Abstract
Ataxia telangiectasia mutated (ATM) deficiency predisposes humans and mice to T lineage lymphomas with recurrent chromosome 14 translocations involving the T cell receptor α/δ (Tcra/d) locus. Such translocations have been thought to result from aberrant repair of DNA double-strand breaks (DSBs) during Tcra locus V(D)J recombination, and to require the Tcra enhancer (Eα) for Tcra rearrangement or expression of the translocated oncogene. We now show that, in addition to the known chromosome 14 translocation, ATM-deficient mouse thymic lymphomas routinely contain a centromeric fragment of chromosome 14 that spans up to the 5′ boundary of the Tcra/d locus, at which position a 500-kb or larger region centromeric to Tcra/d is routinely amplified. In addition, they routinely contain a large deletion of the telomeric end of one copy of chromosome 12. In contrast to prior expectations, the recurrent translocations and amplifications involve V(D)J recombination–initiated breaks in the Tcrd locus, as opposed to the Tcra locus, and arise independently of the Eα. Overall, our studies reveal previously unexpected mechanisms that contribute to the oncogenic transformation of ATM-deficient T lineage cells.
Immature T cell lymphomas represent a significant portion of human lymphoid malignancies (Ferrando and Look, 2000; Armstrong and Look, 2005). Approximately 50% of these tumors harbor recurrent chromosomal translocations (Aifantis et al., 2008), which likely arise through errors in the repair of DNA double-strand breaks (DSBs). In this context, normal mouse and human thymocyte differentiation requires V(D)J recombination, a process that involves the introduction of DSBs at T cell receptor (Tcr) loci by the recombination-activating gene (RAG) 1 and 2 endonuclease (Bassing et al., 2002). If such programmed Tcr DSBs are repaired incorrectly, they can generate chromosomal translocations that involve Tcr loci. In fact, 70% of recurrent translocations in human T cell acute lymphoblastic leukemias (T-ALL) involve TCR loci (Aifantis et al., 2008). Furthermore, RAG-initiated DSBs in and around T cell oncogene loci may also contribute to clonal translocations in T cell lymphoma (Aifantis et al., 2008). In addition, developing T cells are also subject to general DSBs that occur during S phase in association with periods of rapid cellular division. These DSBs apparently can occur in numerous chromosomal locations, and potentially participate in Tcr locus-related translocations (Bassing et al., 2003).
During T cell development in the thymus, TCR variable region gene exons are assembled by V(D)J recombination that is initiated by RAG-generated DSBs between V, D, and J segments and their flanking recombination signal sequences (RSSs; Rooney et al., 2004a). To complete V(D)J recombination, RAG-initiated coding sequence DSBs and RSS DSBs are joined to each other by the classical nonhomologous end-joining pathway (C-NHEJ; Rooney et al., 2004a; Lieber, 2008). The V(D)J recombination process is also influenced by general DSB response factors such as the ataxia telangiectasia mutated (ATM) protein (Liyanage et al., 2000; Bredemeyer et al., 2006; Callén et al., 2007; Huang et al., 2007). In humans and mice, the two distinct lineages of T lymphocytes are distinguished by the surface expression of either αβ or γδ TCRs (Bhandoola et al., 2007). TCRβ, γ, and δ variable region exons are assembled in CD4−/CD8− (double-negative) thymocytes (von Boehmer, 2004). Productive VδDδJδ and VγJγ rearrangements generate TCR δ and γ chains that form cell surface γδ TCR and prescribe γδ T cell expansion and development (Krangel et al., 1998). Productive VβDβJβ rearrangements generate TCRβ chains, which, in conjunction with Notch1 signaling, promote differentiation to the CD4+/CD8+ (double positive; DP) stage, in which Tcra rearrangements occur (Garbe and von Boehmer, 2007). Productive VαJα rearrangements generate TCRα chains that associate with TCRβ and promote further differentiation to CD4+ or CD8+ single-positive (SP) mature T cells.
In both humans and mice, the TCRA and TCRD genes are encoded within the same locus (TCRA/D; Krangel et al., 2004). The mouse Tcra/d locus lies on chromosome 14, with the Cα exons, which lie at the 3′ (telomeric) end of Tcra/d, being preceded upstream by ∼60 Jα segments (Krangel et al., 2004; Fig. S1). The Cδ exons lie upstream of the Jα segments and are flanked downstream by the Vδ5 segment and upstream by a Vδ4 segment, two Dδ segments, and two Jδ segments. Most Vα and Vδ segments lie upstream of Vδ4 with Vα segments lying at the distal (5′ or centromeric) end of the cluster, Vα/Vδ segments in the center, and Vδ segments at the downstream (telomeric) end (Krangel et al., 2004). Because of the unique Tcra/d organization, joining of Vα and Jα segments deletes the Tcrd portion of the locus (including Cδ), permanently committing to the TCRα/β lineage. Vα to Jα rearrangement requires the Tcra enhancer (Eα) at the 3′ end of Tcra/d locus (downstream of Cα) to promote V(D)J recombinational accessibility of Jα segments lying up to 100-kb away (Sleckman et al., 1997). Eα also activates TCRα expression (Sleckman et al., 1997). The human TCRA/D locus is similarly organized and lies on human chromosome 14 (Fig. S1).
DNA DSBs, including V(D)J recombination DSBs, activate the ATM kinase, which then phosphorylates substrates involved in DNA repair and cell cycle control (Kastan et al., 2001). Although not required for V(D)J recombination, ATM helps promote normal DSB repair during this process (Bredemeyer et al., 2006; Callén et al., 2007; Huang et al., 2007). Human ATM mutations cause AT, which is characterized by immunodeficiency, genomic instability, and lymphoid malignancies (Lavin, 2008). Most T cell malignancies in AT patients are T-ALLs, which have not been well characterized cytogenetically. However, older AT patients who harbor clonal expansions of peripheral T cells with inv(14)(q11;q32), t(14;14)(q11;32.1), or rarely, t(X;14)(q28;q11) translocations, sometimes develop T cell prolymphocytic leukemia (T-PLL; Pekarsky et al., 2001). The inv(14)(q11;q32), t(14;14)(q11;32.1), or t(X;14)(q28;q11) translocations appear to involve TCRA/D at 14q11 in humans and the T cell leukemia/lymphoma 1 (TCL1) (14q32.1) and MTCP1 (Xq28) oncogene loci, respectively (Russo et al., 1989; Fig. S1). ATM-deficient mice recapitulate aspects of human AT (Barlow et al., 1996; Elson et al., 1996; Xu et al., 1996; Herzog et al., 1998; Borghesani et al., 2000). In this regard, ATM-deficient mice have impaired thymocyte development with reduced CD3 expression on DP thymocytes and decreased SP thymocyte numbers (Borghesani et al., 2000; Huang et al., 2007), which is consistent with defective Tcra V(D)J recombination. Moreover, ATM-deficient mice succumb to DP thymic lymphomas with RAG-dependent translocations, which often join the Tcra/d on chromosome 14 to the telomeric end of chromosome 12, where mouse Tcl1 is located. Thus, it has been speculated that the inv(14) or t(14;14) in human T-PLL and the t(12;14) in mouse ATM-deficient thymic lymphoma both may activate TCL1 or another dominant oncogene in this chromosomal region (Liyanage et al., 2000; Callén et al., 2009; Fig. S1).
We now characterize recurrent translocations in mouse ATM-deficient thymic lymphomas in depth and also test for roles of Eα in tumor development. Surprisingly, we find that the recurrent chromosome 14 translocations observed in ATM-deficient thymic lymphomas are associated with V(D)J recombination errors at Tcrd, as opposed to Tcra locus. In addition, we show that Eα is completely dispensable for the oncogenic processes leading to these tumors. Finally, we show that most of these tumors amplify a set of genes lying just upstream of the Tcra/d locus. Based on our findings, we propose a model for the generation of the recurrent genomic abnormalities in ATM-deficient thymic lymphomas (Fig. S1).
RESULTS
Recurrent amplification upstream of Tcra/d locus in ATM-deficient thymic lymphomas
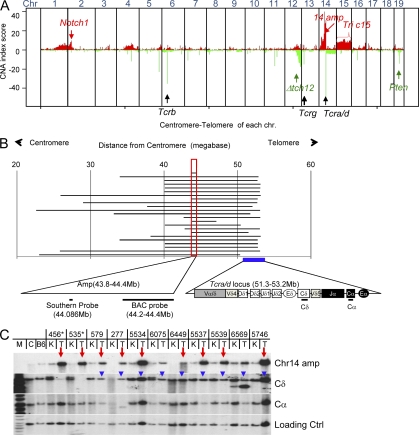
We used comparative genomic hybridization (CGH) to assay for genomic alterations in DNAs prepared from 18 independent thymic lymphomas that arose in mice homozygous for an ATM-inactivating mutation that we have previously described (Borghesani et al., 2000). In addition to copy number losses associated with V(D)J recombination at Tcrβ, γ, α/δ, or immunoglobulin heavy chain (Igh) loci, one of the most striking features of the CGH profiles of these tumors was the common amplification of chromosome 14 sequences. Notably, the telomeric end of the amplification region always began upstream of the Tcra J region segments, frequently within the Tcrd portion of the Tcra/d locus or, in a few cases, upstream (5′) of the Tcra/d locus. In all cases, the amplification region we observed extended well beyond the centromeric end of the Tcra/d locus (Fig. 1 B and Fig. S2). We observed amplification of this general region in all 18 Atm−/− thymic lymphomas analyzed; albeit to differing extents of amplification and with amplicons in given tumors that ranged from ∼500 kb to >20 megabases (Mb; Fig. 1 B). The amplified sequences common to all tumors comprised an ∼500-kb genomic region that lies ∼10 Mb centromeric to the most 5′ Vα of the Tcra/d locus (Fig. 1 B). We independently confirmed amplification of this region in multiple independent Atm−/− thymic lymphomas by performing Southern blotting analyses with probes derived from the commonly amplified 500-kb region (Fig. 1, B and C).
Figure 1.
Amplification of sequences centromeric to the Tcra/d locus in Atm−/− T cell lymphomas. (A) Combined CGH analysis of 18 ATM-deficient thymic lymphomas. Each tumor sample was hybridized and analyzed once with matched kidney control DNA. The y axis represents amplification/deletion index (sum of LN tumor/kidney ratio of all tumors analyzed). Amplification and deletion were scored separately and plotted together. The amplification score at each probe location is plotted in red and deletion score is plotted in green. At some loci, such as the telomeric end of chromosome 12, both deletions and amplifications were found in this collection of tumors, which is termed copy number polymorphism, indicating inconsistent copy number changes in this region. The x axis depicts all 19 mouse chromosomes (1 to 19 arranged from centromere to telomere). The red arrows and text highlight recurrent amplifications of Notch1 and chromosome 14 sequences upstream of the Tcra/d locus, and as well as recurrent trisomy 15 (Tric15). The black arrows and text indicate the position of Tcr gene loci. The green arrows and text highlight recurrent deletions in the telomeric portion of chromosome 12 (Δtch12) and the Pten locus. (B) The diagram shows the length of amplification (defined by LN2 tumor/kidney > 0.667) in each of the ATM-deficient thymic lymphomas. The x axis is the base pair position indicated by distance (Mb) from the chromosome 14 centromere. The common amplification region is marked with a red box and shown in the expanded form at the bottom. The position of the BAC probe used for FISH (RP23_359O5) and probe used for Southern blotting are indicated. The Tcra/d locus is indicated with a filled blue box and expanded at the bottom of the panel to show relative orientation. (C) Southern blot analyses of 11 tumor/kidney DNA pairs. About 10 µg of genomic DNA from kidney (K) and the corresponding tumors (T) was digested with EcoRI and probed with the probe described in B (Chr14 Amp) and with probes that recognize the Tcrd constant region (Cδ) or the Tcra (Cα). The red arrows indicate tumor samples with significant amplification. The blue arrowheads indicate the retention of Cδ in those tumors. The asterisks mark the two samples that show Cδ deletion as predicted by CGH analysis (Fig. 3 B). The loading control probe was derived from an intron of the Mdc1 gene (which lies on mouse chromosome 17). M, DNA Marker; C, control DNA from 129sv mouse strain; B6, control DNA from BL6 mouse strain.
We found several other recurrent copy number changes, in addition to the recurrent amplifications, including hemizygous deletion of the putative Bcl11b tumor suppressor gene (chromosome 12) in 21/23 cases (Fig. 1 A and Fig. S3, A–C), homozygous deletion of the Pten tumor suppressor gene (chromosome 19) in 6/18 cases (Fig. 1 A and Fig. S1 C), and amplification of Notch1 oncogene (chromosome 2) in 5/18 cases (Fig. 1 A and Fig. S2 B). The latter findings led us to sequence the coding exons of Notch1 in 46 independent Atm−/− thymic lymphomas, including 14 analyzed by CGH (including three Notch1-amplified tumors). We found Notch1 mutations in 18/46 cases (39.1%; Fig. S3 D). Of the 20 mutations found (2 out of 18 cases had two independent mutations), 15 were proline/glutamic acid/serine/threonine-rich domain mutations (75%), and 5 were heterodimer domain mutations (25%; Fig. S3 D). Notably, the three Notch1-amplified tumors sequenced (tumor 5534 [Fig. S3 B], 5696, and 6099) had no Notch1 mutations, suggesting that amplification of Notch1 gene might act as an independent means of Notch1 activation in Atm−/− thymic lymphomas.
The most frequent translocation in mouse ATM-deficient thymic lymphomas is a t(12;14) (Liyanage et al., 2000). To characterize potential relationships between the recurrent gene amplification on chromosome 14 and this recurrent translocation, we first used chromosome 12 and 14 paints to assay for chromosomal anomalies in metaphase spreads from individual Atm−/− thymic lymphomas. Of 10 tumors analyzed, 7 had a clonal t(12;14), with clonality defined as the translocation occurring in 50% or more of the metaphases analyzed (Fig. 2 B, Fig. S4 B, and Table S1). Most metaphases also contained, in addition to the t(12;14), a chromosome 14 fragment, referred to as f(14), plus apparently normal chromosomes 14 and 12 (Fig. 2 C, Fig. S2, and Fig. S4 C). To further characterize potential anomalies, we stripped the paints from these metaphases and assayed them via fluorescence in situ hybridization (FISH) with a probe derived from the 3′ (telomeric) end of the Tcra/d locus (Cα-Red; Liyanage et al., 2000) and a probe derived from the centromeric region of chromosome 14 (RP23_145L1; referred to as ch14cen-Green; Fig. 2 C and Fig. S4 C). The t(12;14) retained sequences hybridizing to the Cα probe in the vicinity of the junction, but lacked ch14cen-hybridizing sequences (Fig. 2 C and Fig. S4 C, second row, top), which is consistent with the t(12;14) resulting from fusion of the 3′ region of the Tcra/d locus (including Eα) with chromosome 12. These analyses also revealed an apparently normal chromosome 14 that contained both ch14cen and Cα-hybridizing sequences and a portion of chromosome 14 that contained the ch14cen-hybridizing sequences but lacked Cα-hybridizing sequences (see following paragraph; Fig. 2 C and Fig. S4 C, second row, top). As would be expected, the normal chromosome 12 did not hybridize to either Cα or ch14cen probes (Fig. 2 C and Fig. S4 C, second row, top). Finally, we stripped the slides and applied a probe from the center of the common amplification region (green; RP23_359O5, referred as Amp) and a Cα probe (red). In all eight samples tested, the Cα probe did not hybridize to f(14), whereas the Amp probe gave an intense signal on the f(14), which is consistent with amplification of these sequences in that location; however, no Amp probe signal was detected on the t(12;14) (Fig. 2 C and Fig. S4 C, third row, top; Table S1; see following paragraph). Together, these latter results are consistent with the f(14) being a partner in the t(12;14) translocation (Fig. S2).
Figure 2.
Cytogenetic analysis of ATM-deficient thymic lymphomas. (A) Relative position of the 5′Tcra/d (green), Cα (red), chr 14 Cen (green), and Amp (green) probes in relation to the Tcra/d locus on mouse chromosome 14. The colors of the lines indicating particular probes are consistent with the colors of these probes in C. (B) Representative chromosome 14 (red) and chromosome 12 (green) paint analysis of a metaphase from an ATM-deficient thymic lymphoma shows a t(12;14) translocation (white arrow), two other chromosome 14 or chromosome 14 fragments (red), and a chromosome 12 (green). Other representative analyses are shown in Fig. S4, and a summary of all slides analyzed is shown in Table S1. (C) Combined FISH and paint analyses of chromosomes 14 and 12 (and their derivatives) from ATM-deficient thymic lymphomas. The color dedication of each row is marked by the color of the font indicating the probes at the left. (top) One representative slide from tumor 5534 that was hybridized with chromosome paint, stripped and rehybridized with Cα and ch14cen FISH, and then stripped and rehybridized with Cα and Amp FISH. (middle) Another representative slide from tumor 5534, which was hybridized with chromosome paint, stripped, and hybridized with Cα and 5′Tcra/d probes. (bottom) Another representative slide from tumor 5534, which was first hybridized with chromosome paint, and then stripped and hybridized with Igh (199) FISH probe. Other representative analyses are shown in Fig. S4, and a summary of all slides analyzed is shown in Table S2. (D) Representative FISH analysis of using the Amp BAC (359O5 in green) and Cα BAC (in red) on metaphases from ATM-deficient thymic lymphomas. A white arrow indicates the amplified 359O5 signal and a yellow arrow indicates the isolated Cα. A pink arrow indicates the normal chromosome 14 containing both the Amp and Cα (without amplification) signals. Other representative analyses are shown in Fig. S4, and a summary of all slides analyzed is shown in Table S3. At least 10 (usually 20) independent metaphases were scored for each tumor shown in the representative examples shown in this figure. The figures show results that were found in least 50% (often >85%) of the metaphases scored from each slides.
To unequivocally determine whether Tcra/d locus breaks contribute to the t(12;14) translocation, we prepared another set of metaphase spreads from 7 tumors with t(12:14) translocations and hybridized them with chromosome 12 (green) and chromosome 14 (red) paints first, and then stripped them and hybridized with the Cα probe (red) as well as a 5′Tcra/d probe (green; RP23_304L21 derived from 6 kb upstream of the most 5′Vα). In all seven sets of tumor metaphases analyzed, 5′Tcra/d hybridizing sequences were absent from the t(12;14) (Fig. 2 C and Fig. S4 C, middle), even though Cα-hybridizing sequences were retained near the junction (see previous paragraph; Table S1). Moreover, while Cα-hybridizing sequences were not observed on the 14(f) in any of the seven tumor samples, five of seven samples did contain 5′Tcra/d hybridizing sequences on the 14(f) (Fig. 2 C, Fig. S4 C, middle, and Table S2). Together, these findings suggest the t(12;14) was generated from breaks within the Tcra/d locus that split the locus into the t(12;14) and the 14(f). Finally, to look for a potential reciprocal t(14;12) translocation and to test if the telomeric portion of chromosome 12 is lost during the t(12;14) translocation, we sequentially hybridized six independent sets of metaphases with chromosome 14 plus chromosome 12 paints, and then a FISH probe derived from sequences just 3′ (centromeric) to the Igh locus (199; red), which lies near the telomeric end of chromosome 12. Although the normal chromosome 12 in the metaphases always had a clear signal for the 3′ Igh 199 probe, this probe did not hybridize to the t(12;14) translocation, indicating that the chromosome 12 junctional region within the t(12;14) translocations in ATM-deficient thymic lymphomas generally is located centromeric to the Igh locus (Fig. 2 C and Fig. S4 C, bottom, and Table S3).
ATM-deficient thymic lymphomas derive from cells that have not attempted Tcra rearrangement
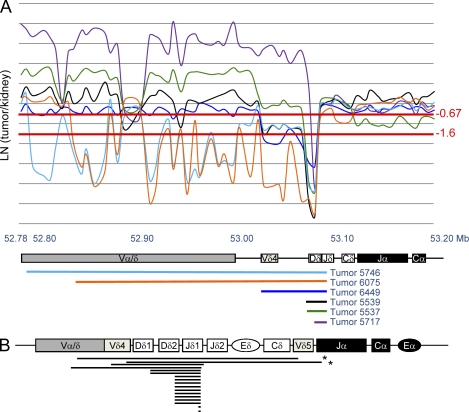
Aberrant TCRA rearrangement has long been proposed as the cause for the recurrent chromosome 14 translocations in ATM-deficient thymic lymphomas (Callén et al., 2007; Huang et al., 2007; Liyanage et al., 2000). As most developing T lymphocytes in mice that rearrange the Tcra locus do so on both alleles and delete the Tcrd locus (Krangel et al., 2004), the hypothesis that recurrent chromosome 14 translocations in Atm−/− thymic lymphomas involve aberrant Tcra locus rearrangements would imply that these tumors should frequently show bi-allelic deletions of, at least, a portion of Jα and complete deletion of the Cδ region. To test this, we analyzed high-density CGH data in and around the vicinity of Tcra/d locus and found that 16 out of 18 ATM-deficient thymic lymphomas, in fact, retained the Cδ region (Fig. 3, A and B), suggesting that the Atm−/− thymic lymphomas derived from immature thymocytes that have not yet attempted Tcra rearrangement. Homozygous deletion of a particular genomic region is defined by an LN2 (CGH signal tumor/control kidney) lower than −1.6 (Fig. 3 A). By this criterion, 16 out of the 18 Atm−/− thymic lymphomas have homozygous deletions between the Vδ4 and Dδ2 segments, but not in other regions in the Tcra/d locus (Fig. 3, A and B), again suggestive of Tcrd but not Tcra rearrangement. To confirm the CGH result, we probed EcoRI-digested DNA from 11 representative Atm−/− thymic lymphomas and corresponding kidneys with a probe from the Cδ region and found that 9 out of 11 tumors retained Cδ, which in comparison to the kidney DNA signal appeared to be present bi-allelically in most tumors (Fig. 1 C). Notably, the Southern blot analysis also confirmed the homozygous deletion of Cδ in the two cases predicted by CGH analysis (456 and 535; marked with an asterisk in Fig. 1 C and Fig. 3 B). As a control, all of the tumors analyzed (with the exception of tumor 277, in which the tumor DNA was substantially degraded) retained the Cα probe (Fig. 1 C). Together, these results indicate that ATM-deficient thymic lymphomas often derive from immature T cells that have not yet attempted Tcra rearrangement.
Figure 3.
ATM-deficient thymic lymphomas usually arise from cells that have not attempted Tcra rearrangement. (A) High-resolution CGH data of the Tcra/d of six representative ATM-deficient thymic lymphomas. Each tumor sample was hybridized and analyzed with matched kidney control DNA. The y axis is the Log2 (tumor/kidney) ratio. The x axis is the megabase position of each probe from the chromosome 14 centromere. The diagram at the bottom shows the relative position of the Tcra/d locus within this region. The homozygous deletion cut-off (Log2 [tumor/kidney] = −1.6) and heterozygous deletion cut-off (Log2 (tumor/kidney) = −0.67) are marked in red. Each tumor is represented by a unique color, and the relative size of their deletions within the analyzed region is schematically represented by a solid line of the same color below the Tcra/d map. (B) Schematic representation of homozygous deleted areas (defined by Log2[tumor/kidney] ratio < −1.6) within the Tcra/d locus for ATM-deficient thymic lymphomas. Each line represents the deleted region in one of the 20 independent tumors analyzed. The analysis includes the six tumor samples shown in A. The asterisks mark two tumors that deleted Cδ based on CGH (also shown in Fig. 1 C).
Molecular characterization of t(12;14) translocations in ATM-deficient thymic lymphomas
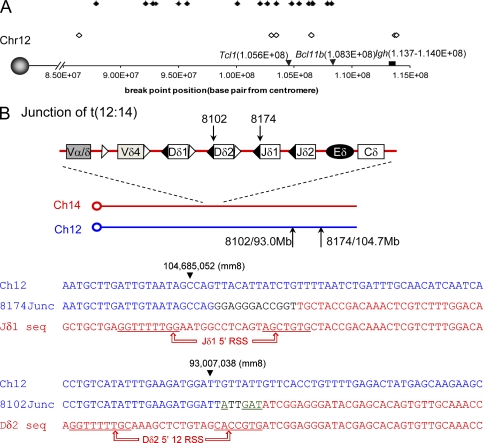
We used the high density CGH profile to analyze the chromosome 12 status in 18 ATM-deficient thymic lymphomas. Consistent with loss of the 3′Igh locus probe (199) from the t(12;14), as determined by FISH (Fig. 2 C and Fig. S4 C, bottom), and the absence of t(14;12) reciprocal translocations, the CGH analyses revealed that 14 of the 18 analyzed tumors had hemizygous deletions of the telomeric portion of chromosome 12 (Fig. 1 A and Fig. S3, A–C) with the deletions ranging from 5–30 Mb (Fig. 4 A). Copy number losses in high-density CGH analyses allowed us to roughly estimate the region of chromosome 12 containing the t(12;14) translocation junction in 23 Atm−/− thymic lymphomas (including 5 Atm−/− tumors from mice that also lacked Eα, referred to as Eα−/−; see last section of Results). We found that t(12;14) translocation junctions in individual tumors were widely spread over an ∼28 Mb region of the telomeric portion of chromosome 12 (Fig. 4 A), regardless of Eα status (Fig. 4 A; filled triangles denote Atm−/− tumors and open triangles denote Eα−/−Atm−/− tumors).
Figure 4.
The t(12;14) translocations in Atm−/− thymic lymphomas have junctions within a broad telomeric region of chromosome 12. (A) CGH mapping of the centromeric boundary of hemizygous deletions at the telomeric end of chromosome 12 in Atm−/− thymic lymphomas. A hemizygous deletion is defined by Log2 (tumor/kidney) ratio below −0.6. The filled diamonds represent Atm−/− tumors and the open diamonds represent the Eα−/−Atm−/− tumors. The x axis is marked with the base pair position from the centromere of chromosome 12 (according to mouse genome mm8). The positions of Tcl1, Bcl11b, and Igh are marked by black arrows or boxes. (B) Diagram and sequence of t(12:14) translocation junctions obtained from two Atm−/− thymic lymphomas (8102 and 8174). Both junctions were independently PCR amplified, cloned (in duplicate), and sequenced from both ends. The arrows on the diagram and arrowheads on the sequences indicate the position of the breakpoints in chromosomes 14 and 12. In the sequence diagrams, sequences corresponding to germ line chromosome 12 sequences are presented in blue and those corresponding to germ line Tcrd sequences (chromosome 14) are shown in red. The black letters in junction 8174 indicate nucleotides that could not be unequivocally matched to either side of the junction (e.g., potential insertions). The underlined, green letters in junction 8102 (GAT and A) are potential palindromic (P) elements. The canonical RSS sequences for the involved Jδ and Dδ segments are underlined.
The TCL1 oncogene, which is involved in some translocations with the TCRA/D locus in T cell lymphomas from human AT patients, maps to mouse chromosome 12. Our CGH data indicate that many of the translocations (14 of 23) we have characterized in mouse Atm−/− lymphomas involve chromosome 12 regions centromeric to Tcl1 and, thus, would delete the Tcl1 gene from the chromosome 12 involved in t(12;14) translocation, as previously found in analyses of a more limited set of tumors (Fig. 4 A; Liyanage et al., 2000). In this context, we verified that the t(12;14) translocations in four of six analyzed Atm−/− thymic lymphomas lacked TCL1 (Fig. S4 A). Likewise, array-based expression analysis of these six ATM-deficient thymic lymphomas indicated that Tcl1 expression is not significantly different among lymphomas and normal thymocytes and similar in all lymphomas, whether or not Tcl1 was retained on the t(12;14) (Fig. S5 B). This is in contrast with the overexpression of TCL1 described in human patients with t(14;14) or inv(14) translocations (Chun et al., 2002). We also found that Bcl11b, which in some experimental backgrounds has been implicated as a haploinsufficient tumor suppressor in mice (Kamimura et al., 2007) and which has been found to be involved in translocations in human T-ALL (MacLeod et al., 2004; Przybylski et al., 2005; Su et al., 2006), is deleted from the chromosome 12 involved in the t(12;14) translocations in 21 of 23 ATM-deficient thymic lymphomas we characterized (Fig. 4 A). In this context, array-based expression analyses of three ATM-deficient thymic lymphomas showed apparently reduced Bcl11b expression levels (15–30% of WT thymocyte levels; Fig. S6 C)
To gain further insight into molecular mechanisms that lead to the recurrent t(12;14) translocations in Atm−/− thymic lymphomas, we further analyzed the CGH data and designed sets of PCR primers that might amplify translocation junctions. One primer in a given set derived from sequences situated just centromeric of the chromosome 12 deletion in particular tumors and the other came from the Dδ/Jδ area on chromosome 14. By this approach, we were able to isolate and sequence putative PCR translocation junction products from two tumors (8102 and 8174; Fig. 4 B). In both cases, the chromosome 14 side of the junction was adjacent to a canonical RSS sequence within the Tcrd locus and, in one case, the junction included a potential P element (Fig. 4 B; P element in green and underlined), strongly indicating that these translocations derived from DSBs generated during V(D)J recombination. However, the two putative chromosome 12 breakpoints, which were ∼10 Mb apart, were rather unremarkable. There was no evidence of a cryptic RSS in this region. The next known coding transcripts on chromosome 12 (predicted by mouse genome version mm8) are 30 kb (8174) and 752 kb (8102) away from the breakpoints.
The Tcra/d and Igh Loci often lie in close proximity in thymocytes
ATM is required for normal end-joining during V(D)J recombination at both the Igh locus in developing B cells and the Tcra/d locus in developing T cells, and ATM is also required to mediate checkpoints that eliminate developing ATM-deficient B and T cells with breaks at their antigen receptor loci (Liyanage et al., 2000; Callén et al., 2007; Huang et al., 2007). Correspondingly, developing B and T cells sometimes suffer unrepaired V(D)J recombination associated DSBs at their Igh or Tcra/d loci, respectively, that persist and can be substrates for translocations (Callén et al., 2009). In this regard, developing T lymphocytes frequently undergo D to J rearrangements at their Igh loci (Züñiga et al., 1982). Thus, the frequent occurrence of unrepaired V(D)J recombination–associated DSBs at the Igh and Tcra/d loci in the progenitors of ATM-deficient thymic lymphomas may be a major contributor to the frequent occurrence t(12;14) translocations in ATM-deficient thymic lymphomas. Indeed, t(12;14) translocations have been observed in nonmalignant ATM-deficient peripheral T cells (Liyanage et al., 2000; Callén et al., 2007, 2009). Loci that are frequently broken in normal cells have, in some cases, been found to translocate at very high frequency (Wang et al., 2009); and such high frequency translocations have been found to also correlate with close proximity of the loci in the interphase nucleus (Wang et al., 2009). To further explore the latter possibility, we used interphase 3D FISH to assay the proximity of the Tcra/d and Igh loci in the interphase nucleus of thymocytes. Although we found only ∼1% of Tcra/d and Igh loci colocalized at a resolution of 0.5 µm (Fig. S7, A–E), we found that Tcra/d colocalized with Igh in 12–19% of thymocytes (Fig. S7, A–E) when measured at a resolution of 1 µm, which appeared significant as only 4–5% of thymocytes showed colocalization of their two Tcra/d loci and <1% of thymocytes showed colocalization of their two Igh loci at the 1-µm level of resolution (Fig. S7, A–E). Thus, the frequent proximity of the Igh and Tcra/d loci in total thymocytes is consistent with proximity also playing a role in promoting recurrent t(12;14) translocations. However, we also must note that it is not yet clear if Igh breaks are indeed intermediates in most t(12;14) translocations in ATM-deficient thymic lymphomas or what the relative proximity of these two loci is in the actual thymocyte progenitors of these tumors.
Atm−/−Eα−/− mice developed thymic lymphomas with clonal translocations involving the Tcrd locus
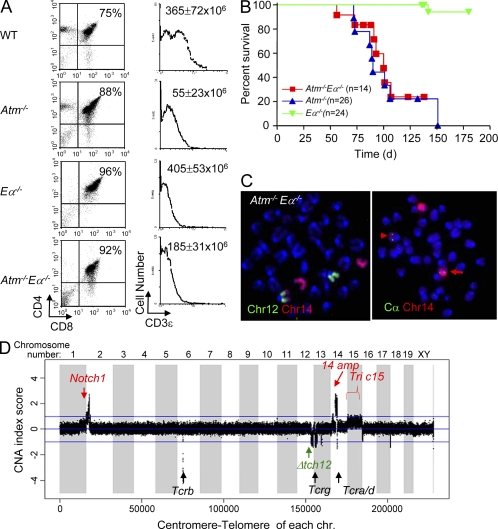
To test putative roles of Eα in thymic lymphomagenesis in the ATM-deficient background, we generated Atm−/−Eα−/− mice by crossbreeding mice harboring these mutations. SP αβ T cells are basically absent in Atm−/−Eα−/− mice (Fig. 5 A), which is consistent with the requirement for Eα for the vast majority of Vα to Jα rearrangements (Krangel et al., 2004). Yet, total thymocyte numbers are increased in Atm−/−Eα−/− mice in comparison to Atm−/− mice, potentially suggesting aberrant Tcra rearrangement in the absence of ATM might contribute to reduced total thymic cellularity in Atm−/− mice (Fig. 5 A). Notably, Atm−/−Eα−/− mice still succumbed to DP thymic lymphomas with average survival of 103 ± 28 d, which is not significantly different from that observed in their Atm−/− counterpart (119 ± 30 d, P value = 0.65; Fig. 5 BA). Cytogenetic analysis of five Atm−/−Eα−/− thymic lymphomas revealed clonal chromosome 14 translocations in all cases analyzed (Fig. 5 C), and CGH analyses revealed characteristic amplifications and t(12;14) translocations that are indistinguishable from those of their Atm−/− counterparts (Fig. 5 D), indicating that neither Eα driven Tcra V(D)J recombination nor Eα driven oncogene expression are essential for the generation of ATM-deficient thymic lymphomas with chromosome 14 translocations/amplifications.
Figure 5.
Ea is not required for ATM-deficient thymic lymphoma with recurrent translocations and amplifications. (A) FACS analyses of CD4 and CD8 expression on WT, Atm−/−, Eα−/−, and Atm−/−Eα−/− thymocytes. The number at the top right quadrant of each dot plot indicates the percentage of CD4+CD8+ DP T cells in total live thymocytes. Surface CD3 levels of gated DP cells (gated on top right quadrant of the dot plot) are plotted on the histogram on the right. The total thymocyte number (average ± standard deviation) from at least three mice of each genotype (at 4–6 wk of age) is marked in the top right corner of the histogram plots. FACS analyses were performed for at least three mice of each genotype independently, and representative plots are shown. (B) Survival analysis of Atm−/−Eα−/− mice and littermate control Atm−/− and Eα−/− mice. (C) Representative chromosome paints and FISH analyses show t(12:14) translocations involving the Tcra/d locus in one Atm−/−Eα−/− thymic lymphoma (6099). The red arrow indicates the normal chromosome 14 and the arrowhead indicates the t(12;14) translocations. Similar results were obtained for all five Atm−/−Eα−/− thymic lymphomas analyzed. 20 metaphases were analyzed from each tumor, and >80% of the metaphases from each tumor had the clonal t(12;14) translocation. (D) Representative CGH analyses of Atm−/−Eα−/− thymic lymphoma DNA. CGH analyses were performed once for each tumor. The analysis shown is for DNA from tumor 5696 and is representative of the results from CGH analyses of five Atm−/−Eα−/− thymic lymphomas. The y axis is amplification/deletion index (LN tumor/kidney ratio of each tumor analyzed). The x axis depicts the 20 mouse chromosomes as indicated (1–19 plus chromosome x and y and centromere to telomere). Red arrows and text indicate recurrent amplifications of sequences corresponding to the Notch1 and Chr14amp probes, as well as trisomy of chr 15 (Tric15). The black arrows and text indicates the various TCR loci. The green arrow and text indicates recurrent deletions in the telomeric portion of chromosome 12 (Δtch12).
DISCUSSION
Most immature T cell lymphomas from ATM-deficient mice have a CD4+CD8+ DP phenotype (Barlow et al., 1996; Borghesani et al., 2000). Thus, these tumors have generally been thought to derive from developing thymocytes that attempted and failed to undergo normal Tcra gene rearrangement (Liyanage et al., 2000; Callén et al., 2009). Correspondingly, recurrent chromosome 14–related translocations in ATM-deficient mouse thymic lymphomas have often been attributed to aberrant Tcra rearrangements and cellular selection for Eα-mediated oncogene overexpression (Liyanage et al., 2000; Callén et al., 2009). In contrast to these expectations, we find that the recurrent t(12;14) translocations in ATM-deficient thymic lymphomas derive from mis-repair of DSBs generated during V(D)J recombination at the Tcrd locus. This conclusion is indicated by our finding of an intact Cδ region in most ATM-deficient thymic lymphomas (Fig. 1 C and Fig. 3) and also by our finding of t(12;14) translocations that have breakpoints within the Tcrd locus (Fig. 4 B). In this context, although Eα is required for normal Tcra rearrangement (Sleckman et al., 1997), our findings clearly demonstrate that Eα is not required for development of ATM-deficient thymic lymphomas harboring clonal t(12;14) translocations and amplifications of the common amplification region upstream of Tcra/d locus.
Together, our findings suggest that aberrant resolution of DSBs generated during Tcrd V(D)J recombination in ATM-deficient double-negative thymocytes leads to chromosome 14 breaks that are aberrantly repaired in the absence of ATM, thereby leading both to recurrent gene amplifications on chromosome 14 and translocations involving chromosome 14 and 12. We note that mechanisms we describe for generating these recurrent forms of genomic instability in ATM-deficient mouse thymic lymphomas, to date, appear uniquely associated with ATM deficiency. Thus, a series of previously characterized thymic lymphomas that arise in other mutant backgrounds (e.g., Notch-ICN overexpression, p53 deficiency, H2AX/p53 deficiency, etc.) lack recurrent amplifications of chromosome 14 sequence and recurrent chromosome 14 or 12 translocations (Bassing et al., 2003, 2008; Li et al., 2008; Giblin et al., 2009). Conceivably, the recurrent translocations found in ATM-deficient thymocytes are associated with the unique combination of checkpoint and V(D)J recombination defects associated with ATM deficiency (Callén et al., 2007).
We find that nearly all ATM-deficient thymic lymphomas harbor amplifications of a common 500-kb region that lies ∼10 Mb upstream of the Tcra/d locus (Fig. 1 and Fig. S2). We further show that this amplification occurs on a residual centromeric fragment of chromosome 14 and not on the recurrent t(12;14) (Fig. 2 and Fig. S4). The fact that this amplification occurs in all 23 tested ATM-deficient thymic lymphomas strongly suggests that it may be linked to some stage of the oncogenic process that leads to the full-blown ATM-deficient thymic lymphomas. The commonly amplified region is relatively compact and appears to hold ∼16 genes (including 10 potential mRNA transcripts, 1 microRNA, and several noncoding transcripts according to University of California at Santa Cruz genome browser mouse gene annotation_2006 Feb version; Fig. S5 A), none of which are known oncogenes. At this time, the precise contribution of this amplified region (and the amplified sequences that are contained within it) to the oncogenic process that generates ATM-deficient thymic lymphomas is unknown; although in many human tumors, gene amplification is associated with tumor progression as opposed to tumor initiation. Indeed, other genetic alterations that we describe in ATM-deficient thymic lymphoma (e.g., Notch1 mutation, Pten deletion, and chromosome 12 deletions) or, potentially, still unknown genetic alterations also likely contribute to the generation of these tumors. Expression array analyses indicated that 3 of 8 tested genes within the common amplification unit were overexpressed by 3–10-fold (Fig. S6 B); however, because passenger genes within amplicons are also often highly overexpressed (Kohl et al., 1983), overexpression does not allow us to pinpoint a potential oncogene within the amplicon. In any case, the genes within the amplified region are candidates to test for genetic activities that can contribute to thymocyte tumor development in mice and, potentially, in humans (see last paragrpah of Discussion).
Why is chromosome 12 a frequent partner for chromosome 14 translocations?
The most common translocations in murine and human ATM-deficient thymic lymphomas, are t(12;14) and t(14;14), respectively. The fact that the vast majority of the genomic sequence of human chromosome 14 maps either to mouse chromosomes 12 or 14 (Fig. S1) suggests that there may potentially be selection of a common target with respect to the human inv(14) or t(14;14) and mouse t(12;14) translocations (Liyanage et al., 2000; Callén et al., 2009; Fig. 2, Fig. S1, Fig. S4, and Table S1). One obvious candidate was TCL1, the proposed oncogenic target of the common inv(14)(q11;q32) or t(14;14)(q11;32.1) (Fig. S1) in T-PLL from AT patients (Russo et al., 1989). TCL1 maps to human chromosome 14q32 and to a position 105 Mb telomeric to the centromere of mouse chromosome 12 (Fig. 4 A and Fig. S1). The simple hypothesis would be that the recurrent t(12;14) translocations in mice might activate Tcl1 on chromosome 12 and be positively selected for transformation (Pekarsky et al., 2001). However, Tcl1 is often deleted in the t(12;14) translocations (Fig. 4 A; Liyanage et al., 2000), and we do not find TCL1 overexpression in ATM-deficient thymic lymphomas (Fig. S5 B). Although there may be other chromosome 12 oncogenes activated via the recurrent t(12;14) translocations, this possibility seems unlikely given that the chromosome 12 junctional region of the t(12;14) translocations occur at various positions within a 28-Mb region, regardless of whether the tumors are from cells in which Eα was present or absent (Fig. 4 A). An alternative possibility is that the t(12;14) translocations might result in hemizygous loss of a putative tumor suppressor gene that lies somewhere near the telomeric end of chromosome 12. In this regard, it is notable that one copy of Bcl11b, a putative haploinsufficient tumor suppressor in p53+/− mouse thymic lymphomas (Kamimura et al., 2007) and potentially also in human T-ALLs (MacLeod et al., 2004, 2008; Przybylski et al., 2005; Su et al., 2006) is deleted in the context of the t(12;14) translocations in the vast majority (21 of 23) of the mouse Atm−/− thymic lymphomas. As might be expected, we observed reduced expression of Bcl11b and several other genes in the deleted region in a subset of tumors analyzed (Fig. S6 C).
Another possibility is that t(12:14) translocation in mouse ATM-deficient thymic lymphomas might not directly contribute to oncogenic transformation but, instead, may reflect a high frequency of the corresponding translocation in tumor progenitor cells caused by mechanistic factors. Such mechanistic factors could include the occurrence of frequent unrepaired RAG-initiated breaks in the Tcra/d and Igh loci in developing thymocytes (Callén et al., 2007, 2009; Huang et al., 2007) and, potentially, the relative proximity of these two loci in total thymocytes (Fig. S7). In this regard, the ATM-deficient background would also promote persistence and frequent translocation of such DSBs (Callén et al., 2007). Although the t(12;14) junctions are spread over a large (28-Mb) area downstream of Igh (Fig. 4 A), it is still conceivable that some or all of them were initiated by RAG JH region DSBs, given findings that persistence of such Igh DSBs in developing ATM-deficient developing B cells persist often leads to substantial levels of downstream chromosomal sequence loss before they enter into translocations in mature B cells (Callén et al., 2007, 2009). Finally, we note that there is a potential precedent for “mechanistically promoted” recurrent translocations; thus, the translocation of activation-induced deaminase (AID)–dependent Igh DSBs to RAG-dependent Igλ DSBs in NHEJ-deficient peripheral B cells is apparently so frequent in tumor progenitors that it leads to these translocations recurrently appearing in peripheral B cell lymphomas that arise from NHEJ-deficient plus p53-deficient B cells (Wang et al., 2008, 2009).
Model for the mechanism of recurrent translocations and amplifications in ATM-deficient thymic lymphomas
Based our current findings and the considerations outlined above, we propose a working model to explain the translocations/amplifications found in ATM-deficient thymocytes (Fig. S2). This model is based, in part from the model that we proposed to explain the recurrent appearance of complex translocations and amplifications, or “complicons,” involving the Igh and c-myc loci in NHEJ/p53-deficient pro–B cell lymphomas (Zhu et al., 2002). This model proposes that Igh/c-myc complicons found in NHEJ/p53 deficient pro–B lymphomas arise by a mechanism in which RAG-generated Igh breaks on chromosome 12 in NHEJ/p53-deficient mouse pro–B cells are fused to chromosome 15 downstream of c-myc leading to dicentric chromosomes that promote c-myc amplification via a breakage-fusion-bridge (BFB) mechanism (Zhu et al., 2002). By analogy, in ATM-deficient developing thymocytes, unrepaired Tcrd locus breaks may lead to the generation of dicentric chromosomes that then go through a BFB cycle, facilitated by the checkpoint deficiency associated with ATM deficiency to allow amplification of the region upstream of Tcra/d (Fig. S2). The fact that most amplicons begin within or upstream of Tcrd is consistent with this possibility, as sequences proximal to initiating DSBs are sometimes deleted during the translocation/amplification process (Zhu et al., 2002). Dicentrics could be chromosome 14 fusions (after replication through Tcrd DSBs facilitated by the G1 checkpoint deficiency in ATM-deficient cells [Fig. S2]) or fusions to DSBs on other chromosomes (Zhu et al., 2002; Rooney et al., 2004b). In this regard, in preliminary studies we indeed observed an increased frequency of chromosome 14 dicentrics in IL-7–stimulated thymocytes from ATM-deficient mice (freq = 0.95 ± 0.24%) versus WT mice (freq = 0 ± 0%; P value = 0.017 by Student’s t test, between WT and Atm−/− mice; Fig. S7 F). Involved chromosomal breaks outside of Tcrd might also be RAG-initiated or they may be generated by other factors (Fig. S2). In this model, chromosome 12, because of frequent and persistent unrepaired RAG-initiated breaks in the ATM-deficient background, might serve either as a partner to generate a dicentric or, because of frequent breaks and relative proximity, simply as a frequent recipient for persistent chromosome 14 breaks generating t(12;14) as a mechanistic “passenger” (Wang et al., 2009; Fig. S2). Finally, this model would not rule out the possibility that frequent chromosome 14 to chromosome 12 translocations that occurred in this context might be selected for those harboring large chromosome 12 telomeric deletions, which might contribute to tumorigenesis, for example, by deletion of a tumor suppressor.
Implications for T cell lymphoma in AT patients
Although the mature T cell lymphomas from older AT patients contain characteristic translocations between the TCRA/D locus and a TCL1 family member (Russo et al., 1989), the nature of the potential oncogenic events that contribute to the common immature T cell leukemia (T-ALL) in AT patients is still largely unknown. Developmentally, ATM-deficient murine thymic lymphomas resemble the developmental stage of human T-ALL. Our current finding of recurrent gene amplifications in ATM-deficient mouse thymic lymphomas suggests a previously unconsidered potential oncogenic mechanism (e.g., gene amplification) that may contribute to the generation of T-ALLs in human AT patients. In preliminary studies aimed at further elucidating this possibility, we performed CGH array analyses on a T-ALL sample from an AT patient, which showed an amplification of sequences lying at human chromosome 14q22 (Fig. S3 E), which is the region of this human chromosome that corresponds to the amplified region we identified in mouse ATM-deficient thymic lymphomas. However, because of the unavailability of appropriate materials, we were not able perform a more detailed characterization of these human T-ALL samples. Finally, our CGH analyses of ATM-deficient mouse T cell lymphomas revealed frequent activation of Notch1 (via mutation or amplification) and deletion of Pten (Fig. 1 and Fig. S3). Both of these events are common in human T-ALL (Weng et al., 2004). Therefore, further characterization of the oncogenic events leading to ATM-deficient thymic lymphomas, including definition of the target of the common gene amplification, may provide further insights into the etiology of human T-ALLs.
MATERIALS AND METHODS
Mice.
We have previously described the ATM-deficient mice used in this study (Borghesani et al., 2000). To generate the Atm−/−Eα−/− cohort, Eα−/− mice, which we also described previously (Sleckman et al., 1997), were bred with Atm+/− mice to generate mice homozygous for both mutations. Animal protocols were approved by the Institutional Animal Care and Use Committee of Children’s Hospital Boston.
Southern blotting.
Genomic DNA was isolated from tumors or normal tissues from control mice, and Southern blotting was performed with 10 µg of EcoRI-digested genomic DNA. The Cα probe was a 438-bp fragment derived from intron1 of Cα, which was prepared via PCR amplification from genomic DNA using 5′-GAAGTGGCAGAGCATCTCGGTC-3′ and 5′-GTGGCATCACAGGGAACGTCTAAA-3′ primers. The Cδ probe was a 508-bp fragment derived from intron1-exon3 and PCR amplified with 5′-CCTCTTGCCCTCAGCTTACC-3′ and 5′-CTTGGCAAACAGCAGTCGTAG-3′ primers. The Amp probe is a 916-bp PCR fragment (described in more detail in the Results section) that was generated with 5′-AAGTGGCCGTGGATACTGA-3′ and 5′-GTATACAGGTGCAGCCGTTAAA-3′ primers.
Thymocyte development.
Thymocyte populations were analyzed by flow cytometry, as previously described (Sleckman et al., 1997; Borghesani et al., 2000). Conditions for isolation and activation of splenic T cells were described previously (Franco et al., 2006). In brief, ∼1 × 106 cells were used for each flow cytometric staining. FITC-conjugated anti–mouse CD8α, PE-conjugated anti–mouse CD4, PerCP-Cy5.5–conjugated anti–mouse CD3ε, and APC-conjugated anti–mouse TCRβ or CD3ε were purchased from BD, and the staining was performed as suggested by the manufacturer. Flow cytometry data were collected on a FACSCalibur (BD) and analyzed by Win MDI2.8 (developed by the Scripps Institute).
Cytogenetic analysis of chromosome 14 and the Tcra/d locus.
To generate metaphases, freshly isolated ATM-deficient T cell lymphomas were made into single-cell suspensions and cultured in RPMI with 15% fetal bovine serum in the presence of 5 ng/ml recombinant mouse IL-7 (R&D Systems) and 100 ng/ml colcemid (Invitrogen) for 6 h. Metaphases and two-color FISH assays for Tcra/d locus breaks were performed essentially as previously described, with the indicated BAC probes (Franco et al., 2006). Chromosome paints were purchased from Applied Spectral Imaging. In brief, metaphases were first probed with fluorochrome-conjugated paints to mouse chromosome 12 and 14, following the manufacturer’s instructions. Metaphase images were captured using a Nikon Eclipse microscope equipped with a charge-coupled device camera (Applied Spectral Imaging) and a 63× objective lens. More than 10 (often 20) metaphases were analyzed for each independent sample. After image recording, selected slides were dehydrated through serial ethanol washes to strip the chromosome paints, and then reprobed with fluorochrome-labled BAC probes. To prepare the probes, BACs were labeled with either biotin (Biotin-Nick Translation Mix; Roche) or digoxigenin (Dig-Nick Translation Mix; Roche), as per manufacturer instructions. 200 ng of BAC DNA was precipitated with mouse Cot1 DNA (Invitrogen; ratio of BAC DNA to Cot1 DNA, 1:20), resuspended in 15 µl of hybridization solution (50% formamide, 2X SSC, 10% dextran sulfate, and 0.15% SDS), and co-denatured on slides for 5 min at 76°C. Slides were then incubated at 37°C for 16 h, washed twice in 50% formamide/2X SSC for 5 min at 45°C, washed twice in 2X SSC for 5 min at 45°C, and incubated with avidin-Cy3 and anti–digoxigenin-FITC (Roche; 1:250 dilution) in 2XSSC/0.05% Tween-20 for 1 h at room temperature. After three washes in 2XSSC/0.05% Tween-20, slides were mounted in Vectashield with DAPI (Vector Laboratories). Images were captured as outlined for chromosome paint.
Sequence of the coding exon of Notch 1 in ATM-deficient thymic lymphomas.
Individual DNA samples (5 µg) from 50 different ATM-deficient thymic lymphomas were subjected to exon sequencing as previously described (O’Neil et al., 2006). Tumors had <10% contamination with normal cells as judged by Southern blotting with Tcrb locus probes (Khor and Sleckman, 2005) downstream of Jβ1 and Jβ2. We obtained clear sequence results from 46 out of 50 samples.
Cytogenetic analysis of chromosome 14 dicentric formation in developing thymocytes.
To measure the chromosome 14 dicentric formation, freshly isolated thymocytes were cultured in RPMI with 15% fetal bovine serum in the presence of 5 ng/ml recombinant mouse IL-7 (R&D Systems) and 100 ng/ml colcemid (Invitrogen) for 6 h. Metaphase chromosome paints were performed as described previously (Franco et al., 2006).
Interphase FISH analyses of thymocytes from WT and ATM−/− mice.
Three-dimensional FISH was performed similarly to a previously described procedure (Callén et al., 2007). In brief, freshly isolated thymocytes were transferred to a glass slide coated with Cell Tak (BD), fixed for 10 min with 4% paraformaldehyde, permeabilized in 0.1% saponin/0.1% Triton X-100 in PBS for 10 min, and immersed in liquid nitrogen for three freeze/thaw cycles. Slides were incubated in 0.1 M HCl, blocked with 3% BSA and 100 mg ml−1 RNase A, and repermeabilized in 0.5% saponin/0.1% Triton X-100 in PBS. Genomic DNA was denatured at 73°C and hybridized with FISH probes labeled with either biotin or digoxin (Roche). Detecting antibodies were streptavidin-Alexa Fluor 488 (Invitrogen) and mouse anti–Dig-cy3 (Jackson Immuno Research Laboratories) and were used at 1:200 dilution. Images of ∼50 serial optical sections spaced by 0.2 µm were captured with Marianas spinning disk confocal microscope (63×) with a charge-coupled device detector (Intelligent Imaging Innovations) and analyzed with Slidebook software (Intelligent Imaging Innovations).
Array CGH analyses.
For CGH, 5 µg of genomic DNA from ATM-deficient mouse thymic lymphomas was profiled against 5 µg of matched normal kidney DNA from the same mouse. Genomic DNA processing, labeling and hybridization to 244k mouse Agilent CGH 60-mer oligo arrays was performed at the Arthur and Rochelle Belfer Cancer Genomics Center at the Dana-Farber Cancer Institute. Detailed features of the 244k mouse genome array can be found at http://www.agilent.com. Fluorescence ratios of scanned images were normalized and copy number profiles were generated by circular binary segmentation, which determines significance of change points in raw data through permutation (Olshen et al., 2004). Thresholds for copy number alterations were set at log2 = ±0.6 for trisomy or hemizygous deletion.
Online supplemental material.
Fig. S1 diagrams the syntenic regions of human chromosome 14 and mouse chromosomes 12 and 14. Fig. S2 describes the prior and current models for potential mechanisms and roles of chromosome 12 and 14 translocations in ATM-deficient thymic lymphomas. Fig. S3 shows the CGH results and Notch1 mutation analyses of ATM-deficient T cell malignancies. Fig. S4 shows additional examples of cytogenetic analyses of ATM-deficient thymic lymphomas. Fig. S5 describes that t(12;14) translocations in ATM-deficient thymic lymphomas are associated with deletion of telomeric sequence of chromosome 12, but not overexpression of TCL1. Fig. S6 describes the genes amplified or deleted in recurrent translocations in ATM-deficient thymic lymphomas. Fig. S7 elucidates the spatial colocalization of the Igh and Tcra/d loci in thymocytes. Tables S1–3 presents the summary of cytogenetic analysis of ATM-deficient thymic lymphomas. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20100285/DC1.
Acknowledgments
We thank Drs. Alexei Protopopov and Yonghong Xiao for assistance on the CGH analysis. We thank Yuko Fujiwara, Lisa Acquaviva, and Tiffany Borjeson for their assistance in animal care. We thank Dr. Marek Ossowicz for his assistance in these studies.
This work was supported by Leukemia and Lymphoma Society of American SCOR grant 7391-07 and National Institutes of Health RO1 AI020047 to F.W. Alt, National Institutes of Health CA109901 to F.W. Alt and A.T. Look, and National Institutes of Health R01 CA125195 to C.H. Bassing. F.W. Alt is an investigator of the Howard Hughes Medical Institute and S. Zha was a fellow and is now a special fellow of the Leukemia and Lymphomas Society of America.
The authors claim no commercial interest in this study.
Footnotes
Abbreviations used:
- AID
- activation-induced deaminase
- ATM
- ataxia telangiectasia mutated
- BFB
- breakage-fusion-bridge
- CGH
- comparative genomic hybridization
- C-NHEJ
- classical nonhomologous end-joining
- DP
- double positive
- DSB
- double-strand break
- FISH
- fluorescence in situ hybridization
- RAG
- recombination-activating gene
- RSS
- recombination signal sequence
- T-ALL
- T cell acute lymphoblastic leukemia
- T-PLL
- T cell prolymphocytic leukemia
- TCL1
- T cell leukemia/lymphoma 1
References
- Aifantis I., Raetz E., Buonamici S. 2008. Molecular pathogenesis of T-cell leukaemia and lymphoma. Nat. Rev. Immunol. 8:380–390 10.1038/nri2304 [DOI] [PubMed] [Google Scholar]
- Armstrong S.A., Look A.T. 2005. Molecular genetics of acute lymphoblastic leukemia. J. Clin. Oncol. 23:6306–6315 10.1200/JCO.2005.05.047 [DOI] [PubMed] [Google Scholar]
- Barlow C., Hirotsune S., Paylor R., Liyanage M., Eckhaus M., Collins F., Shiloh Y., Crawley J.N., Ried T., Tagle D., Wynshaw-Boris A. 1996. Atm-deficient mice: a paradigm of ataxia telangiectasia. Cell. 86:159–171 10.1016/S0092-8674(00)80086-0 [DOI] [PubMed] [Google Scholar]
- Bassing C.H., Swat W., Alt F.W. 2002. The mechanism and regulation of chromosomal V(D)J recombination. Cell. 109(Suppl):S45–S55 10.1016/S0092-8674(02)00675-X [DOI] [PubMed] [Google Scholar]
- Bassing C.H., Suh H., Ferguson D.O., Chua K.F., Manis J., Eckersdorff M., Gleason M., Bronson R., Lee C., Alt F.W. 2003. Histone H2AX: a dosage-dependent suppressor of oncogenic translocations and tumors. Cell. 114:359–370 10.1016/S0092-8674(03)00566-X [DOI] [PubMed] [Google Scholar]
- Bassing C.H., Ranganath S., Murphy M., Savic V., Gleason M., Alt F.W. 2008. Aberrant V(D)J recombination is not required for rapid development of H2ax/p53-deficient thymic lymphomas with clonal translocations. Blood. 111:2163–2169 10.1182/blood-2007-08-104760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhandoola A., von Boehmer H., Petrie H.T., Zúñiga-Pflücker J.C. 2007. Commitment and developmental potential of extrathymic and intrathymic T cell precursors: plenty to choose from. Immunity. 26:678–689 10.1016/j.immuni.2007.05.009 [DOI] [PubMed] [Google Scholar]
- Borghesani P.R., Alt F.W., Bottaro A., Davidson L., Aksoy S., Rathbun G.A., Roberts T.M., Swat W., Segal R.A., Gu Y. 2000. Abnormal development of Purkinje cells and lymphocytes in Atm mutant mice. Proc. Natl. Acad. Sci. USA. 97:3336–3341 10.1073/pnas.050584897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredemeyer A.L., Sharma G.G., Huang C.Y., Helmink B.A., Walker L.M., Khor K.C., Nuskey B., Sullivan K.E., Pandita T.K., Bassing C.H., Sleckman B.P. 2006. ATM stabilizes DNA double-strand-break complexes during V(D)J recombination. Nature. 442:466–470 10.1038/nature04866 [DOI] [PubMed] [Google Scholar]
- Callén E., Jankovic M., Difilippantonio S., Daniel J.A., Chen H.T., Celeste A., Pellegrini M., McBride K., Wangsa D., Bredemeyer A.L., et al. 2007. ATM prevents the persistence and propagation of chromosome breaks in lymphocytes. Cell. 130:63–75 10.1016/j.cell.2007.06.016 [DOI] [PubMed] [Google Scholar]
- Callén E., Bunting S., Huang C.Y., Difilippantonio M.J., Wong N., Khor B., Mahowald G., Kruhlak M.J., Ried T., Sleckman B.P., Nussenzweig A. 2009. Chimeric IgH-TCRalpha/delta translocations in T lymphocytes mediated by RAG. Cell Cycle. 8:2408–2412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun H.H., Castellví-Bel S., Wang Z., Nagourney R.A., Plaeger S., Becker-Catania S.G., Naeim F., Sparkes R.S., Gatti R.A. 2002. TCL-1, MTCP-1 and TML-1 gene expression profile in non-leukemic clonal proliferations associated with ataxia-telangiectasia. Int. J. Cancer. 97:726–731 10.1002/ijc.10102 [DOI] [PubMed] [Google Scholar]
- Elson A., Wang Y., Daugherty C.J., Morton C.C., Zhou F., Campos-Torres J., Leder P. 1996. Pleiotropic defects in ataxia-telangiectasia protein-deficient mice. Proc. Natl. Acad. Sci. USA. 93:13084–13089 10.1073/pnas.93.23.13084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrando A.A., Look A.T. 2000. Clinical implications of recurring chromosomal and associated molecular abnormalities in acute lymphoblastic leukemia. Semin. Hematol. 37:381–395 10.1016/S0037-1963(00)90018-0 [DOI] [PubMed] [Google Scholar]
- Franco S., Gostissa M., Zha S., Lombard D.B., Murphy M.M., Zarrin A.A., Yan C., Tepsuporn S., Morales J.C., Adams M.M., et al. 2006. H2AX prevents DNA breaks from progressing to chromosome breaks and translocations. Mol. Cell. 21:201–214 10.1016/j.molcel.2006.01.005 [DOI] [PubMed] [Google Scholar]
- Garbe A.I., von Boehmer H. 2007. TCR and Notch synergize in alphabeta versus gammadelta lineage choice. Trends Immunol. 28:124–131 10.1016/j.it.2007.01.004 [DOI] [PubMed] [Google Scholar]
- Giblin W., Chatterji M., Westfield G., Masud T., Theisen B., Cheng H.L., DeVido J., Alt F.W., Ferguson D.O., Schatz D.G., Sekiguchi J. 2009. Leaky severe combined immunodeficiency and aberrant DNA rearrangements due to a hypomorphic RAG1 mutation. Blood. 113:2965–2975 10.1182/blood-2008-07-165167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog K.H., Chong M.J., Kapsetaki M., Morgan J.I., McKinnon P.J. 1998. Requirement for Atm in ionizing radiation-induced cell death in the developing central nervous system. Science. 280:1089–1091 10.1126/science.280.5366.1089 [DOI] [PubMed] [Google Scholar]
- Huang C.Y., Sharma G.G., Walker L.M., Bassing C.H., Pandita T.K., Sleckman B.P. 2007. Defects in coding joint formation in vivo in developing ATM-deficient B and T lymphocytes. J. Exp. Med. 204:1371–1381 10.1084/jem.20061460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamimura K., Ohi H., Kubota T., Okazuka K., Yoshikai Y., Wakabayashi Y., Aoyagi Y., Mishima Y., Kominami R. 2007. Haploinsufficiency of Bcl11b for suppression of lymphomagenesis and thymocyte development. Biochem. Biophys. Res. Commun. 355:538–542 10.1016/j.bbrc.2007.02.003 [DOI] [PubMed] [Google Scholar]
- Kastan M.B., Lim D.S., Kim S.T., Yang D. 2001. ATM—a key determinant of multiple cellular responses to irradiation. Acta Oncol. 40:686–688 10.1080/02841860152619089 [DOI] [PubMed] [Google Scholar]
- Khor B., Sleckman B.P. 2005. Intra- and inter-allelic ordering of T cell receptor beta chain gene assembly. Eur. J. Immunol. 35:964–970 10.1002/eji.200425806 [DOI] [PubMed] [Google Scholar]
- Kohl N.E., Kanda N., Schreck R.R., Bruns G., Latt S.A., Gilbert F., Alt F.W. 1983. Transposition and amplification of oncogene-related sequences in human neuroblastomas. Cell. 35:359–367 10.1016/0092-8674(83)90169-1 [DOI] [PubMed] [Google Scholar]
- Krangel M.S., Hernandez-Munain C., Lauzurica P., McMurry M., Roberts J.L., Zhong X.P. 1998. Developmental regulation of V(D)J recombination at the TCR alpha/delta locus. Immunol. Rev. 165:131–147 10.1111/j.1600-065X.1998.tb01236.x [DOI] [PubMed] [Google Scholar]
- Krangel M.S., Carabana J., Abbarategui I., Schlimgen R., Hawwari A. 2004. Enforcing order within a complex locus: current perspectives on the control of V(D)J recombination at the murine T-cell receptor alpha/delta locus. Immunol. Rev. 200:224–232 10.1111/j.0105-2896.2004.00155.x [DOI] [PubMed] [Google Scholar]
- Lavin M.F. 2008. Ataxia-telangiectasia: from a rare disorder to a paradigm for cell signalling and cancer. Nat. Rev. Mol. Cell Biol. 9:759–769 10.1038/nrm2514 [DOI] [PubMed] [Google Scholar]
- Li X., Gounari F., Protopopov A., Khazaie K., von Boehmer H. 2008. Oncogenesis of T-ALL and nonmalignant consequences of overexpressing intracellular NOTCH1. J. Exp. Med. 205:2851–2861 10.1084/jem.20081561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber M.R. 2008. The mechanism of human nonhomologous DNA end joining. J. Biol. Chem. 283:1–5 10.1074/jbc.R700039200 [DOI] [PubMed] [Google Scholar]
- Liyanage M., Weaver Z., Barlow C., Coleman A., Pankratz D.G., Anderson S., Wynshaw-Boris A., Ried T. 2000. Abnormal rearrangement within the alpha/delta T-cell receptor locus in lymphomas from Atm-deficient mice. Blood. 96:1940–1946 [PubMed] [Google Scholar]
- MacLeod R.A., Nagel S., Drexler H.G. 2004. BCL11B rearrangements probably target T-cell neoplasia rather than acute myelocytic leukemia. Cancer Genet. Cytogenet. 153:88–89 10.1016/j.cancergencyto.2004.02.020 [DOI] [PubMed] [Google Scholar]
- O’Neil J., Calvo J., McKenna K., Krishnamoorthy V., Aster J.C., Bassing C.H., Alt F.W., Kelliher M., Look A.T. 2006. Activating Notch1 mutations in mouse models of T-ALL. Blood. 107:781–785 10.1182/blood-2005-06-2553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olshen A.B., Venkatraman E.S., Lucito R., Wigler M. 2004. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 5:557–572 10.1093/biostatistics/kxh008 [DOI] [PubMed] [Google Scholar]
- Pekarsky Y., Hallas C., Croce C.M. 2001. Molecular basis of mature T-cell leukemia. JAMA. 286:2308–2314 10.1001/jama.286.18.2308 [DOI] [PubMed] [Google Scholar]
- Przybylski G.K., Dik W.A., Wanzeck J., Grabarczyk P., Majunke S., Martin-Subero J.I., Siebert R., Dölken G., Ludwig W.D., Verhaaf B., et al. 2005. Disruption of the BCL11B gene through inv(14)(q11.2q32.31) results in the expression of BCL11B-TRDC fusion transcripts and is associated with the absence of wild-type BCL11B transcripts in T-ALL. Leukemia. 19:201–208 10.1038/sj.leu.2403619 [DOI] [PubMed] [Google Scholar]
- Rooney S., Chaudhuri J., Alt F.W. 2004a. The role of the non-homologous end-joining pathway in lymphocyte development. Immunol. Rev. 200:115–131 10.1111/j.0105-2896.2004.00165.x [DOI] [PubMed] [Google Scholar]
- Rooney S., Sekiguchi J., Whitlow S., Eckersdorff M., Manis J.P., Lee C., Ferguson D.O., Alt F.W. 2004b. Artemis and p53 cooperate to suppress oncogenic N-myc amplification in progenitor B cells. Proc. Natl. Acad. Sci. USA. 101:2410–2415 10.1073/pnas.0308757101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo G., Isobe M., Gatti R., Finan J., Batuman O., Huebner K., Nowell P.C., Croce C.M. 1989. Molecular analysis of a t(14;14) translocation in leukemic T-cells of an ataxia telangiectasia patient. Proc. Natl. Acad. Sci. USA. 86:602–606 10.1073/pnas.86.2.602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleckman B.P., Bardon C.G., Ferrini R., Davidson L., Alt F.W. 1997. Function of the TCR alpha enhancer in alphabeta and gammadelta T cells. Immunity. 7:505–515 10.1016/S1074-7613(00)80372-6 [DOI] [PubMed] [Google Scholar]
- Su X.Y., Della-Valle V., Andre-Schmutz I., Lemercier C., Radford-Weiss I., Ballerini P., Lessard M., Lafage-Pochitaloff M., Mugneret F., Berger R., et al. 2006. HOX11L2/TLX3 is transcriptionally activated through T-cell regulatory elements downstream of BCL11B as a result of the t(5;14)(q35;q32). Blood. 108:4198–4201 10.1182/blood-2006-07-032953 [DOI] [PubMed] [Google Scholar]
- von Boehmer H. 2004. Selection of the T-cell repertoire: receptor-controlled checkpoints in T-cell development. Adv. Immunol. 84:201–238 10.1016/S0065-2776(04)84006-9 [DOI] [PubMed] [Google Scholar]
- Wang J.H., Alt F.W., Gostissa M., Datta A., Murphy M., Alimzhanov M.B., Coakley K.M., Rajewsky K., Manis J.P., Yan C.T. 2008. Oncogenic transformation in the absence of Xrcc4 targets peripheral B cells that have undergone editing and switching. J. Exp. Med. 205:3079–3090 10.1084/jem.20082271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.H., Gostissa M., Yan C.T., Goff P., Hickernell T., Hansen E., Difilippantonio S., Wesemann D.R., Zarrin A.A., Rajewsky K., et al. 2009. Mechanisms promoting translocations in editing and switching peripheral B cells. Nature. 460:231–236 10.1038/nature08159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng A.P., Ferrando A.A., Lee W., Morris J.P., IV, Silverman L.B., Sanchez-Irizarry C., Blacklow S.C., Look A.T., Aster J.C. 2004. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 306:269–271 10.1126/science.1102160 [DOI] [PubMed] [Google Scholar]
- Xu Y., Ashley T., Brainerd E.E., Bronson R.T., Meyn M.S., Baltimore D. 1996. Targeted disruption of ATM leads to growth retardation, chromosomal fragmentation during meiosis, immune defects, and thymic lymphoma. Genes Dev. 10:2411–2422 10.1101/gad.10.19.2411 [DOI] [PubMed] [Google Scholar]
- Zhu C., Mills K.D., Ferguson D.O., Lee C., Manis J., Fleming J., Gao Y., Morton C.C., Alt F.W. 2002. Unrepaired DNA breaks in p53-deficient cells lead to oncogenic gene amplification subsequent to translocations. Cell. 109:811–821 10.1016/S0092-8674(02)00770-5 [DOI] [PubMed] [Google Scholar]
- Züñiga M.C., D’Eustachio P., Ruddle N.H. 1982. Immunoglobulin heavy chain gene rearrangement and transcription in murine T cell hybrids and T lymphomas. Proc. Natl. Acad. Sci. USA. 79:3015–3019 10.1073/pnas.79.9.3015 [DOI] [PMC free article] [PubMed] [Google Scholar]