Abstract
Expression of the regulatory T (T reg) cell–associated transcription factor Foxp3 can be induced by signals from the T cell receptor (TCR), interleukin-2 (IL-2), and transforming growth factor (TGF)-β. These signals are integrated by a network involving phosphatidylinositol 3 kinase (PI3K), protein kinase B (PKB; here referred to as Akt), and the mammalian target of rapamycin (mTOR). New studies show that the Foxo proteins Foxo1 and Foxo3a, which are inactivated by Akt, drive Foxp3 expression. These studies therefore explain the negative regulation of Foxp3 by PI3K signaling, and add Foxo proteins to the growing list of nuclear factors capable of modulating Foxp3 expression.
The immune system affords protection from environmental pathogens, but this service comes at a price: uncontrolled responses can damage the host by causing immune pathology, and responses to self-antigens can result in autoimmunity.T reg cells are therefore essential for a balanced immune system. The forkhead–winged helix transcription factor Foxp3 coordinates the T reg cell gene expression program, and its absence causes death by lymphoproliferation and multiorgan autoimmunity in humans with immunodysregulation, polyendocrinopathy, and enteropathy X-linked syndrome and in Foxp3-deficient mice (Brunkow et al., 2001; Fontenot et al., 2003; Khattri et al., 2003).
Intrathymically induced T reg (itTreg) cells are thought to arise by a two-step process, in which TCR signaling induces competence for Foxp3 expression and the expression of the high-affinity IL-2 receptor-α chain (CD25). IL-2, or other cytokines that activate STAT5, then induce Foxp3 expression (Burchill et al., 2008; Lio and Hsieh, 2008; Wirnsberger et al., 2009). The contributions of TGF-β to itTreg cell differentiation (Liu et al., 2008) and to the maintenance of functional T reg cells in peripheral lymphoid organs (Marie et al., 2005; Li et al., 2006; Pesu et al., 2008) remain to be fully elucidated. Runx transcription factors are involved in the induction and in the maintenance of Foxp3 expression (Bruno et al., 2009; Kitoh et al., 2009; Klunker et al., 2009; Rudra et al., 2009), and microRNAs contribute to both the development (Cobb et al., 2006) and the maintenance of T reg cells (Chong et al., 2008; Liston et al., 2008; Zhou et al., 2008).
Extrathymically induced T reg (etTreg) cells can be formed in peripheral lymphoid organs (Apostolou and von Boehmer, 2004; Kretschmer et al., 2005; Curotto de Lafaille and Lafaille, 2009). Although the physiological importance of etTreg cells is much less certain than that of itTreg cells, etTreg cells have been intensely studied because they can easily be generated in vitro and bear therapeutic promise. Major inducers of Foxp3 expression in peripheral T cells in vitro include TCR signaling in the presence of TGF-β (Chen et al., 2003), the downstream TGF-β signal transducers Smad2 and Smad3, and retinoic acid (Benson et al., 2007; Coombes et al., 2007; Mucida et al., 2007; Sun et al., 2007). Genetic and pharmacological evidence indicates that the PI3K–Akt–mTOR signaling network interferes with Foxp3 induction in vitro, as well as in vivo (Haxhinasto et al. 2008; Sauer et al., 2008; unpublished data), but the mechanisms that link PI3K–Akt–mTOR signaling to Foxp3 expression have until recently been unknown.
Conserved noncoding sequences integrate signals influencing Foxp3 expression
Like other metazoan genes, the expression of Foxp3 is regulated by multiple transcription factors, by chromatin, and by cis-regulatory elements. TCR activation induces the binding of transcription factors such as NFAT, AP1, CREB, and ATF to the Foxp3 promoter and enhancer elements (Kim and Leonard, 2007; Tone et al., 2008). T reg cell development is impaired in T cells lacking signaling molecules needed for NF-κB activation (e.g., PKC-θ, Bcl10, CARMA1, and MALT1), and c-Rel is a critical NF-κB component in this context (Isomura et al., 2009; Long et al., 2009; Ruan et al., 2009; Zheng et al., 2010).
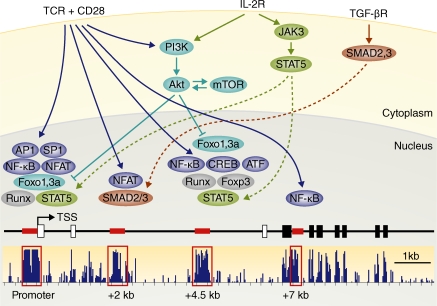
In addition to the promoter, at least three conserved noncoding sequence (CNS) elements contribute to the regulation of the Foxp3 locus (Kim and Leonard, 2007; Tone et al., 2008; Huehn et al., 2009; Zheng et al., 2010). Because the nomenclatures used in these studies differ, we will refer to these elements by their position relative to the Foxp3 transcription start site (TSS; Fig. 1). Two CNS +2 kb and +4.5 kb in the 5′ untranslated region (referred to as CNS2 and 3 in Tone et al., 2008 and Kim and Leonard, 2007, and as CNS1 and 2 in Zheng et al., 2010). A further CNS is at +7 kb, just downstream of the first coding exon (CNS3 in Zheng et al., 2010). The CNS at +7 kb plays a role in itTreg and etTreg cells, as its deletion reduces the frequency of T reg cells generated in the thymus and in the periphery (Zheng et al., 2010). In contrast, the CNS at +2 kb is not required for itTreg cell differentiation. Consistent with a role in inducible Foxp3 expression (Zheng et al., 2010), this CNS contains binding sites for NFAT, an effector of TCR signaling, and for SMAD proteins, which mediate TGF-β signaling (Kim and Leonard, 2007; Tone et al., 2008; Zheng et al., 2010). Finally, the CNS at +4.5 kb is important for the maintenance, rather than the induction, of Foxp3 expression (Zheng et al., 2010). This CNS contains a region rich in CpG dinucleotides, the targets for mammalian DNA methylation, and is methylated in conventional T cells and demethylated in itTreg cells. Demethylation of this region by inhibition or genetic deletion of methyltransferase enzymes leads to stable Foxp3 expression (Huehn et al., 2009). Loss of DNA methylation promotes the binding to the CNS at +4.5 kb of the transcription factors CREB, ATF (Kim and Leonard, 2007), the NF-kB component c-Rel (Long et al., 2009), Runx–Cbfb (Bruno et al., 2009; Rudra et al., 2009), and, interestingly, Foxp3 itself. Hence, the CNS at +4.5 kb could form part of a positive feedback loop, by which Foxp3 maintains its own expression (Zheng et al., 2010).
Figure 1.
Signals, factors, and regulatory elements that control Foxp3 expression. The figure shows the 5′ part of the mouse Foxp3 locus, as well as signals and factors known to regulate its expression. Conservation is indicated in blue as a track from the University of California Santa Cruz Genome Browser (http://genome.ucsc.edu; Rhead et al., 2010). Conserved noncoding sequences with known functions are indicated in red. Black rectangles indicate exons (open for noncoding, filled for coding). TSS indicates the transcription start site.
Foxos link PI3K–Akt–mTOR to Foxp3
Efficient Foxp3 induction requires proteins that limit PI3K activity, such as Cbl-b (Wohlfert et al., 2006) and PTEN (Sauer et al., 2008). Conversely, constitutive Akt activity interferes with Foxp3 induction (Haxhinasto et al., 2008). mTORC2 activates Akt by phosphorylating it on Ser473, and loss or inhibition of mTORC2 inactivates Akt and consequently promotes Foxp3 induction (Delgoffe et al., 2009). Harada et al. (this issue) and Ouyang et al. (2010) now align Foxo1 and Foxo3a in the signaling pathway connecting PI3K–Akt–mTOR to Foxp3.
In agreement with previous work (Wohlfert et al., 2006), Harada et al. (2010) found that Cbl-b–deficient CD4 T cells expressed less Foxp3 in response to TGF-β in vitro and in an adoptive transfer model in vivo. Cbl-b deficiency was thought to impair cellular responses to TGF-β by affecting Smad2 phosphorylation (Wohlfert et al., 2006). Surprisingly, Harada et al. (2010) found intact TGF-β responses in Cbl-b–deficient CD4 T cells. Instead, they analyzed Foxo transcription factors, which have important functions in the homeostasis of the immune system (Hedrick, 2009). The activity of Foxo proteins is regulated at multiple levels, most strikingly by Akt-mediated phosphorylation. Phosphorylation by Akt inactivates Foxo proteins by excluding them from the nucleus. In this way, Foxo localization reflects PI3K signaling (Hedrick, 2009). The link to Cbl-b is previous work showing that Cbl-b promotes ubiquitinylation of p85, the regulatory subunit of PI3K, and thereby affects the activity of Akt (Fang and Liu, 2001). Consistent with increased PI3K–Akt signaling in Cbl-b–deficient T cells, Harada et al. (2010) noted increased phosphorylation of Foxo3a in Cbl-b–deficient CD4 T cells. Foxo3a-deficient CD4 T cells showed reduced Foxp3 induction in response to TGF-β. In contrast, deletion of Foxo3a did not have any effect on the numbers of Foxp3-expressing itTreg cells (Harada et al., 2010), suggesting a preferential involvement of Foxo3a in etTreg cell, as opposed to itTreg cell differentiation. This is consistent with observations showing that the increased PI3K signaling that results from the loss of Cbl-b impaired inducible Foxp3 expression, but did not substantially impact itTreg cell numbers (Wohlfert et al., 2006). However, both inducible and itTreg differentiation were affected by constitutively active Akt (Haxhinasto et al., 2008).
Ouyang et al. (2010) demonstrate that the apparently selective requirement of Foxo3a for Foxp3 induction in etTreg cells is caused, at least in part, by redundancy between Foxo3a and Foxo1. Consistent with the finding that the loss of Foxo3a alone did not affect itTreg cell development (Harada et al., 2010), Ouyang et al. (2010) found that even a single allele of Foxo1 (Foxo1+/−Foxo3a−/−) or Foxo3a (Foxo1−/−Foxo3a+/−) was sufficient for normal numbers of itTreg and etTreg cells. In contrast, complete loss of Foxo1 and Foxo3a in the T cell lineage abrogated inducible Foxp3 expression. Foxo1−/−Foxo3a−/− CD4 T cells and thymocytes failed to up-regulate Foxp3 expression not only in response to activation in the presence of TGF-β, but also when subjected to 18 h of TCR signaling followed by the removal of TCR signal, an in vitro protocol that may be more akin to in vivo generation of etTreg cells (Kretschmer et al., 2005; Polansky et al., 2008). This treatment results in a loss of PI3K–Akt–mTOR activity and the de novo expression of Foxp3 in wild-type cells (Sauer et al., 2008).
Importantly, Ouyang et al. (2010) found that T cell–specific deletion of Foxo1 and Foxo3a also led to lower numbers of thymic and peripheral T reg cells at 3 wk of age. This numerical deficit disappeared by 6 wk of age, but the T reg cells that were generated were not normal. Foxo1−/−Foxo3a−/− T reg cells produced inflammatory cytokines (interferon-γ and IL-17) after activation and showed impaired regulatory function in vitro and in vivo. Foxo1−/−Foxo3a−/− splenic T reg cells showed normal expression of Foxp3, but many T reg signature genes were deregulated (Ouyang et al., 2010). It remains to be seen whether these genes are directly regulated by Foxo factors, or if their expression is affected by the increased proliferation of Foxo1−/−Foxo3a−/− T reg cells (Ouyang et al., 2010).
Harada et al. (2010) identified Foxo binding motifs in the Foxp3 promoter region and used chromatin immunoprecipitation (ChIP) assays to demonstrate the binding of Foxo proteins to the Foxp3 promoter. Ouyang et al. (2010) found several conserved Foxo consensus sequences in the Foxp3 locus, and their ChIP experiments indicate binding to the promoter and to the CNS region at +4.5 kb (referred to as CNS3 in their paper) in T reg cells, but not in conventional T cells. Reporter gene assays indicate the functional importance of a site close to the Foxp3 transcription start site (Harada et al., 2010; Ouyang et al., 2010). In light of these data, which suggest a role for Foxo proteins in the regulation of Foxp3, it is puzzling that Foxp3 expression was apparently normal in Foxo1−/−Foxo3a−/− splenic T reg cells (Ouyang et al., 2010).
Conclusions
The emerging scenario is that PI3K–Akt–mTOR signaling prevents the induction, but does not interfere with the maintenance, of Foxp3 expression via Foxo factors. It is therefore likely that with Foxo1 and Foxo3a, Harada et al. (2010) and Ouyang et al. (2010) have identified a molecular link between PI3K–Akt–mTOR signaling and Foxp3 induction. Time will tell if there are other negative regulators of Foxp3 expression downstream of PI3K–Akt–mTOR signaling.
Given the promise of inducible Foxp3 expression in the immunotherapy of autoimmunity and immune pathology (Daniel et al., 2009), Foxo factors, especially the kinases that control their activity, represent potential drug targets (Bruno and Merkenschlager, 2008).
Acknowledgments
Related work in the authors’ laboratories is supported by the Medical Research Council, UK (M. Merkenschlager) and National Institutes of Health grant R37 AI 53102 (H. von Boehmer).
The authors declare that they have no conflict of interest.
References
- Apostolou I., von Boehmer H. 2004. In vivo instruction of suppressor commitment in naive T cells. J. Exp. Med. 199:1401–1408 10.1084/jem.20040249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson M.J., Pino-Lagos K., Rosemblatt M., Noelle R.J. 2007. All-trans retinoic acid mediates enhanced T reg cell growth, differentiation, and gut homing in the face of high levels of co-stimulation. J. Exp. Med. 204:1765–1774 10.1084/jem.20070719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunkow M.E., Jeffery E.W., Hjerrild K.A., Paeper B., Clark L.B., Yasayko S.A., Wilkinson J.E., Galas D., Ziegler S.F., Ramsdell F. 2001. Disruption of a new forkhead/winged-helix protein, scurfin, results in the fatal lymphoproliferative disorder of the scurfy mouse. Nat. Genet. 27:68–73 10.1038/83784 [DOI] [PubMed] [Google Scholar]
- Bruno L., Merkenschlager M. 2008. Directing T cell differentiation and function with small molecule inhibitors. Cell Cycle. 71:2296–2298 [DOI] [PubMed] [Google Scholar]
- Bruno L., Mazzarella L., Hoogenkamp M., Hertweck A., Cobb B.S., Sauer S., Hadjur S., Leleu M., Naoe Y., Telfer J.C., et al. 2009. Runx proteins regulate Foxp3 expression. J. Exp. Med. 206:2329–2337 10.1084/jem.20090226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burchill M.A., Yang J., Vang K.B., Moon J.J., Chu H.H., Lio C.W., Vegoe A.L., Hsieh C.S., Jenkins M.K., Farrar M.A. 2008. Linked T cell receptor and cytokine signaling govern the development of the regulatory T cell repertoire. Immunity. 28:112–121 10.1016/j.immuni.2007.11.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Jin W., Hardegen N., Lei K.J., Li L., Marinos N., McGrady G., Wahl S.M. 2003. Conversion of peripheral CD4+CD25- naive T cells to CD4+CD25+ regulatory T cells by TGF-beta induction of transcription factor Foxp3. J. Exp. Med. 198:1875–1886 10.1084/jem.20030152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong M.M., Rasmussen J.P., Rudensky A.Y., Rundensky A.Y., Littman D.R. 2008. The RNAseIII enzyme Drosha is critical in T cells for preventing lethal inflammatory disease. J. Exp. Med. 205:2005–2017 10.1084/jem.20081219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb B.S., Hertweck A., Smith J., O’Connor E., Graf D., Cook T., Smale S.T., Sakaguchi S., Livesey F.J., Fisher A.G., Merkenschlager M. 2006. A role for Dicer in immune regulation. J. Exp. Med. 203:2519–2527 10.1084/jem.20061692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coombes J.L., Siddiqui K.R., Arancibia-Cárcamo C.V., Hall J., Sun C.M., Belkaid Y., Powrie F. 2007. A functionally specialized population of mucosal CD103+ DCs induces Foxp3+ regulatory T cells via a TGF-β and retinoic acid–dependent mechanism. J. Exp. Med. 204:1757–1764 10.1084/jem.20070590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curotto de Lafaille M.A., Lafaille J.J. 2009. Natural and adaptive foxp3+ regulatory T cells: more of the same or a division of labor? Immunity. 30:626–635 10.1016/j.immuni.2009.05.002 [DOI] [PubMed] [Google Scholar]
- Daniel C., Nolting J., von Boehmer H. 2009. Mechanisms of self-nonself discrimination and possible clinical relevance. Immunotherapy. 1:631–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgoffe G.M., Kole T.P., Zheng Y., Zarek P.E., Matthews K.L., Xiao B., Worley P.F., Kozma S.C., Powell J.D. 2009. The mTOR kinase differentially regulates effector and regulatory T cell lineage commitment. Immunity. 30:832–844 10.1016/j.immuni.2009.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang D., Liu Y.-C. 2001. Proteolysis-independent regulation of PI3K by Cbl-b-mediated ubiquitination in T cells. Nat. Immunol. 2:870–875 10.1038/ni0901-870 [DOI] [PubMed] [Google Scholar]
- Fontenot J.D., Gavin M.A., Rudensky A.Y. 2003. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat. Immunol. 4:330–336 10.1038/ni904 [DOI] [PubMed] [Google Scholar]
- Harada Y., Harada Y., Elly C., Ying G., Paik J.H., Depinho R.A., Liu Y.C. 2010. Transcription factors Foxo3a and Foxo1 couple the E3 ligase Cbl-b to the induction of Foxp3 expression in induced regulatory T cells. J. Exp. Med. 207:1381–1391 10.1084/jem20100004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haxhinasto S., Mathis D., Benoist C. 2008. The AKT-mTOR axis regulates de novo differentiation of CD4+Foxp3+ cells. J. Exp. Med. 205:565–574 10.1084/jem.20071477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrick S.M. 2009. The cunning little vixen: Foxo and the cycle of life and death. Nat. Immunol. 10:1057–1063 10.1038/ni.1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huehn J., Polansky J.K., Hamann A. 2009. Epigenetic control of FOXP3 expression: the key to a stable regulatory T-cell lineage? Nat. Rev. Immunol. 9:83–89 10.1038/nri2474 [DOI] [PubMed] [Google Scholar]
- Isomura I., Palmer S., Grumont R.J., Bunting K., Hoyne G., Wilkinson N., Banerjee A., Proietto A., Gugasyan R., Wu L., et al. 2009. c-Rel is required for the development of thymic Foxp3+ CD4 regulatory T cells. J. Exp. Med. 206:3001–3014 10.1084/jem.20091411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khattri R., Cox T., Yasayko S.A., Ramsdell F. 2003. An essential role for Scurfin in CD4+CD25+ T regulatory cells. Nat. Immunol. 4:337–342 10.1038/ni909 [DOI] [PubMed] [Google Scholar]
- Kim H.P., Leonard W.J. 2007. CREB/ATF-dependent T cell receptor-induced FoxP3 gene expression: a role for DNA methylation. J. Exp. Med. 204:1543–1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitoh A., Ono M., Naoe Y., Ohkura N., Yamaguchi T., Yaguchi H., Kitabayashi I., Tsukada T., Nomura T., Miyachi Y., et al. 2009. Indispensable role of the Runx1-Cbfbeta transcription complex for in vivo-suppressive function of FoxP3+ regulatory T cells. Immunity. 31:609–620 10.1016/j.immuni.2009.09.003 [DOI] [PubMed] [Google Scholar]
- Klunker S., Chong M.M., Mantel P.Y., Palomares O., Bassin C., Ziegler M., Rückert B., Meiler F., Akdis M., Littman D.R., Akdis C.A. 2009. Transcription factors RUNX1 and RUNX3 in the induction and suppressive function of Foxp3+ inducible regulatory T cells. J. Exp. Med. 206:2701–2715 10.1084/jem.20090596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretschmer K., Apostolou I., Hawiger D., Khazaie K., Nussenzweig M.C., von Boehmer H. 2005. Inducing and expanding regulatory T cell populations by foreign antigen. Nat. Immunol. 6:1219–1227 10.1038/ni1265 [DOI] [PubMed] [Google Scholar]
- Li M.O., Sanjabi S., Flavell R.A. 2006. Transforming growth factor-β controls development, homeostasis, and tolerance of T cells by regulatory T cell-dependent and -independent mechanisms. Immunity. 25:455–471 10.1016/j.immuni.2006.07.011 [DOI] [PubMed] [Google Scholar]
- Lio C.W., Hsieh C.S. 2008. A two-step process for thymic regulatory T cell development. Immunity. 28:100–111 10.1016/j.immuni.2007.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liston A., Lu L.F., O’Carroll D., Tarakhovsky A., Rudensky A.Y. 2008. Dicer-dependent microRNA pathway safeguards regulatory T cell function. J. Exp. Med. 205:1993–2004 10.1084/jem.20081062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Zhang P., Li J., Kulkarni A.B., Perruche S., Chen W. 2008. A critical function for TGF-beta signaling in the development of natural CD4+CD25+Foxp3+ regulatory T cells. Nat. Immunol. 9:632–640 10.1038/ni.1607 [DOI] [PubMed] [Google Scholar]
- Long M., Park S.G., Strickland I., Hayden M.S., Ghosh S. 2009. Nuclear factor-kappaB modulates regulatory T cell development by directly regulating expression of Foxp3 transcription factor. Immunity. 31:921–931 10.1016/j.immuni.2009.09.022 [DOI] [PubMed] [Google Scholar]
- Marie J.C., Letterio J.J., Gavin M., Rudensky A.Y. 2005. TGF-β1 maintains suppressor function and Foxp3 expression in CD4+CD25+ regulatory T cells. J. Exp. Med. 201:1061–1067 10.1084/jem.20042276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mucida D., Park Y., Kim G., Turovskaya O., Scott I., Kronenberg M., Cheroutre H. 2007. Reciprocal TH17 and regulatory T cell differentiation mediated by retinoic acid. Science. 317:256–260 10.1126/science.1145697 [DOI] [PubMed] [Google Scholar]
- Ouyang W., Beckett O., Ma Q., Paik J.-h., DePinho R.A., Li M.O. 2010. Foxo proteins cooperatively control the differentiation of Foxp3+ regulatory T cells. Nat. Immunol. 10.1038/ni.1884 [DOI] [PubMed] [Google Scholar]
- Pesu M., Watford W.T., Wei L., Xu L., Fuss I., Strober W., Andersson J., Shevach E.M., Quezado M., Bouladoux N., et al. 2008. T-cell-expressed proprotein convertase furin is essential for maintenance of peripheral immune tolerance. Nature. 455:246–250 10.1038/nature07210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polansky J.K., Kretschmer K., Freyer J., Floess S., Garbe A., Baron U., Olek S., Hamann A., von Boehmer H., Huehn J. 2008. DNA methylation controls Foxp3 gene expression. Eur. J. Immunol. 38:1654–1663 10.1002/eji.200838105 [DOI] [PubMed] [Google Scholar]
- Rhead B., Karolchik D., Kuhn R.M., Hinrichs A.S., Zweig A.S., Fujita P.A., Diekhans M., Smith K.E., Rosenbloom K.R., Raney B.J., et al. 2010. The UCSC Genome Browser database: update 2010. Nucleic Acids Res. 38:D613–D619 10.1093/nar/gkp939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan Q., Kameswaran V., Tone Y., Li L., Liou H.C., Greene M.I., Tone M., Chen Y.H. 2009. Development of Foxp3(+) regulatory t cells is driven by the c-Rel enhanceosome. Immunity. 31:932–940 10.1016/j.immuni.2009.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudra D., Egawa T., Chong M.M., Treuting P., Littman D.R., Rudensky A.Y. 2009. Runx-CBFbeta complexes control expression of the transcription factor Foxp3 in regulatory T cells. Nat. Immunol. 10:1170–1177 10.1038/ni.1795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer S., Bruno L., Hertweck A., Finlay D., Leleu M., Spivakov M., Knight Z.A., Cobb B.S., Cantrell D., O’Connor E., et al. 2008. T cell receptor signaling controls Foxp3 expression via PI3K, Akt, and mTOR. Proc. Natl. Acad. Sci. USA. 105:7797–7802 10.1073/pnas.0800928105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C.M., Hall J.A., Blank R.B., Bouladoux N., Oukka M., Mora J.R., Belkaid Y. 2007. Small intestine lamina propria dendritic cells promote de novo generation of Foxp3 T reg cells via retinoic acid. J. Exp. Med. 204:1775–1785 10.1084/jem.20070602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tone Y., Furuuchi K., Kojima Y., Tykocinski M.L., Greene M.I., Tone M. 2008. Smad3 and NFAT cooperate to induce Foxp3 expression through its enhancer. Nat. Immunol. 9:194–202 10.1038/ni1549 [DOI] [PubMed] [Google Scholar]
- Wirnsberger G., Mair F., Klein L. 2009. Regulatory T cell differentiation of thymocytes does not require a dedicated antigen-presenting cell but is under T cell-intrinsic developmental control. Proc. Natl. Acad. Sci. USA. 106:10278–10283 10.1073/pnas.0901877106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlfert E.A., Gorelik L., Mittler R., Flavell R.A., Clark R.B. 2006. Cutting edge: deficiency in the E3 ubiquitin ligase Cbl-b results in a multifunctional defect in T cell TGF-beta sensitivity in vitro and in vivo. J. Immunol. 176:1316–1320 [DOI] [PubMed] [Google Scholar]
- Zheng Y., Josefowicz S., Chaudhry A., Peng X.P., Forbush K., Rudensky A.Y. 2010. Role of conserved non-coding DNA elements in the Foxp3 gene in regulatory T-cell fate. Nature. 463:808–812 10.1038/nature08750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Jeker L.T., Fife B.T., Zhu S., Anderson M.S., McManus M.T., Bluestone J.A. 2008. Selective miRNA disruption in T reg cells leads to uncontrolled autoimmunity. J. Exp. Med. 205:1983–1991 10.1084/jem.20080707 [DOI] [PMC free article] [PubMed] [Google Scholar]