured with or without antigen stimulation for 2 or 4 h at 37°C. The antigen stimulation treatments involved peptide-loaded HLA class I multimer binding. The cells were pelleted and assayed for CD69 up-regulation by antibody staining and FACS. T cell activation was measured by the increase of the mean fluorescence of CD69 staining with gated JurkatCD8-TK3 cells. SKW3-TK3 cell activation assays were conducted similarly, with the exception of pulsing the APCs (C1R-B*3501) first with peptides at the concentration indicated in the figures for 1 h at 37°C before adding an equivalent number of SKW3-TK3 cells.
Cytotoxicity assays.
The primary TK3 CTL clone was tested in duplicate for cytotoxicity in standard 5-h chromium release assays. In brief, CTLs were assayed against 51Cr-labeled autologous PHA blast targets (effector/target = 2:1) that were pretreated with various concentrations of synthetic peptide or left untreated. Peptides were synthesized by Mimotopes Ltd. Toxicity testing of all peptides was performed before use by adding peptide to 51Cr-labeled PHA blasts in the absence of CTL effectors. A β scintillation counter (Topcount Microplate; PerkinElmer) was used to measure 51Cr levels in assay supernatant samples. The mean spontaneous lysis for target cells in the culture medium was always <20%, and the variation from the mean specific lysis was <10%.
SPR measurement and analysis.
All SPR experiments were conducted at 25°C on the BIAcore 3000 instrument (GE Healthcare) with HBS buffer (10 mM Hepes, pH 7.4, 150 mM NaCl, and 0.005% surfactant P20). The HBS buffer was supplemented with 1% bovine serum albumin to prevent nonspecific binding. The human TCR-specific mAb 12H8 (Borg et al., 2005) was coupled to research-grade CM5 chips with standard amine coupling. For each experiment, the TCRs (TK3, TK3Gln55His, or TK3Gln55Ala) were passed over the flow cell until ∼200–400 response units were captured by the antibody. One flow cell was left empty to be used as a control cell for each experiment. HLA-B*3501, either in complex with the wild-type or variant peptides, were injected over all four flow cells at a rate of 20 µl/min with a concentration range of 0.78–200 µM. The final response was calculated by subtraction of the response of the antibody alone from that of the antibody–TK3WT, –TK3Gln55His, or –TK3Gln55Ala complexes. The antibody surface was regenerated between each analyte injection with Actisep (Sterogene). All experiments were conducted at least in duplicate.
BIAevaluation (version 3.1) was used for data analysis, and the 1:1 Langmuir binding model was modified to include an additional parameter for the drifting baseline for the TCR capture by the 12H8 antibody to calculate the kinetic constants.
Crystallization.
Crystals of the TK3WT–HLA-B*3501HPVG complex were grown by the hanging-drop, vapor-diffusion method at 20°C with a protein/reservoir drop ratio of 1:1, at a concentration of 6 mg/ml in 10 mM Tris, pH 8, and 150 mM NaCl. Large rhombus-shaped crystals grew using 15% PEG 3350, 0.2 M Li2SO4, and 0.1 M Na-citrate, pH 5.6. The crystals of TK3Gln55His and TK3Gln55Ala in complex with HLA-B*3501HPVG were obtained in the same conditions.
Data collection and structure determination.
The TCR–pMHC crystals were soaked in a cryoprotectant solution containing mother liquor solution with a PEG concentration increased to 30% (wt/vol), and were flash frozen in liquid nitrogen. The data were collected on the 3BM1 beamline at the Australian Synchrotron, Clayton using a charge-coupled device detector at 100 K (Quantum 210; Area Detector Systems Corporation). Data were processed using XDS software and scaled using XSCALE software (Kabsch, 1993).
The TK3–HLA-B*3501HPVG crystal (and the other complexes) belonged to the space group P1 (Table S1), with unit cell dimensions consistent with one complex in the asymmetric unit. The structure was determined by molecular replacement using the PHASER program (Read, 2001) with the CF34 TCR as the search model for the TCR (PDB accession no. 3FFC; Gras et al., 2009) and the HLA-B*3501HPVG complex for the MHC model without the peptide (PDB accession no. 2FYY; Miles et al., 2006). Manual model building was conducted using COOT software (Emsley and Cowtan, 2004), followed by maximum-likelihood refinement with the REFMAC 5 program (CCP4 suite; Collaborative Computational Project, Number 4, 1994; Murshudov et al., 1997). Translation, liberation, and screw-rotation displacement refinement was also used during the refinement process to model anisotropic displacements of defined domains. The TCR was numbered according to the ImMunoGeneTics unique numbering system (Lefranc, 2003), whereby the CDR1 loops start at residue 27, the CDR2 loops start at residue 56, and the CDR3 loops start at residue 105. The final model was validated using the PDB validation web site and the final refinement statistics are summarized in Table S1. All molecular graphics representations were created using PyMOL (DeLano, 2002).
PDB accession numbers.
The coordinates of the TK3WT–HLA-B*3501HPVG, TK3Gln55His–HLA-B*3501HPVG, and TK3Gln55Ala–HLA-B*3501HPVG complexes have been deposited under PDB accession nos. 3MV7, 3MV8, and 3MV9, respectively.
Online supplemental material.
Table S1 summarizes data collection and refinement statistics for the structural studies. Table S2 lists the TCR–MHC contacts in the three TCR–pMHC complexes. Table S3 lists the TCR–peptide contacts in the three TCR–pMHC complexes. Fig. S1 shows SPR data for the TK3WT, TK3Gln55His, and TK3Gln55Ala TCRs with HLA-B*3501 bound to HPVG and its natural variants. Fig. S2 shows conformational changes in the pMHC upon ligation with TK3WT, and structural differences between the TK3WT–HLA-B*3501HPVG and TK3Gln55His–HLA-B*3501HPVG complexes. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20100603/DC1.
Acknowledgments
This work was supported by the Australian Research Council (ARC; Federation Fellowship to J. Rossjohn) and the National Health and Medical Research Council (NHMRC) of Australia (Career Development Award to L.C. Sullivan; Senior Research Fellowships to S.R. Burrows, A.W. Purcell, and R. Khanna; C.J. Martin Overseas Biomedical Fellowship to J.J. Miles; and Dora Lush Scholarship to R.M. Brennan), and by grants from the NHMRC and ARC.
The authors have no conflicting financial interests.
Footnotes
Abbreviations used:
- BSA
- buried surface area
- CDR
- complementarity-determining region
- pMHC
- peptide-MHC
- SNP
- single nucleotide polymorphism
- SPR
- surface plasmon resonance
- vdw
- van der Waals
References
- Acha-Orbea H., Mitchell D.J., Timmermann L., Wraith D.C., Tausch G.S., Waldor M.K., Zamvil S.S., McDevitt H.O., Steinman L. 1988. Limited heterogeneity of T cell receptors from lymphocytes mediating autoimmune encephalomyelitis allows specific immune intervention. Cell. 54:263–273 10.1016/0092-8674(88)90558-2 [DOI] [PubMed] [Google Scholar]
- Archbold J.K., Macdonald W.A., Gras S., Ely L.K., Miles J.J., Bell M.J., Brennan R.M., Beddoe T., Wilce M.C., Clements C.S., et al. 2009. Natural micropolymorphism in human leukocyte antigens provides a basis for genetic control of antigen recognition. J. Exp. Med. 206:209–219 10.1084/jem.20082136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argaet V.P., Schmidt C.W., Burrows S.R., Silins S.L., Kurilla M.G., Doolan D.L., Suhrbier A., Moss D.J., Kieff E., Sculley T.B., Misko I.S. 1994. Dominant selection of an invariant T cell antigen receptor in response to persistent infection by Epstein-Barr virus. J. Exp. Med. 180:2335–2340 10.1084/jem.180.6.2335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arstila T.P., Casrouge A., Baron V., Even J., Kanellopoulos J., Kourilsky P. 1999. A direct estimate of the human alphabeta T cell receptor diversity. Science. 286:958–961 10.1126/science.286.5441.958 [DOI] [PubMed] [Google Scholar]
- Beddoe T., Chen Z., Clements C.S., Ely L.K., Bushell S.R., Vivian J.P., Kjer-Nielsen L., Pang S.S., Dunstone M.A., Liu Y.C., et al. 2009. Antigen ligation triggers a conformational change within the constant domain of the alphabeta T cell receptor. Immunity. 30:777–788 10.1016/j.immuni.2009.03.018 [DOI] [PubMed] [Google Scholar]
- Bell M.J., Brennan R., Miles J.J., Moss D.J., Burrows J.M., Burrows S.R. 2008. Widespread sequence variation in Epstein-Barr virus nuclear antigen 1 influences the antiviral T cell response. J. Infect. Dis. 197:1594–1597 10.1086/587848 [DOI] [PubMed] [Google Scholar]
- Blake N., Lee S., Redchenko I., Thomas W., Steven N., Leese A., Steigerwald-Mullen P., Kurilla M.G., Frappier L., Rickinson A. 1997. Human CD8+ T cell responses to EBV EBNA1: HLA class I presentation of the (Gly-Ala)-containing protein requires exogenous processing. Immunity. 7:791–802 10.1016/S1074-7613(00)80397-0 [DOI] [PubMed] [Google Scholar]
- Borg N.A., Ely L.K., Beddoe T., Macdonald W.A., Reid H.H., Clements C.S., Purcell A.W., Kjer-Nielsen L., Miles J.J., Burrows S.R., et al. 2005. The CDR3 regions of an immunodominant T cell receptor dictate the ‘energetic landscape’ of peptide-MHC recognition. Nat. Immunol. 6:171–180 10.1038/ni1155 [DOI] [PubMed] [Google Scholar]
- Bour H., Michielin O., Bousso P., Cerottini J.C., MacDonald H.R. 1999. Dramatic influence of V beta gene polymorphism on an antigen-specific CD8+ T cell response in vivo. J. Immunol. 162:4647–4656 [PubMed] [Google Scholar]
- Brzezinski J.L., Deka R., Menon A.G., Glass D.N., Choi E. 2005. Variability in TRBV haplotype frequency and composition in Caucasian, African American, Western African and Chinese populations. Int. J. Immunogenet. 32:413–420 10.1111/j.1744-313X.2005.00550.x [DOI] [PubMed] [Google Scholar]
- Burrows S.R., Silins S.L., Moss D.J., Khanna R., Misko I.S., Argaet V.P. 1995. T cell receptor repertoire for a viral epitope in humans is diversified by tolerance to a background major histocompatibility complex antigen. J. Exp. Med. 182:1703–1715 10.1084/jem.182.6.1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrows J.M., Wynn K.K., Tynan F.E., Archbold J., Miles J.J., Bell M.J., Brennan R.M., Walker S., McCluskey J., Rossjohn J., et al. 2007. The impact of HLA-B micropolymorphism outside primary peptide anchor pockets on the CTL response to CMV. Eur. J. Immunol. 37:946–953 10.1002/eji.200636588 [DOI] [PubMed] [Google Scholar]
- Cho S.H., Son J.W., Koh Y.Y., Min K.U., Kim Y.Y., Kim Y.K. 2001. Linkage between bronchial responsiveness to methacholine and gene markers of IL-4 cytokine gene cluster and T-cell receptor alpha/delta gene complex in Korean nuclear families. Clin. Exp. Allergy. 31:103–109 [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50:760–763 [DOI] [PubMed] [Google Scholar]
- Dai S., Huseby E.S., Rubtsova K., Scott-Browne J., Crawford F., Macdonald W.A., Marrack P., Kappler J.W. 2008. Crossreactive T cells spotlight the germline rules for alphabeta T cell-receptor interactions with MHC molecules. Immunity. 28:324–334 10.1016/j.immuni.2008.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis M.M., Bjorkman P.J. 1988. T-cell antigen receptor genes and T-cell recognition. Nature. 334:395–402 10.1038/334395a0 [DOI] [PubMed] [Google Scholar]
- DeLano W.L. 2002. PyMOL Molecular Viewer. http://www.pymol.org/(accessed July 15, 2004)
- Dolan A., Addison C., Gatherer D., Davison A.J., McGeoch D.J. 2006. The genome of Epstein-Barr virus type 2 strain AG876. Virology. 350:164–170 10.1016/j.virol.2006.01.015 [DOI] [PubMed] [Google Scholar]
- Emsley P., Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126–2132 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- Feng D., Bond C.J., Ely L.K., Maynard J., Garcia K.C. 2007. Structural evidence for a germline-encoded T cell receptor-major histocompatibility complex interaction ‘codon’. Nat. Immunol. 8:975–983 10.1038/ni1502 [DOI] [PubMed] [Google Scholar]
- Germain R.N., Margulies D.H. 1993. The biochemistry and cell biology of antigen processing and presentation. Annu. Rev. Immunol. 11:403–450 10.1146/annurev.iy.11.040193.002155 [DOI] [PubMed] [Google Scholar]
- Godfrey D.I., Rossjohn J., McCluskey J. 2008. The fidelity, occasional promiscuity, and versatility of T cell receptor recognition. Immunity. 28:304–314 10.1016/j.immuni.2008.02.004 [DOI] [PubMed] [Google Scholar]
- Gras S., Kjer-Nielsen L., Burrows S.R., McCluskey J., Rossjohn J. 2008. T-cell receptor bias and immunity. Curr. Opin. Immunol. 20:119–125 10.1016/j.coi.2007.12.001 [DOI] [PubMed] [Google Scholar]
- Gras S., Burrows S.R., Kjer-Nielsen L., Clements C.S., Liu Y.C., Sullivan L.C., Bell M.J., Brooks A.G., Purcell A.W., McCluskey J., Rossjohn J. 2009. The shaping of T cell receptor recognition by self-tolerance. Immunity. 30:193–203 10.1016/j.immuni.2008.11.011 [DOI] [PubMed] [Google Scholar]
- Hallmayer J., Faraco J., Lin L., Hesselson S., Winkelmann J., Kawashima M., Mayer G., Plazzi G., Nevsimalova S., Bourgin P., et al. 2009. Narcolepsy is strongly associated with the T-cell receptor alpha locus. Nat. Genet. 41:708–711 10.1038/ng.372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibberd M.L., Millward B.A., Wong F.S., Demaine A.G. 1992. T-cell receptor constant beta chain polymorphisms and susceptibility to type 1 diabetes. Diabet. Med. 9:929–933 10.1111/j.1464-5491.1992.tb01733.x [DOI] [PubMed] [Google Scholar]
- Hockertz M.K., Paty D.W., Beall S.S. 1998. Susceptibility to relapsing-progressive multiple sclerosis is associated with inheritance of genes linked to the variable region of the TcR beta locus: use of affected family-based controls. Am. J. Hum. Genet. 62:373–385 10.1086/301700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holst J., Szymczak-Workman A.L., Vignali K.M., Burton A.R., Workman C.J., Vignali D.A. 2006. Generation of T-cell receptor retrogenic mice. Nat. Protoc. 1:406–417 10.1038/nprot.2006.61 [DOI] [PubMed] [Google Scholar]
- Hülsmeyer M., Fiorillo M.T., Bettosini F., Sorrentino R., Saenger W., Ziegler A., Uchanska-Ziegler B. 2004. Dual, HLA-B27 subtype-dependent conformation of a self-peptide. J. Exp. Med. 199:271–281 10.1084/jem.20031690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hundhausen T., Laus R., Müller-Ruchholtz W. 1992. New parental cell lines for generating human hybridomas. J. Immunol. Methods. 153:21–29 10.1016/0022-1759(92)90301-9 [DOI] [PubMed] [Google Scholar]
- Kabsch W. 1993. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Cryst. 26:795–800 10.1107/S0021889893005588 [DOI] [Google Scholar]
- Lawlor D.A., Zemmour J., Ennis P.D., Parham P. 1990. Evolution of class-I MHC genes and proteins: from natural selection to thymic selection. Annu. Rev. Immunol. 8:23–63 10.1146/annurev.iy.08.040190.000323 [DOI] [PubMed] [Google Scholar]
- Lee S.P., Brooks J.M., Al-Jarrah H., Thomas W.A., Haigh T.A., Taylor G.S., Humme S., Schepers A., Hammerschmidt W., Yates J.L., et al. 2004. CD8 T cell recognition of endogenously expressed Epstein-Barr virus nuclear antigen 1. J. Exp. Med. 199:1409–1420 10.1084/jem.20040121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefranc M.P. 2003. IMGT, the international ImMunoGeneTics database. Nucleic Acids Res. 31:307–310 10.1093/nar/gkg085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limou S., Le Clerc S., Coulonges C., Carpentier W., Dina C., Delaneau O., Labib T., Taing L., Sladek R., Deveau C., et al. ; ANRS Genomic Group 2009. Genomewide association study of an AIDS-nonprogression cohort emphasizes the role played by HLA genes (ANRS Genomewide Association Study 02). J. Infect. Dis. 199:419–426 10.1086/596067 [DOI] [PubMed] [Google Scholar]
- Macdonald W., Williams D.S., Clements C.S., Gorman J.J., Kjer-Nielsen L., Brooks A.G., McCluskey J., Rossjohn J., Purcell A.W. 2002. Identification of a dominant self-ligand bound to three HLA B44 alleles and the preliminary crystallographic analysis of recombinant forms of each complex. FEBS Lett. 527:27–32 10.1016/S0014-5793(02)03149-6 [DOI] [PubMed] [Google Scholar]
- Macdonald W.A., Purcell A.W., Mifsud N.A., Ely L.K., Williams D.S., Chang L., Gorman J.J., Clements C.S., Kjer-Nielsen L., Koelle D.M., et al. 2003. A naturally selected dimorphism within the HLA-B44 supertype alters class I structure, peptide repertoire, and T cell recognition. J. Exp. Med. 198:679–691 10.1084/jem.20030066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackelprang R., Livingston R.J., Eberle M.A., Carlson C.S., Yi Q., Akey J.M., Nickerson D.A. 2006. Sequence diversity, natural selection and linkage disequilibrium in the human T cell receptor alpha/delta locus. Hum. Genet. 119:255–266 10.1007/s00439-005-0111-z [DOI] [PubMed] [Google Scholar]
- Miles J.J., Borg N.A., Brennan R.M., Tynan F.E., Kjer-Nielsen L., Silins S.L., Bell M.J., Burrows J.M., McCluskey J., Rossjohn J., Burrows S.R. 2006. TCR alpha genes direct MHC restriction in the potent human T cell response to a class I-bound viral epitope. J. Immunol. 177:6804–6814 [DOI] [PubMed] [Google Scholar]
- Moffatt M.F., Hill M.R., Cornélis F., Schou C., Faux J.A., Young R.P., James A.L., Ryan G., le Souef P., Musk A.W., et al. 1994. Genetic linkage of T-cell receptor alpha/delta complex to specific IgE responses. Lancet. 343:1597–1600 10.1016/S0140-6736(94)93057-0 [DOI] [PubMed] [Google Scholar]
- Moffatt M.F., Schou C., Faux J.A., Cookson W.O. 1997. Germline TCR-A restriction of immunoglobulin E responses to allergen. Immunogenetics. 46:226–230 10.1007/s002510050266 [DOI] [PubMed] [Google Scholar]
- Murshudov G.N., Vagin A.A., Dodson E.J. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53:240–255 10.1107/S0907444996012255 [DOI] [PubMed] [Google Scholar]
- Read R.J. 2001. Pushing the boundaries of molecular replacement with maximum likelihood. Acta Crystallogr. D Biol. Crystallogr. 57:1373–1382 10.1107/S0907444901012471 [DOI] [PubMed] [Google Scholar]
- Robinson J., Waller M.J., Parham P., de Groot N., Bontrop R., Kennedy L.J., Stoehr P., Marsh S.G. 2003. IMGT/HLA and IMGT/MHC: sequence databases for the study of the major histocompatibility complex. Nucleic Acids Res. 31:311–314 10.1093/nar/gkg070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph M.G., Stanfield R.L., Wilson I.A. 2006. How TCRs bind MHCs, peptides, and coreceptors. Annu. Rev. Immunol. 24:419–466 10.1146/annurev.immunol.23.021704.115658 [DOI] [PubMed] [Google Scholar]
- Scott-Browne J.P., White J., Kappler J.W., Gapin L., Marrack P. 2009. Germline-encoded amino acids in the alphabeta T-cell receptor control thymic selection. Nature. 458:1043–1046 10.1038/nature07812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seboun E., Robinson M.A., Doolittle T.H., Ciulla T.A., Kindt T.J., Hauser S.L. 1989. A susceptibility locus for multiple sclerosis is linked to the T cell receptor beta chain complex. Cell. 57:1095–1100 10.1016/0092-8674(89)90046-9 [DOI] [PubMed] [Google Scholar]
- Snudden D.K., Smith P.R., Lai D., Ng M.H., Griffin B.E. 1995. Alterations in the structure of the EBV nuclear antigen, EBNA1, in epithelial cell tumours. Oncogene. 10:1545–1552 [PubMed] [Google Scholar]
- Subrahmanyan L., Eberle M.A., Clark A.G., Kruglyak L., Nickerson D.A. 2001. Sequence variation and linkage disequilibrium in the human T-cell receptor beta (TCRB) locus. Am. J. Hum. Genet. 69:381–395 10.1086/321297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tellam J., Connolly G., Green K.J., Miles J.J., Moss D.J., Burrows S.R., Khanna R. 2004. Endogenous presentation of CD8+ T cell epitopes from Epstein-Barr virus–encoded nuclear antigen 1. J. Exp. Med. 199:1421–1431 10.1084/jem.20040191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner S.J., Doherty P.C., McCluskey J., Rossjohn J. 2006. Structural determinants of T-cell receptor bias in immunity. Nat. Rev. Immunol. 6:883–894 10.1038/nri1977 [DOI] [PubMed] [Google Scholar]
- Tynan F.E., Borg N.A., Miles J.J., Beddoe T., El-Hassen D., Silins S.L., van Zuylen W.J., Purcell A.W., Kjer-Nielsen L., McCluskey J., et al. 2005a. High resolution structures of highly bulged viral epitopes bound to major histocompatibility complex class I. Implications for T-cell receptor engagement and T-cell immunodominance. J. Biol. Chem. 280:23900–23909 10.1074/jbc.M503060200 [DOI] [PubMed] [Google Scholar]
- Tynan F.E., Burrows S.R., Buckle A.M., Clements C.S., Borg N.A., Miles J.J., Beddoe T., Whisstock J.C., Wilce M.C., Silins S.L., et al. 2005b. T cell receptor recognition of a ‘super-bulged’ major histocompatibility complex class I-bound peptide. Nat. Immunol. 6:1114–1122 10.1038/ni1257 [DOI] [PubMed] [Google Scholar]
- Tynan F.E., Elhassen D., Purcell A.W., Burrows J.M., Borg N.A., Miles J.J., Williamson N.A., Green K.J., Tellam J., Kjer-Nielsen L., et al. 2005c. The immunogenicity of a viral cytotoxic T cell epitope is controlled by its MHC-bound conformation. J. Exp. Med. 202:1249–1260 10.1084/jem.20050864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tynan F.E., Reid H.H., Kjer-Nielsen L., Miles J.J., Wilce M.C., Kostenko L., Borg N.A., Williamson N.A., Beddoe T., Purcell A.W., et al. 2007. A T cell receptor flattens a bulged antigenic peptide presented by a major histocompatibility complex class I molecule. Nat. Immunol. 8:268–276 10.1038/ni1432 [DOI] [PubMed] [Google Scholar]
- Vessey S.J., Bell J.I., Jakobsen B.K. 1996. A functionally significant allelic polymorphism in a T cell receptor V beta gene segment. Eur. J. Immunol. 26:1660–1663 10.1002/eji.1830260739 [DOI] [PubMed] [Google Scholar]
- Wang W.Y., Chien Y.C., Jan J.S., Chueh C.M., Lin J.C. 2002. Consistent sequence variation of Epstein-Barr virus nuclear antigen 1 in primary tumor and peripheral blood cells of patients with nasopharyngeal carcinoma. Clin. Cancer Res. 8:2586–2590 [PubMed] [Google Scholar]
- Zernich D., Purcell A.W., Macdonald W.A., Kjer-Nielsen L., Ely L.K., Laham N., Crockford T., Mifsud N.A., Bharadwaj M., Chang L., et al. 2004. Natural HLA class I polymorphism controls the pathway of antigen presentation and susceptibility to viral evasion. J. Exp. Med. 200:13–24 10.1084/jem.20031680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.S., Wang H.H., Hu L.F., Li A., Zhang R.H., Mai H.Q., Xia J.C., Chen L.Z., Zeng Y.X. 2004. V-val subtype of Epstein-Barr virus nuclear antigen 1 preferentially exists in biopsies of nasopharyngeal carcinoma. Cancer Lett. 211:11–18 10.1016/j.canlet.2004.01.035 [DOI] [PubMed] [Google Scholar]