Abstract
The transcription factor Foxp3 is essential for optimal regulatory T (T reg) cell development and function. Here, we show that CD4+ T cells from Cbl-b RING finger mutant knockin or Cbl-b–deficient mice show impaired TGF-β–induced Foxp3 expression. These T cells display augmented Foxo3a phosphorylation, but normal TGF-β signaling. Expression of Foxo3a rescues Foxp3 expression in Cbl-b–deficient T cells, and Foxo3a deficiency results in defective TGF-β–driven Foxp3 induction. A Foxo3a-binding motif is present in a proximal region of the Foxp3 promoter, and is required for Foxo3a association. Foxo1 exerts similar effects as Foxo3a on Foxp3 expression. This study reveals that Foxo factors promote transcription of the Foxp3 gene in induced T reg cells, and thus provides new mechanistic insight into Foxo-mediated T cell regulation.
CD4+CD25+ regulatory T (T reg) cells are a unique subset of T cells that play a dominant role in maintaining immune tolerance (Sakaguchi et al., 2008). The expression of the transcription factor Foxp3 is the genetic hallmark of T reg cells, and Foxp3 critically controls the development and inhibitory function of T reg cells (Fontenot et al., 2003; Hori et al., 2003; Khattri et al., 2003). Foxp3 functions via forming complexes with other transcription factors such as NFAT and Runx1 (Wu et al., 2006; Ono et al., 2007). T reg cells can be divided into two subsets, naturally occurring T reg (nT reg) cells, and induced T reg (iT reg) cells. nT reg cells develop via thymic selection and constitute the circulating CD4+CD25+ T reg cells in the peripheral lymph tissues in normal mice. iT reg cells are induced from naive CD4+ T cells upon TCR and TGF-β stimulation in vitro (Chen et al., 2003), or by tolerogenic antigen administration in vivo (Kretschmer et al., 2005). In both T reg cell populations, the induction of Foxp3 is subjected to epigenetic control, including histone modification, although to a different extent (Floess et al., 2007). Transcription factors such as NFAT or AP-1 play key roles in promoting Foxp3 expression (Mantel et al., 2006). A recent study identified an enhancer region in the Foxp3 gene that contains the binding sites for both NFAT and Smad3 in close proximity, thus providing an explanation for the synergistic effect of TCR and TGF-β signaling in Foxp3 expression (Tone et al., 2008). However, the detailed mechanisms governing the transcriptional regulation of the Foxp3 gene remain largely undiscovered.
The Foxo subfamily of transcription factors includes at least four members (Foxo1, Foxo3a, Foxo4, and Foxo6), and these factors are important regulators of metabolism, organ development, cell cycle, or apoptosis in diverse systems (Burgering, 2008). They can act as either transcriptional activators or repressors by forming complexes with different transcriptional modulators. Their function is tightly regulated by the upstream phosphoinositide 3-kinase (PI3K) and Akt pathway, which in turn induces the phosphorylation of Foxo factors and their nuclear export into the cytoplasm (Brunet et al., 1999). In the immune system, Foxo1 deficiency is linked to T cell homeostasis and tolerance, partly via modulating IL-7 receptor expression (Kerdiles et al., 2009; Ouyang et al., 2009). However, the functional role of Foxo3a in T cells is controversial. In an early study, Foxo3a-deficient mice displayed autoimmunity and defective NF-κB activation in T cells (Lin et al., 2004). A recent study indicates that Foxo3a plays an indirect role in T cell regulation via modulating IL-6 cytokine production from dendritic cells (Dejean et al., 2009). Therefore, the exact function of Foxo3a in T cells remains to be elucidated.
Cbl-b is an E3 ubiquitin ligase which is essential for T cell activation and tolerance induction (Liu et al., 2005). Loss of Cbl-b results in excessive IL-2 production and proliferation of T cells (Bachmaier et al., 2000; Chiang et al., 2000). Cbl-b promotes ubiquitin conjugation to the regulatory p85 subunit of PI3K, and affects downstream PI3K–Akt signaling (Fang and Liu, 2001). In addition, Cbl-b is up-regulated in anergic T cells, and it plays an essential role in T cell anergy induction by inhibiting critical signal transduction pathways (Heissmeyer et al., 2004; Jeon et al., 2004). Cbl-b interferes with TGF-β–mediated Foxp3 expression (Wohlfert et al., 2006). However, the exact mechanism underlying Cbl-b in Foxp3 expression is still lacking.
To further understand Cbl-b–mediated regulation of T cell function, we performed detailed studies on the induction of Foxp3 expression in T cells from mice lacking Cbl-b and Cbl-b knockin mice expressing a RING finger mutant form in the Cbl-b locus using both in vitro and in vivo systems. The phosphorylation of Foxo3a and Foxo1 is up-regulated in these mutant T cells. Further molecular and genetic studies demonstrate that Foxo3a and Foxo1 act as transcription factors promoting Foxp3 gene expression.
RESULTS
Cbl-b regulates Foxp3 expression in vitro and in vivo
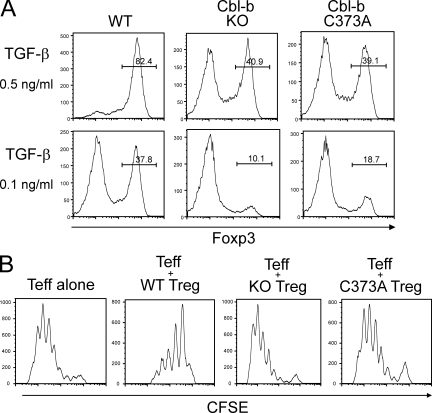
A previous study documented that Cbl-b–deficient CD4+ T cells are resistant to TGF-β–induced conversion to Foxp3+ cells (Wohlfert et al., 2006). Here, we performed mechanistic analyses to investigate whether the E3 ligase activity of Cbl-b is involved in the Foxp3 expression. Consistent with the previous study, Cbl-b–deficient naive CD4+ T cells stimulated with TGF-β displayed reduced Foxp3 expression (Fig. 1 A). Importantly, CD4+ T cells from mice expressing Cbl-b with a C373A mutation in the critical RING finger domain (Kojo et al., 2009), showed similar impairment to TGF-β–induced Foxp3 expression as Cbl-b–deficient cells. The suppressive function of these in vitro induced T reg cells was analyzed by co-culture with CFSE-labeled naive T cells. Although wild-type induced T reg cells were effective in inhibiting T effector cell division, T reg cells from Cbl-b–deficient or RING finger mutant knockin mice were much less potent in proliferative suppression (Fig. 1 B).
Figure 1.
The requirement of Cbl-b E3 ligase activity in the regulation of Foxp3+ iT reg cells. (A) Naive CD4+CD62L+CD25− T cells from wild-type (WT), Cbl-b–deficient (KO), and Cbl-b C373A knockin mice were stimulated with anti-CD3 and anti-CD28 in the presence of indicated concentrations of TGF-β. Development of Foxp3+ iT reg cells was assessed by FACS analysis after 5 d. The percentages of Foxp3-expressing cells are shown. Data are representative of five independent experiments. (B) The cells in A were co-cultured with CFSE-labeled CD4+CD25− T eff cells at a 1:1 ratio in the presence of irradiated T cell–depleted splenocytes and anti-CD3. CFSE dilution was assessed 4 d later by FACS analysis. Data are representative of at least three independent experiments.
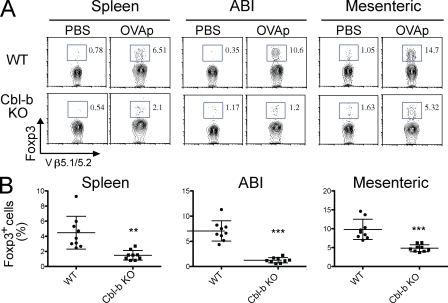
We further examined the generation of T reg cells in vivo by performing adoptive transfer of TCR transgenic T cells and soluble antigen tolerization. Administration of OVA peptide induced the generation of Foxp3+ T cells in mice receiving wild-type OTII transgenic T cells in both spleen and lymph nodes (Fig. 2 A); however, such increase was largely abrogated in recipients of Cbl-b–deficient OTII T cells. The experiment was repeated three times and significant differences were observed in the generation of Foxp3+ T cells between the two groups of mice (Fig. 2 B).
Figure 2.
Cbl-b deficiency attenuates Foxp3+ iT reg cell differentiation in vivo. Naive OTII T cells (Vβ 5.1/5.2+) from WT and Cbl-b KO mice (CD45.2) were adoptively transferred to B6 SJL (CD45.1) mice, and the recipients were immunized with 10 µg of OVA323-339 peptide (OVAp). 5 d after immunization, Foxp3 expression in CD45.2+TCR Vβ 5.1/5.2+ T cells were measured. ABI, axillary, branchial, and inguinal lymph nodes. (A) Representative FACS plots showing the percentage of Foxp3-expressing 5.1/5.2+ donor OTII T cells. Data are representative of three independent experiments. (B) Statistical representation of Foxp3 induction in donor OTII T cells. Each dot represents an individual mouse. Small horizontal lines indicate the mean, and error bars indicate standard deviations. **, P < 0.01; ***, P < 0.001. Data are from three independent experiments.
Foxo3a as a target in Cbl-b–regulated signaling
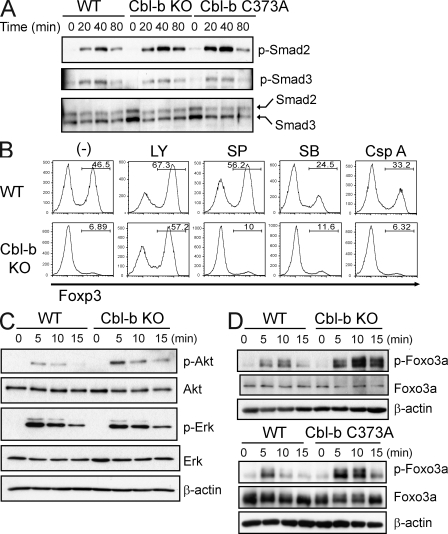
A previous study suggested that Cbl-b directly affects TGF-β signaling via modulating Smad2 phosphorylation (Wohlfert et al., 2006). We failed to observe an effect of Cbl-b deficiency on the phosphorylation of either Smad2 or Smad3 under different stimulation conditions (Fig. 3 A and Fig. S1), upon repeated experiments. In addition, upon TGF-β stimulation T cells from Cbl-b C373A knockin mice showed similar levels of Smad2 or Smad3 phosphorylation as wild-type T cells (Fig. 3 A). We next sought to identify potential signaling pathways that may be altered by Cbl-b ablation. For this purpose, we used a panel of chemical inhibitors for different signaling molecules in the T reg cell differentiation assay. Of these inhibitors, the PI3K inhibitor LY294002 rescued the defective Foxp3 expression in Cbl-b–deficient T cells (Fig. 3 B). This result was further substantiated by the observation that Akt phosphorylation was up-regulated in Cbl-b–deficient T cells, whereas the amount of phospho-Erk was not altered (Fig. 3 C), which is consistent with our previous observation (Fang and Liu, 2001).
Figure 3.
PI3K–Akt–Foxo3a activation in Cbl-b–mediated regulation of Foxp3+ iT reg cells. (A) Phosphorylation of Smad proteins in naive CD4+ T cells from WT, Cbl-b KO, and Cbl-b C373A mice. Naive CD4+ T cells were stimulated with anti-CD3 and anti-CD28 in the presence of TGF-β for indicated time periods. Whole-cell lysates were immunoblotted with anti–phospho-(p)-Smad2, or anti–p-Smad3, and reprobed with anti-Smad2/3. Smad2, 60 kD; Smad3, 52 kD. Data are representative of three independent experiments. (B) Naive CD4+ T cells from WT and Cbl-b KO mice were stimulated with anti-CD3 and anti-CD28 together with TGF-β in the absence or presence of the PI3K inhibitor LY294002 (LY), the JNK inhibitor SP600125 (SP), the p38 inhibitor SB203580 (SB), or the calcineurin inhibitor cyclosporine A (Csp A). Foxp3 expression was assessed by FACS analysis after 5 d. The percentages of Foxp3-expressing cells are shown. Data are representative of three independent experiments. (C) Naive CD4+ T cells were stimulated with anti-CD3 and anti-CD28 for the indicated time periods. Whole-cell lysates were immunoblotted with anti–p-Akt, anti-Akt, anti–p-Erk, anti-Erk2, and anti–β-actin. Akt, 56 kD; Erk, 42/44 kD; β-actin, 45 kD. Data are representative of at least three independent experiments. (D) Regulation of Foxo3a phosphorylation by Cbl-b E3 ligase. Naive CD4+ T cells from WT and Cbl-b KO mice (top) or WT and Cbl-b C373A mice (bottom) were stimulated with anti-CD3 and anti-CD28 for indicated time periods. Whole-cell lysates were immunoblotted with anti–p-Foxo3a, anti-Foxo3a, and anti–β-actin. Foxo3a, 95 kD; β-actin, 45 kD. Data are representative of at least three independent experiments.
Foxo proteins such as Foxo3a have been shown to be downstream targets of PI3K–Akt signal pathway (Brunet et al., 1999). We examined the status of Foxo3a phosphorylation in wild-type and mutant T cells. Cbl-b deficiency resulted in a marked increase of phosphorylated Foxo3a upon anti-CD3 plus anti-CD28 stimulation (Fig. 3 D). This increase was also observed in Cbl-b C373A knockin T cells (Fig. 3 D), suggesting that the E3 ligase activity of Cbl-b is required for the negative regulation of Foxo3a phosphorylation.
Foxo3a regulates Foxp3 expression
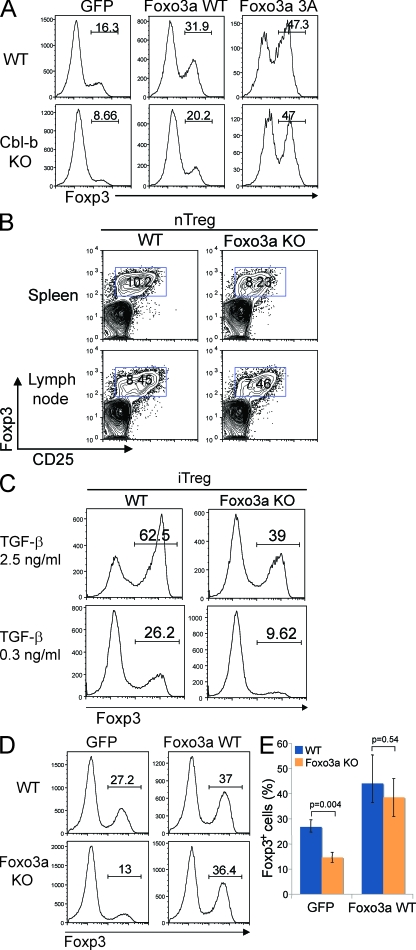
The increased phosphorylation of Foxo3a in Cbl-b mutant T cells prompted us to investigate whether it has a biological role in regulating Foxp3 expression. First, we retrovirally transduced into wild-type CD4 T cells with wild-type Foxo3a or a constitutively active Foxo3a mutant in which all three conserved phosphorylation sites were mutated to alanines (Brunet et al., 1999). The effect on Foxp3 expression was then examined by performing in vitro iT reg cell differentiation assays. Introduction of wild-type Foxo3a increased Foxp3 expression (Fig. 4 A, top), and this effect was further augmented by the transduction of the constitutively active Foxo3a mutant. To determine whether Foxo3a acts as a downstream target of Cbl-b, we examined whether Foxo3a can rescue the defective Foxp3 expression in Cbl-b–deficient T cells. Retroviral transduction of wild-type Foxo3a increased the Foxp3 expression in Cbl-b–deficient T cells to a certain degree (Fig. 4 A, bottom). Notably, the constitutively active Foxo3a mutant augmented Foxp3 expression to the same level in wild-type and Cbl-b–deficient T cells. These results suggest that Foxo3a is a downstream target of Cbl-b.
Figure 4.
Foxo3a regulates Foxp3+ iT reg cell differentiation. (A) Naive CD4+ T cells from WT and Cbl-b KO mice were stimulated with anti-CD3 and anti-CD28 for 2 d and retrovirally transduced with control-IRES-GFP (GFP), Foxo3a WT-IRES-GFP (Foxo3a WT), or Foxo3a 3A-IRES-GFP (Foxo3a 3A). After infection, TGF-β was added and Foxp3 expression was assessed 3 d later by FACS analysis. The percentages of Foxp3-expressing cells in gated GFP-positive cells are shown. Data are representative of four independent experiments. (B) nT reg cells in the spleen and lymph nodes of Foxo3a KO mice. The spleen and the lymph node cells from WT and Foxo3a KO mice were stained with anti-CD4, anti-CD25, and anti-Foxp3. The percentages of Foxp3-expressing cells in CD4 T cells are shown. Data are representative of three independent experiments. (C) Naive CD4+ T cells were stimulated with anti-CD3 and anti-CD28 in the presence of indicated concentrations of TGF-β for 3 d. Foxp3 expression was assessed by FACS analysis. The percentages of Foxp3-expressing cells in gated CD4 T cells are shown. Data are representative of five independent experiments. (D) Naive CD4 T cells from WT and Foxo3a KO mice were stimulated with anti-CD3 and anti-CD28 for 2 d and retrovirally transduced with control-IRES-GFP (GFP) or Foxo3a WT-IRES-GFP (Foxo3a WT). 1 d after infection, TGF-β was added, and Foxp3 expression was assessed 3 d later by FACS analysis. A representative of three repeated experiments. (E) The percentages of Foxp3-expressing cells in GFP-positive cells as shown in D are calculated and shown as mean ± SD of three independent experiments. Statistical significance of the data were evaluated by unpaired two-tailed Student’s t test.
We then sought to obtain further genetic evidence of the functional role of Foxo3a in T reg cell differentiation by analyzing T cells from Foxo3a-deficient mice. Ablation of Foxo3a gene did not affect the development of nT reg cells, as wild-type and Foxo3a-deficient mice showed similar percentage of CD25+Foxp3+ CD4+ T cells in both spleen and lymph nodes (Fig. 4 B). We next examined whether Foxo3a deficiency affects Foxp3 expression induced by TGF-β in iT reg cells. Ablation of Foxo3a expression resulted in a marked reduction of Foxp3 expression in iT reg cells (Fig. 4 C), similar to that observed in Cbl-b–deficient T cells.
A reconstitution experiment was then performed using retroviral transduction of Foxo3a in Foxo3a-deficient T cells. Re-introduction of wild-type Foxo3a largely restored the Foxp3 expression in Foxo3a-deficient T cells to a degree similar to wild-type control cells (Fig. 4, D and E). Thus, Foxo3a plays an intrinsic role in regulating Foxp3 expression in iT reg cells.
Foxo3a directly binds to Foxp3 promoter
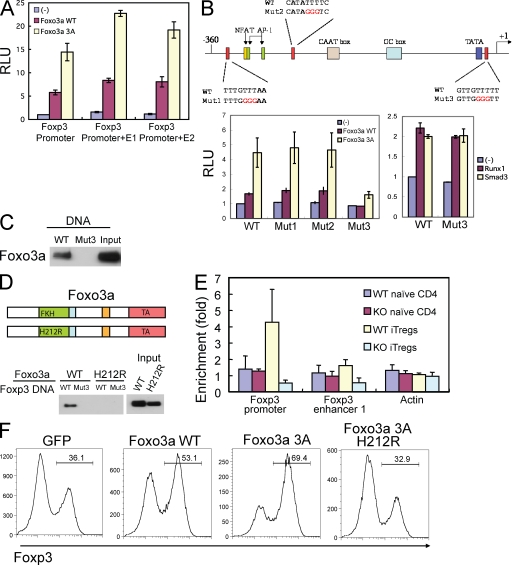
The transcription unit of the Foxp3 gene includes the basic promoter and at least two enhancers (E1 and E2; von Boehmer and Nolting, 2008). To examine whether Foxo3a has a direct effect on Foxp3 expression, we constructed luciferase reporter plasmids containing either the basic promoter, or a combination with one of the two enhancers, and tested the effect of Foxo3a in driving luciferase expression in a transient transfection assay. Foxo3a acted primarily at the basic promoter, as addition of either enhancer only showed marginal effects (Fig. 5 A). Notably, the phosphorylation-defective, constitutively active mutant of Foxo3a was much more potent in driving Foxp3 promoter activity.
Figure 5.
Foxo3a directly binds to the Foxp3 promoter. (A) 293T cells were transfected with luciferase reporter plasmids containing the Foxp3 promoter, the promoter and the Foxp3 enhancer 1 (E1), or the promoter and the Foxp3 enhancer 2 (E2), together with an empty, Foxo3a WT, or Foxo3a 3A expression vector. Bars show the mean relative luciferase unit (RLU) ± SD as arbitrary light units of three independent experiments. (B, top) Schematic structure of the Foxp3 promoter region and mutated sequence of the Foxp3 promoter. (bottom) Jurkat cells were transfected with luciferase reporter plasmids containing the Foxp3 promoter and the Foxp3 enhancer 1 or the Foxp3 promoter with Foxo3a binding site mutated and the Foxp3 enhancer 1 (WT, Mut1, Mut2, and Mut3) together with an empty, Foxo3a WT, Foxo3a 3A, Runx1, or Smad3 expression vectors. 24 h after transfection, these cells were stimulated with anti-CD3 and anti-CD28 for 8 h. Bars show the mean RLU ± SD as arbitrary light units of three independent experiments. (C) Pull-down assay of Foxo3a binding to a WT or mutated (Mut3) sequence of the Foxp3 promoter. Cell lysates from naive CD4 T cells stimulated with anti-CD3, anti-CD28, and TGF-β were mixed with biotinylated DNA probes. The labeled DNA probes were precipitated with streptavidin-agarose beads, and the precipitates were subjected to SDS-PAGE, followed by immunoblotting with anti-Foxo3a. Input represents 5% of the total amount used for precipitation. Foxo3a, 95 kD. Data are representative of three independent experiments. (D, top) Schematic structure of Foxo3a WT and H212R mutant. (bottom) 293T cells were transfected with myc-tagged Foxo3a WT or H212R mutant. The cell lysates were mixed with biotinylated DNA probes encoding WT or Mut3 Foxp3 promoters and pull-down assay was performed as in C. Foxo3a was detected by immunoblotting with anti-myc to detect myc-tagged Foxo3a proteins. Foxo3a, 95 kD. Data are representative of three independent experiments. (E) Chromatin immunoprecipitation analysis of Foxo3a binding to the Foxp3 promoter region. Naive CD4 T cells were unstimulated or stimulated with anti-CD3 and anti-CD28 together with TGF-β. Cell lysates were immunoprecipitated with anti-Foxo3a or control IgG. Immunoprecipitates from WT and Foxo3a KO T cells were analyzed by quantitative real-time PCR, using primers corresponding to Foxp3 promoter, its enhancer 1, and a nonspecific Actin promoter as a control. The results were presented as fold of template enrichment in immunoprecipitates of anti-Foxo3a relative to those of control IgG (mean and SD of three independent experiments). (F) Naive CD4+ T cells were stimulated with anti-CD3 and anti-CD28 for 2 d and retrovirally transduced with control-IRES-GFP (GFP), Foxo3a WT-IRES-GFP (Foxo3a WT), Foxo3a 3A-IRES-GFP (Foxo3a 3A), or Foxo3a 3A H212R-IRES-GFP (Foxo3a 3A H212R). After infection, TGF-β was added and Foxp3 expression was assessed 3 d later by FACS analysis. The percentages of Foxp3-expressing cells in GFP positive cells are shown. Data are representative of at least three independent experiments.
Foxo subfamily of transcription factors binds to a consensus DNA sequence of (T/C/G)(T/C/G)T(G/A)TTTT(A/G/T; Paik et al., 2007). Inspection of the basic Foxp3 promoter revealed three putative binding sites for Foxo3a (Fig. 5 B, top). We then generated individual mutations at these sites and found that one of the sites proximal to the TATA box (Mut3) is critical for Foxo3a-driven luciferase expression, as mutation at this site almost completely abrogated the luciferase induction even with the constitutively active Foxo3a mutant (Fig. 5 B, bottom left). However, mutations of the other two upstream potential binding sites did not have any effects (Fig. 5 B, bottom left). Importantly, mutation at the Mut3 Foxo3a binding site did not interfere with the general promoter activity because Smad3 or Runx1-driven luciferase activity was not altered by this mutation (Fig. 5 B, bottom right), suggesting that Foxo3a acts specifically and independently at the Foxp3 promoter. It should be noted that compared with the induction of Foxp3 reporter activity in 293T cells (Fig. 5 A), the Foxp3-driven luciferase activity is relatively low in Jurkat T cells (Fig. 5 B). This may reflect the fact that Jurkat T cells have high Akt activity, which is caused by the deficiency of Pten (Shan et al., 2000).
To investigate whether Foxo3a directly binds to the third putative binding motif in the Foxp3 promoter, we performed DNA pull-down assay using biotin-labeled DNA probes. The wild-type motif (-TTGTTTT-) precipitated Foxo3a protein from TGF-β–stimulated mouse T cell lysates, whereas such association was completely eliminated by the mutation of this motif (-TTGGGGT; Fig. 5 C). We then determined whether the DNA-binding domain in Foxo3a is responsible for such interaction. It was found that mutation at the critical histidine-212 to arginine in the Foxo3a DNA binding domain completely blocked the association between the consensus DNA motif and Foxo3a (Fig. 5 D).
To further investigate whether Foxo3a binds to Foxp3 promoter in cells, we performed chromatin immunoprecipitation assay. Anti-Foxo3a immunoprecipitated the DNA fragments corresponding to the proximal region of Foxp3 promoter, but not the enhancer 1 region in iT reg cells, and such precipitation was abolished in Foxo3a-deficient T cells (Fig. 5 E). Importantly, a functional DNA binding domain of Foxo3a is required for the induction of Foxp3 protein, as the H212R mutant was unable to promote Foxp3 expression in iT reg cells, even with the constitutively active mutation (Fig. 5 F). These results collectively indicate that Foxo3a induces Foxp3 gene expression by directly binding to Foxp3 basic promoter.
Foxo1 has similar function in Foxp3 gene induction
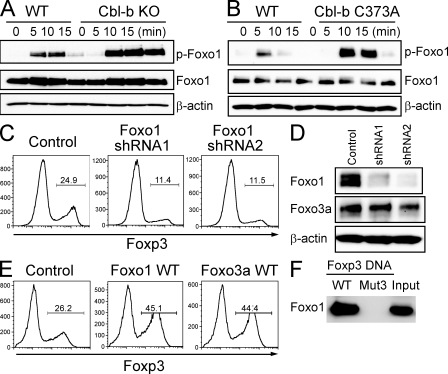
Because Foxo1, a close homologue of Foxo3a, is also involved in T cell regulation (Kerdiles et al., 2009; Ouyang et al., 2009), we next examined whether it also promotes Foxp3 transcription. First we observed the up-regulation of Foxo1 phosphorylation in Cbl-b–deficient CD4+ T cells (Fig. 6 A) and in Cbl-b RING finger mutant knockin T cells (Fig. 6 B). We then performed shRNA-mediated Foxo1 knockdown in primary mouse CD4+ T cells and examined in vitro TGF-β–induced Foxp3 induction. Retroviral transduction of two Foxo1 shRNAs impaired Foxp3 expression as compared with the control cells (Fig. 6 C). The effect of shRNAs on Foxo1 expression was determined by immunoblotting, showing that both shRNAs were effective in reducing Foxo1 expression (Fig. 6 D). Similarly, retroviral transduction of Foxo1 in mouse CD4+ T cells augmented Foxp3 expression to the similar extent as Foxo3a (Fig. 6 E). Finally, we demonstrated that Foxo1 bound to the wild-type, but not the mutated, Foxo3a binding motif in the DNA pull-down assay (Fig. 6 F). The results suggest that Foxo1 plays a role similar to that of Foxo3a in Foxp3 induction.
Figure 6.
Foxo1 regulates Foxp3 expression. (A and B) Naive CD4+ T cells from WT and Cbl-b KO mice (A) or WT and Cbl-b C373A mice (B) were stimulated with anti-CD3 and anti-CD28 for indicated time periods. Whole-cell lysates were immunoblotted with anti–p-Foxo1, anti-Foxo1 (75 kD), and anti–β-actin (45 kD). Data are representative of three independent experiments. (C) Naive CD4+ T cells were stimulated with anti-CD3 and anti-CD28 for 2 d and transduced with retroviruses encoding Foxo1-specific shRNAs (shRNA1 or shRNA2) or with empty LMP vector (control). After 24 h, TGF-β was added and Foxp3 expression was assessed 3 d later by FACS analysis. Cells were treated with puromycin for last 48 h. The percentages of Foxp3-expressing cells in GFP-positive cells are shown. Data are representative of three independent experiments. (D) Cell lysates in (C) were immunoblotted with anti-Foxo1 (75 kD), anti-Foxo3a (95 kD), and anti-β-actin (45 kD). Data are representative of three independent experiments. (E) Naive CD4 T cells were stimulated with anti-CD3 and anti-CD28 for 2 d and retrovirally transduced with control-IRES-GFP (GFP), Foxo1 WT-IRES-GFP (Foxo1 WT), or Foxo3a WT-IRES-GFP (Foxo3a WT). After infection, TGF-β was added and Foxp3 expression was assessed 3 d later by FACS analysis. The percentages of Foxp3-expressing cells in GFP-positive cells are shown. Data are representative of four independent experiments. (F) Pull-down assay of Foxo1 binding to WT or mutated (Mut3) sequence of the Foxp3 promoter. DNA pull-down assay was performed using biotinylated DNA probes. The precipitates were subjected to SDS-PAGE, followed by immunoblotting with anti-Foxo1 (75 kD). Data are representative of at least three independent experiments.
DISCUSSION
By using both in vitro and in vivo genetic and biochemical approaches, we have provided convincing evidence that Foxo3a acts as a transcription factor influencing Foxp3 gene expression. The phosphorylation of Foxo3a is up-regulated in Cbl-b–deficient or the ligase-inactive RING finger mutant T cells. Foxo3a expression increases Foxp3 expression and rescues the defective Foxp3 expression in Cbl-b mutant T cells. T cells deficient in Foxo3a are resistant to TGF-β–induced Foxp3 induction, whereas retroviral reconstitution of Foxo3a restores such defect. Finally, a functional Foxo binding motif is present in the basic promoter of Foxp3, and a direct protein–DNA interaction is necessary for Foxp3 gene transcription. In addition, Foxo1 has a function similar to that of Foxo3a. We thus conclude that transcription factors Foxo3a and Foxo1 couple the Cbl-b E3 ligase to the induction of Foxp3 in iT reg cells.
Our current findings seem to be at odds with a previous publication showing that Cbl-b regulates TGF-β–induced phosphorylation of Smad2, which impairs Foxp3 expression in iT reg cells (Wohlfert et al., 2006). However, our repeated experiments convincingly told us that this might not be the case. Although we cannot exclude the possibility that Cbl-b may play an indirect role in regulating TGF-β signaling, at present we would favor the pathway that Cbl-b regulates Foxp3 expression via modulating PI3K–Akt–Foxo3a signaling as documented in this study. The present study is consistent with our original finding that Cbl-b promotes ubiquitination of p85, which is the regulatory subunit of PI3-K, and affects downstream Akt phosphorylation (Fang and Liu, 2001). More importantly, we extended the previous studies by showing that the E3 ligase activity of Cbl-b is critical for such regulation, as T cells containing the ligase-inactive RING finger mutant exhibited a similar phenotype as Cbl-b–deficient T cells in regard to Foxo3a/Foxo1 phosphorylation and in vitro Foxp3 induction. Therefore, the present study identifies a unique mechanism by which the Cbl-b E3 ligase regulates the development and function of iT reg cells. It should be noted that this study is consistent with the previous observation that Cbl-b only regulates the Foxp3 expression in iT reg cells, but not in nT reg cells (Wohlfert et al., 2006). One possible explanation is that Cbl-b plays a dominant role in peripheral mature T cells, but not during early thymic development (Bachmaier et al., 2000; Chiang et al., 2000). Another possibility is the overlapping role of Cbl-b with c-Cbl; deficiency of either isoform may not lead to a significant effect in early stage nT reg cells development. Future studies are needed to test such possibilities.
One of the downstream targets of Akt is the mammalian target of rapamycin (mTOR; Hay and Sonenberg, 2004). Two recent studies documented that mTOR forms the PI3K–Akt–mTOR axis in regulating Foxp3 expression (Haxhinasto et al., 2008; Sauer et al., 2008). The involvement of mTOR in the differentiation of iT reg cells was further supported by a more recent publication showing that T cells lacking mTOR kinase differentiate into iT reg cells with only TCR stimulation in the absence of TGF-β (Delgoffe et al., 2009). Therefore, the possibility arises that Cbl-b regulates Foxp3 expression via modulating the mTOR pathway. However, detailed examination of the mTOR-deficient T cells showed that only the mTOR complex 2 (TORC2), which is less sensitive to rapamycin, but not the rapamycin-sensitive TORC1, is involved in Foxp3 expression (Delgoffe et al., 2009). It is suggested that TORC2 exerts its function via modulating the activation of Akt (Guertin et al., 2006) to form a positive regulatory loop. A similar mechanism may operate here if Foxo3a is a downstream target of both Akt and mTOR pathways in Cbl-b–regulated Foxp3 expression in iT reg cells.
Although an earlier study suggested that the basic Foxp3 promoter contains NFAT binding site (Mantel et al., 2006), a recent study indicated that a functional NFAT binding site lies in an enhancer region, with close proximity to the Smad3 binding motif (Tone et al., 2008). This raises the issue of whether the transcriptional activity of the basic promoter is regulated by other transcription factors. Several recent studies have documented that the Runx transcription factors are critically involved in induction and suppressive function of T reg cells (Bruno et al., 2009; Kitoh et al., 2009; Klunker et al., 2009; Rudra et al., 2009), with the identification of direct binding sites, including in the Foxp3 basic promoter (Klunker et al., 2009; Rudra et al., 2009). We reported that TIEG1, a TGF-β–induced early gene product, is involved in Foxp3 gene expression, via ubiquitination-dependent modification by the E3 ligase Itch (Venuprasad et al., 2008). Interestingly, the putative Foxo3a binding site close to the TATA box seems to play a dominant role in driving Foxp3 gene transcription. This unusual binding for Foxo3a was also found in the promoter of Atrogen-1 gene (Sandri et al., 2004), even though the functional significance remains unclear. It is still possible that Foxo3a binds to other sites/regions of Foxp3 gene locus. Nevertheless, the description of Foxo3a as a transcription factor directly acting at the basic promoter thus provides additional mechanisms for the transcriptional control of the Foxp3 gene.
The fact that the highly related Foxo family members serve as a common target of PI3K–Akt pathway suggests that they are functionally redundant, which is supported by genetic studies that conditional deletion of Foxo1, 3a, and 4 results in tumor formation and alteration of common downstream targets (Paik et al., 2007). Although genetic deletion of Foxo1 or Foxo3a resulted in different phenotypes in T cells (Dejean et al., 2009; Kerdiles et al., 2009; Lin et al., 2004; Ouyang et al., 2009), the possibility exists that these Foxo members function in a redundant manner. We indeed observed that Foxo1 phosphorylation is also regulated in Cbl-b mutant T cells and shRNA knockdown of Foxo1 reduces Foxp3 expression in iT reg cells. Although the functional role Foxo1 or Foxo3a remains unclear in the development of nT reg cells (Dejean et al., 2009; Ouyang et al., 2009 and this study), it is quite possible that the overlapping function of the Foxo family members affects the development of both nT reg cells and iT reg cells. Nevertheless, the present study provided compelling evidence that Foxo transcription factors play an intrinsic role in T cells by regulating the Foxp3 expression at the transcription level.
In this study, we emphasize that Cbl-b regulates iT reg cells via modulating the PI3K–Akt–Foxo pathway in an E3 ligase–dependent manner. Previous studies have shown that Cbl-b may target different substrates in different cell types or under different physiological conditions. For example, Cbl-b is shown to promote the ubiquitination of PLC-γ1 and/or PKCθ in anergic T cells, which blocks T cell activation upon subsequent effective T cell stimulation (Heissmeyer et al., 2004; Jeon et al., 2004). However, in anergic natural killer T cells, Cbl-b primarily induces the ubiquitination of CARMA1, and affects NF-κB–mediated IFN-γ production (Kojo et al., 2009). We propose that Cbl-b acts as a gatekeeper of lymphocyte activation by targeting different substrates to ensure that the immune tolerance is properly maintained. Understanding such detailed mechanisms may benefit the therapeutic intervention in human immune diseases.
MATERIALS AND METHODS
Mice.
C57BL/6 mice were obtained from The Jackson Laboratory. B6 SJL (CD45.1) mice were obtained from Taconic. Cbl-b–deficient mice on B6 background were obtained from H. Gu (Columbia University, New York, NY). Cbl-b C373A knockin mice were provided by W. Langdon (University of West Australia, Crawley, WA, Australia; Kojo et al., 2009), OVA323-329-specific TCR transgenic OT-II mice (Jeon et al., 2004), and Foxo3a-deficient mice (Castrillon et al., 2003) were previously described. All mice were housed in specific pathogen–free conditions. The experiment protocols were approved by members of the Institutional Animal Care and Use Committee of the La Jolla Institute for Allergy and Immunology.
Separation of cells.
CD4+CD62L+CD25− naive T cells and CD4+CD25− effector T (T eff) cells were separated by MACS beads (Miltenyi Biotec) according to the manufacturer’s instruction. The purity of separated cell population was >93% for CD4+CD62L+CD25− T cells and >95% for CD4+CD25− T eff cells. T cell–depleted splenocytes were obtained by depleting T cells by using anti-CD4 and anti-CD8 microbeads (Miltenyi Biotec).
In vitro T reg cells generation.
CD4+CD62L+CD25− T cells (2 × 105) from wild-type C57BL/6, Cbl-b–deficient, Cbl-b C373A, wild-type FVB/NJ, or Foxo3a-deficient mice were stimulated with plate-bound anti-CD3 (145-2C11) and soluble anti-CD28 (PV-1; provided by R. Abe, Tokyo University of Science, Noda, Chiba, Japan) together with indicated concentrations of rhTGF-β (PeproTech) in 96-well flat-bottomed plates for 3 d, or cells were cultured with rhIL-2 for additional 2 d. For inhibitor experiments, LY294002 (for PI3K, 10 µM), SP600125 (for JNK, 10 µM), SB203580 (for p38, 10 µM), or cyclosporine A (for calcineurin, 20 ng/ml) was added to the culture. All inhibitors were purchased from Calbiochem.
T reg cell suppression assay.
Naive CD4+CD25− T cells (105 cells) were labeled with 5 µM CFSE (Invitrogen) for 10 min at 37°C in PBS/0.1% BSA, and were then cocultured with in vitro induced T reg cells (105 cells) from WT, Cbl-b–deficient, or Cbl-b C373A mice in the presence of T cell–depleted splenocytes (5 × 105 cells) and soluble anti-CD3. 4 d later, cells were harvested, and CFSE dilution was measured by FACS analysis.
In vivo induction of iT reg cells.
Naive CD4+CD62L+CD25− T cells were purified from spleen of WT or Cbl-b–deficient OTII mice, and 1–2 × 106 cells were injected i.v. to B6.SJL (CD45.1) mice. The next day, the recipient mice were immunized with soluble chicken OVA323-339 peptide (10 µg; AnaSpec). 5 d after administration of OVA peptide, cells were collected from secondary lymphoid tissues (axillary, branchial, inguinal, and mesenteric lymph nodes and spleen).
Flow cytometry.
Antibody against TCRVβ 5.1/5.2 was purchased from BD, antibodies against CD4 and CD25 were purchased from BioLegend, and antibodies against Foxp3, CD62L, and CD45.2 were purchased from eBioscience. Intracellular Foxp3 staining was performed with Cytofix/Cytoperm reagent (BD).
Immunoprecipitation and immunoblotting.
Antibodies to phospho-Smad2, phospho-Smad3, phospho-Akt (Ser473), Akt, phospho-Erk1/2, phospho-Foxo3a (Ser253), Foxo3a, phospho-Foxo1 (Ser256), and Foxo1 were purchased from Cell Signaling Technology. Antibodies to Smad2/3, Erk2, and Myc were purchased from Santa Cruz Biotechnology. Anti–β-actin was obtained from MP Biomedicals. Cells were lysed with 1 x NP-40 lysis buffer (1% NP-40, 20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 5 mM EDTA, 50 mM NaF, 2 mM Na3VO4,and 10 µg/ml each of aprotinin and leupeptin) or were lysed with 1 x SDS sample buffer (50 mM Tris-HCl, pH 6.8, 100 M DTT, 2% SDS, and 10% glycerol). Samples were subjected to 8–10% SDS-PAGE and electrotransferred onto polyvinylidene difluoride membranes (Millipore). Membranes were probed with the indicated primary antibodies, followed by HRP-conjugated secondary antibodies. Membranes were then washed and visualized with the enhanced chemiluminescence detection system (ECL; GE Healthcare). When necessary, membranes were stripped by incubation in stripping buffer (Thermo Fisher Scientific) for 15 min with constant agitation, washed, and then reprobed with various other antibodies.
Plasmids.
The Foxp3 promoter (−490/+184), enhancer 1 (+1987/+2736), and enhancer 2 (+3,665/+4,823) regions were amplified by PCR from genomic DNA of B6 mice by using the following primers: Foxp3 promoter forward, 5′-CTTCCCATTCACATGGCAGGCTTC-3′; Foxp3 promoter reverse, 5′-CAAAGTCCTTACCTGGAGTGGCTG-3′; Foxp3 enhancer1 forward, 5′-TTGTGCTTTGTAATGCATGTG-3′; Foxp3 enhancer1 reverse, 5′-GGACTGATGCTGCTAGGTG-3′; Foxp3 enhancer 2 forward, 5′-TTGTCCCAGGAGAGC-3′; and Foxp3 enhancer 2 reverse, 5′-CCCATATGGCTGGAC-3′. The PCR products were cloned into the pGL3 basic vector (Promega Biotech). Mutations of three putative Foxo3a binding sites in the Foxp3 promoter region were generated by oligonucleotide-directed site-specific mutagenesis. The following primers and their complementary strands were used: Mut1, 5′- CTGACTCTACACACTTTTGGGGAAGAAATTGTGGTTTCTC-3′; Mut2, 5′-GTGAGGGGAAGAAATCATAGGGTCAGATGACTTGTAAAGGG-3′; and Mut3, 5′-CGGTATAAAAGCAAAGTTGGGGTTGATAATGTGGCAGTTTCC-3′.
The Foxo3a wild-type, a mutant with three phosphorylation sites mutated to alanines (Foxo3a 3A), and Foxo1 cDNAs were subcloned in the pcDNA3 myc (Invitrogen) and pMX-IRES-GFP vectors, respectively. The DNA binding-defective mutant of Foxo3a 3A was generated by making point mutation at histidine 212 to arginine (H212R). To construct Foxo1 shRNA vector, oligonucleotides were cloned in the LMP vector according to manufacture’s protocol (Open Biosystems). Oligonucleotide sequences were the following: Foxo1 shRNA1, 5′-TGCTGTTGACAGTGAGCGACGTGCCCTACTTCAAGGATAATAGTGAAGCCACAGATGTATTATCCTTGAAGTAGGGCACGCTGCCTACTGCCTCGGA-3′; Foxo1 shRNA2, 5′-TGCTGTTGACAGTGAGCGCCCATGGACAACAACAGTAAATTAGTGAAGCCACAGATGTAATTTACTGTTGTTGTCCATGGATGCCTACTGCCTCGGA-3′.
Retroviral transduction.
Plat-E cells were transfected with 3 µg of pMX-IRE-GFP vector or LMP vector with 9 µl of TransIT-LT1 (Mirus). At 48 h, the culture supernatant containing retrovirus was collected. Naive CD4+CD62L+CD25− T cells were stimulated with plate-bound anti-CD3 and soluble anti-CD28. At 48 h, CD4 T cells were infected with retrovirus together with 5 µg/ml polybrene by centrifuging cells at 2,000 rpm for 60 min at room temperature. After infection, TGF-β was added and T cells were cultured for additional 3 d. Cells were cultured with 100 U/ml human rIL-2 for the last 2 d.
Reporter assay.
293T or Jurkat T cells were transfected with indicated amounts of Foxp3 promoter luciferase reporter plasmid together with 0.5 µg of Foxo3a WT, Foxo3a 3A, Smad3, or Runx1 expression vectors, and 0.05 µg of β-gal expression vector by using 3 µl of TransIT-LT1 for 293T cells, or electroporation for Jurkat T cells. 24 h later, cells were resuspended in 100 µl of lysis buffer (100 mM potassium phosphate buffer, pH 7.8, 0.2% Triton X-100, and 1 mM DTT) and incubated at room temperature for 10 min. After centrifugation, 30 µl of the supernatant was used with 100 µl of luciferase assay reagent (BD). Luminescence was measured with a Monolight 2010 (Analytical Luminescence Laboratory).
DNA pull-down assay.
A DNA fragment containing wild-type or mutant Foxo-binding site was amplified by PCR using a 5′-biotin-labeled forward primer. Primer sequences were as follows: DNA pull-down forward, 5′-ATTAGAAGAGCGAGGTCTGCGGC-3’; DNA pull-down reverse, 5′-CTTGGTGAAGTGGACTGCTAGAG-3’. The biotinylated probes (1.5 µg) was mixed with cell lysates in 400 µl of a binding buffer (25 mM Hepes, pH7.9, 50 mM NaCl, 1 mM MgCl2, 1 mM DTT, 2 µg polydI-dC) for 2 h at 4°C. Then 20 µl of streptavidin-agarose beads (Invitrogen) was added and incubated for an additional 1 h. The streptavidin-agarose beads were washed five times with the binding buffer, and then SDS-sample buffer was added. The complexes were subjected to SDS-PAGE, followed by immunoblotting with anti-Foxo3a, anti-Foxo1, or anti-Myc.
Chromatin immunoprecipitation assay.
Cells were cross-linked by addition of fresh 1% formaldehyde in PBS for 10 min at room temperature, followed by quenching with 135 mM glycine. Fixed cells were resuspended in a lysis buffer (10 mM Hepes, pH 7.9, 0.5% IGEPAL-CA630, 1.5 mM MgCl2, and 10 mM KCl) with protease inhibitor for 15 min on ice. After centrifugation, the cell pellet was resuspended in nuclear lysis buffer (50 mM Tris, pH 8.1, 10 mM EDTA, 1% SDS) with protease inhibitor for 10 min on ice and sonicated 7 times for 30 s, and then the lysates were cleaned by centrifugation. The sheared DNA was diluted 10-fold in a dilution buffer (0.01% SDS, 16.7 mM Tris-Hcl, pH 8.1, 1.2 mM EDTA, and 167 mM NaCl). The chromatin solution was incubated with 4 µg of control rabbit IgG (Santa Cruz Biotechnology, Inc.), anti-Foxo3a (Santa Cruz Biotechnology, Inc.) or anti-Foxo3a (Cell Signaling Technology), and 20 µl of fully suspended protein A magnetic beads (Millipore) overnight at 4°C. The beads were washed sequentially with low-salt wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, pH 8.1, 150 mM NaCl), high-salt wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, pH 8.1, and 500 mM NaCl), LiCi wash buffer (0.25 M LiCl, 1% IGEPAL-CA630, 1 mM EDTA, 10 mM Tris-HCl, pH 8.1, and 1% deoxycholic acid), and TE butter. Precipitates were extracted and cross-linking was reversed with 1% SDS, 0.1 M NaHCO3, 200 µg/ml proteinase K by heating at 62°C for 2 h with shaking. The DNA mixture was purified using a PCR purification kit (QIAGEN). The purified DNA was used for real-time PCR with iTaq SYBR green supermix with ROX (Bio-Rad Laboratories). Primer sequences were as follows: Foxp3 promoter forward, 5′-GGATTATTAGAAGAGCGAGGTCTGC-3’; Foxp3 promoter reverse, 5′-ACTCGCTCACCTTGGTGAAGTG-3’; Foxp3 enhancer 1 forward, 5′-TGTTGGCTTCCAGTCTCCTT-3′; Foxp3 enhancer 1 reverse, 5′-TTGAGGCTAGGTTGTTCCAGA-3′; Actin forward, 5′-CCCGCGTGTCCCTCAA-3′; Actin reverse, 5′-TCACGTGGATATCGAGCACTTAA-3′.
Online supplemental material.
Fig. S1 shows data describing Smad2/3 phosphorylation. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20100004/DC1.
Acknowledgments
We thank Drs. W. Langdon for providing Cbl-b RING finger mutant knockin mice and R. Abe for providing anti-CD28 (PV-1).
This work is supported by grants from National Institutes of Health to Y-C. Liu.
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- iT reg cell
- induced T reg cell
- mTOR
- mammalian target of rapamycin
- nT reg cell
- naturally occuring T reg cell
- PI3K
- phosphoinositide 3-kinase
- T eff
- effector T cell
- T reg cell
- regulatory T cell
References
- Bachmaier K., Krawczyk C., Kozieradzki I., Kong Y.Y., Sasaki T., Oliveira-dos-Santos A., Mariathasan S., Bouchard D., Wakeham A., Itie A., et al. 2000. Negative regulation of lymphocyte activation and autoimmunity by the molecular adaptor Cbl-b. Nature. 403:211–216 10.1038/35003228 [DOI] [PubMed] [Google Scholar]
- Brunet A., Bonni A., Zigmond M.J., Lin M.Z., Juo P., Hu L.S., Anderson M.J., Arden K.C., Blenis J., Greenberg M.E. 1999. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 96:857–868 10.1016/S0092-8674(00)80595-4 [DOI] [PubMed] [Google Scholar]
- Bruno L., Mazzarella L., Hoogenkamp M., Hertweck A., Cobb B.S., Sauer S., Hadjur S., Leleu M., Naoe Y., Telfer J.C., et al. 2009. Runx proteins regulate Foxp3 expression. J. Exp. Med. 206:2329–2337 10.1084/jem.20090226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgering B.M. 2008. A brief introduction to FOXOlogy. Oncogene. 27:2258–2262 10.1038/onc.2008.29 [DOI] [PubMed] [Google Scholar]
- Castrillon D.H., Miao L., Kollipara R., Horner J.W., DePinho R.A. 2003. Suppression of ovarian follicle activation in mice by the transcription factor Foxo3a. Science. 301:215–218 10.1126/science.1086336 [DOI] [PubMed] [Google Scholar]
- Chen W., Jin W., Hardegen N., Lei K.J., Li L., Marinos N., McGrady G., Wahl S.M. 2003. Conversion of peripheral CD4+CD25- naive T cells to CD4+CD25+ regulatory T cells by TGF-beta induction of transcription factor Foxp3. J. Exp. Med. 198:1875–1886 10.1084/jem.20030152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang Y.J., Kole H.K., Brown K., Naramura M., Fukuhara S., Hu R.J., Jang I.K., Gutkind J.S., Shevach E., Gu H. 2000. Cbl-b regulates the CD28 dependence of T-cell activation. Nature. 403:216–220 10.1038/35003235 [DOI] [PubMed] [Google Scholar]
- Dejean A.S., Beisner D.R., Ch’en I.L., Kerdiles Y.M., Babour A., Arden K.C., Castrillon D.H., DePinho R.A., Hedrick S.M. 2009. Transcription factor Foxo3 controls the magnitude of T cell immune responses by modulating the function of dendritic cells. Nat. Immunol. 10:504–513 10.1038/ni.1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgoffe G.M., Kole T.P., Zheng Y., Zarek P.E., Matthews K.L., Xiao B., Worley P.F., Kozma S.C., Powell J.D. 2009. The mTOR kinase differentially regulates effector and regulatory T cell lineage commitment. Immunity. 30:832–844 10.1016/j.immuni.2009.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang D., Liu Y.-C. 2001. Proteolysis-independent regulation of PI3K by Cbl-b-mediated ubiquitination in T cells. Nat. Immunol. 2:870–875 10.1038/ni0901-870 [DOI] [PubMed] [Google Scholar]
- Floess S., Freyer J., Siewert C., Baron U., Olek S., Polansky J., Schlawe K., Chang H.D., Bopp T., Schmitt E., et al. 2007. Epigenetic control of the foxp3 locus in regulatory T cells. PLoS Biol. 5:e38 10.1371/journal.pbio.0050038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontenot J.D., Gavin M.A., Rudensky A.Y. 2003. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat. Immunol. 4:330–336 10.1038/ni904 [DOI] [PubMed] [Google Scholar]
- Guertin D.A., Stevens D.M., Thoreen C.C., Burds A.A., Kalaany N.Y., Moffat J., Brown M., Fitzgerald K.J., Sabatini D.M. 2006. Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCalpha, but not S6K1. Dev. Cell. 11:859–871 10.1016/j.devcel.2006.10.007 [DOI] [PubMed] [Google Scholar]
- Haxhinasto S., Mathis D., Benoist C. 2008. The AKT-mTOR axis regulates de novo differentiation of CD4+Foxp3+ cells. J. Exp. Med. 205:565–574 10.1084/jem.20071477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay N., Sonenberg N. 2004. Upstream and downstream of mTOR. Genes Dev. 18:1926–1945 10.1101/gad.1212704 [DOI] [PubMed] [Google Scholar]
- Heissmeyer V., Macián F., Im S.H., Varma R., Feske S., Venuprasad K., Gu H., Liu Y.C., Dustin M.L., Rao A. 2004. Calcineurin imposes T cell unresponsiveness through targeted proteolysis of signaling proteins. Nat. Immunol. 5:255–265 10.1038/ni1047 [DOI] [PubMed] [Google Scholar]
- Hori S., Nomura T., Sakaguchi S. 2003. Control of regulatory T cell development by the transcription factor Foxp3. Science. 299:1057–1061 10.1126/science.1079490 [DOI] [PubMed] [Google Scholar]
- Jeon M.S., Atfield A., Venuprasad K., Krawczyk C., Sarao R., Elly C., Yang C., Arya S., Bachmaier K., Su L., et al. 2004. Essential role of the E3 ubiquitin ligase Cbl-b in T cell anergy induction. Immunity. 21:167–177 10.1016/j.immuni.2004.07.013 [DOI] [PubMed] [Google Scholar]
- Kerdiles Y.M., Beisner D.R., Tinoco R., Dejean A.S., Castrillon D.H., DePinho R.A., Hedrick S.M. 2009. Foxo1 links homing and survival of naive T cells by regulating L-selectin, CCR7 and interleukin 7 receptor. Nat. Immunol. 10:176–184 10.1038/ni.1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khattri R., Cox T., Yasayko S.A., Ramsdell F. 2003. An essential role for Scurfin in CD4+CD25+ T regulatory cells. Nat. Immunol. 4:337–342 10.1038/ni909 [DOI] [PubMed] [Google Scholar]
- Kitoh A., Ono M., Naoe Y., Ohkura N., Yamaguchi T., Yaguchi H., Kitabayashi I., Tsukada T., Nomura T., Miyachi Y., et al. 2009. Indispensable role of the Runx1-Cbfbeta transcription complex for in vivo-suppressive function of FoxP3+ regulatory T cells. Immunity. 31:609–620 10.1016/j.immuni.2009.09.003 [DOI] [PubMed] [Google Scholar]
- Klunker S., Chong M.M., Mantel P.Y., Palomares O., Bassin C., Ziegler M., Rückert B., Meiler F., Akdis M., Littman D.R., Akdis C.A. 2009. Transcription factors RUNX1 and RUNX3 in the induction and suppressive function of Foxp3+ inducible regulatory T cells. J. Exp. Med. 206:2701–2715 10.1084/jem.20090596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojo S., Elly C., Harada Y., Langdon W.Y., Kronenberg M., Liu Y.C. 2009. Mechanisms of NKT cell anergy induction involve Cbl-b-promoted monoubiquitination of CARMA1. Proc. Natl. Acad. Sci. USA. 106:17847–17851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretschmer K., Apostolou I., Hawiger D., Khazaie K., Nussenzweig M.C., von Boehmer H. 2005. Inducing and expanding regulatory T cell populations by foreign antigen. Nat. Immunol. 6:1219–1227 10.1038/ni1265 [DOI] [PubMed] [Google Scholar]
- Lin L., Hron J.D., Peng S.L. 2004. Regulation of NF-kappaB, Th activation, and autoinflammation by the forkhead transcription factor Foxo3a. Immunity. 21:203–213 10.1016/j.immuni.2004.06.016 [DOI] [PubMed] [Google Scholar]
- Liu Y.C., Penninger J., Karin M. 2005. Immunity by ubiquitylation: a reversible process of modification. Nat. Rev. Immunol. 5:941–952 10.1038/nri1731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantel P.Y., Ouaked N., Rückert B., Karagiannidis C., Welz R., Blaser K., Schmidt-Weber C.B. 2006. Molecular mechanisms underlying FOXP3 induction in human T cells. J. Immunol. 176:3593–3602 [DOI] [PubMed] [Google Scholar]
- Ono M., Yaguchi H., Ohkura N., Kitabayashi I., Nagamura Y., Nomura T., Miyachi Y., Tsukada T., Sakaguchi S. 2007. Foxp3 controls regulatory T-cell function by interacting with AML1/Runx1. Nature. 446:685–689 10.1038/nature05673 [DOI] [PubMed] [Google Scholar]
- Ouyang W., Beckett O., Flavell R.A., Li M.O. 2009. An essential role of the Forkhead-box transcription factor Foxo1 in control of T cell homeostasis and tolerance. Immunity. 30:358–371 10.1016/j.immuni.2009.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik J.H., Kollipara R., Chu G., Ji H., Xiao Y., Ding Z., Miao L., Tothova Z., Horner J.W., Carrasco D.R., et al. 2007. FoxOs are lineage-restricted redundant tumor suppressors and regulate endothelial cell homeostasis. Cell. 128:309–323 10.1016/j.cell.2006.12.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudra D., Egawa T., Chong M.M., Treuting P., Littman D.R., Rudensky A.Y. 2009. Runx-CBFbeta complexes control expression of the transcription factor Foxp3 in regulatory T cells. Nat. Immunol. 10:1170–1177 10.1038/ni.1795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi S., Yamaguchi T., Nomura T., Ono M. 2008. Regulatory T cells and immune tolerance. Cell. 133:775–787 10.1016/j.cell.2008.05.009 [DOI] [PubMed] [Google Scholar]
- Sandri M., Sandri C., Gilbert A., Skurk C., Calabria E., Picard A., Walsh K., Schiaffino S., Lecker S.H., Goldberg A.L. 2004. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell. 117:399–412 10.1016/S0092-8674(04)00400-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer S., Bruno L., Hertweck A., Finlay D., Leleu M., Spivakov M., Knight Z.A., Cobb B.S., Cantrell D., O’Connor E., et al. 2008. T cell receptor signaling controls Foxp3 expression via PI3K, Akt, and mTOR. Proc. Natl. Acad. Sci. USA. 105:7797–7802 10.1073/pnas.0800928105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan X., Czar M.J., Bunnell S.C., Liu P., Liu Y., Schwartzberg P.L., Wange R.L. 2000. Deficiency of PTEN in Jurkat T cells causes constitutive localization of Itk to the plasma membrane and hyperresponsiveness to CD3 stimulation. Mol. Cell. Biol. 20:6945–6957 10.1128/MCB.20.18.6945-6957.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tone Y., Furuuchi K., Kojima Y., Tykocinski M.L., Greene M.I., Tone M. 2008. Smad3 and NFAT cooperate to induce Foxp3 expression through its enhancer. Nat. Immunol. 9:194–202 10.1038/ni1549 [DOI] [PubMed] [Google Scholar]
- Venuprasad K., Huang H., Harada Y., Elly C., Subramaniam M., Spelsberg T., Su J., Liu Y.C. 2008. The E3 ubiquitin ligase Itch regulates expression of transcription factor Foxp3 and airway inflammation by enhancing the function of transcription factor TIEG1. Nat. Immunol. 9:245–253 10.1038/ni1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Boehmer H., Nolting J. 2008. What turns on Foxp3? Nat. Immunol. 9:121–122 10.1038/ni0208-121 [DOI] [PubMed] [Google Scholar]
- Wohlfert E.A., Gorelik L., Mittler R., Flavell R.A., Clark R.B. 2006. Cutting edge: deficiency in the E3 ubiquitin ligase Cbl-b results in a multifunctional defect in T cell TGF-beta sensitivity in vitro and in vivo. J. Immunol. 176:1316–1320 [DOI] [PubMed] [Google Scholar]
- Wu Y., Borde M., Heissmeyer V., Feuerer M., Lapan A.D., Stroud J.C., Bates D.L., Guo L., Han A., Ziegler S.F., et al. 2006. FOXP3 controls regulatory T cell function through cooperation with NFAT. Cell. 126:375–387 10.1016/j.cell.2006.05.042 [DOI] [PubMed] [Google Scholar]