Abstract
Ranviruses (Iridoviridae) are increasingly associated with mortality events in amphibians, fish, and reptiles. They have been recently associated with mass mortality events in Brazilian farmed tadpoles of the American bullfrog Rana catesbeiana Shaw. 1802. The objectives of the present study were to further characterize the virus isolated from sick R. catesbeiana tadpoles and confirm the etiology in these outbreaks. Sick tadpoles were collected in 3 farms located in Goiás State, Brazil, from 2003 to 2005 and processed for virus isolation and characterization, microbiology, histopathology, and parasitology. The phylogenetic relationships of Rana catesbeiana ranavirus (RCV-BR) with other genus members was investigated by PCR with primers specific for the major capsid protein gene (MCP) and the RNA polymerase DNA-dependent gene (Pol II). Sequence analysis and multiple alignments for MCP products showed >99% amino acid identity with other ranaviruses, while Pol II products showed 100% identity. Further diagnostics of the pathology including histology and transmission electron microscopy confirmed the viral etiology of these mass deaths. As for as we know, this is the first report of a ranaviral infection affecting aquatic organisms in Brazil. Additionally, our results suggest that American bullfrogs may have served as a vector of transmission of this virus, which highlights the potential threat of amphibian translocation in the world distribution of pathogens.
Keywords: Iridovirus, FV3, Ranavirus, Rana catesbeiana
INTRODUCTION
Brazilian frog farming is a flourishing industry that mainly focuses on producing the American bullfrog Rana catesbiana Shaw, 1802. Ranaculture has been recently plagued by mass mortality events. These acute outbreaks were identified as a severe condition in tadpoles at early developmental stages and resulted in significant economic loses (Galli et al. 2006, Mazzoni 2006). The clinical signs of disease were apparently non-specific and common to several disorders. Nutritional, toxic, or management causes, associated or not with various genera and species of bacteria, have been suspected as possible etiologies (Hipólito & Bach 2002). However, viruses of the family Iridoviridae, genus Ranavirus have been increasingly signaled as possible etiological agents in both wild and cultured aquatic organisms (Williams et al. 2005).
Ranaviruses belong to the family Iridoviridae, which comprises 4 other genera: Lymphocystisvirus, Chloriridovirus, Iridovirus, and Megalocytivirus. Viral size ranges from 120 to 300 nm in diameter with icosahedrical symmetry (Webby & Kalmarkoff 1998). The genome is composed of a unique DNA double-stranded linear molecule with variable size ranging from 102 to 212 kbp depending on the species. The virus particle is formed by 3 structures: the external protein capsid, a polypeptide-lipid intermediate membrane, and a central nucleus containing a DNA-protein complex. The DNA is both circularly permuted and terminally redundant (Goorha & Murti 1982), leading to a genome map that is circular while the actual molecule is still linear (Houts el al. 1974) with >20% of its cytosine CpG sequences methylated (Willis & Granoff 1980). All iridovirus family members express the major capsid protein (MCP) of about 50 kDa, whose molecular properties have been extensively used for viral characterization and identification (Webby & Kalmarkoff 1998, Chinchar et al. 2005, Williams et al. 2005). Complete ranavirus MCP sequences are available from frog virus 3 (FV3) (Mao el al. 1996, Tan et al. 2004), tiger frog virus (He et al. 2002), Bohle iridovirus (Marsh el al. 2002), epizootic hematopoietic necrosis virus (Marsh et al. 2002), Ambystoma tigrinum virus (Jancovich et al. 2003), and Rana grylio virus (RGV 9506, RGV 9507, and RGV 9508; Zhang et al. 2006). Complete genome sequences from several members of the genus are also available, including FV3, the type species of the family (Tan et al 2004). Diseases produced by ranaviruses have been increasingly reported to affect 3 different taxonomic classes of cold-blooded vertebrates: teleost fish, amphibians, and reptiles. They are considered to be emerging pathogens that produce economic damage to commercially raised species and cause population declines in the wild (Williams et al. 2005, Fox et al. 2006). Ranaviruses were reported to be associated with mass mortality episodes affecting Rana grylio tadpoles (Zhang et al. 2001), R. tigrina rugulosa tadpoles (Weng et al. 2002), and R. catesbeiana tadpoles in the USA showing abdominal swelling (Majji et al. 2006).
Ranaviruses were suspected as etiological agents in mass mortalities affecting farmed tadpoles Rana catesbeiana in Brazil (Galli et al. 2006). Galli el al. (2006) reported high sequence identity with FV3 and other Ranavirus genus members for MCP and the immediately early protein (IE-ICP18) gene products, raising the question about the source and origin of the Brazilian virus isolate, given that bullfrog populations, signaled as endemic FV3 carriers (Wolf et al. 1968), were imported from North America in 1970 (Mathias & Scoll 2004), so a pathogen translocation through the bullfrog trade is possible. However, ranaviruses have been identified in almost all continents and previous reports in South America showed their presence in local species (Zupanovic et al. 1998, Fox et al. 2006).
The MCP gene has been used extensively to assess the phylogeny and taxonomy of iridoviruses. Based on the MCP sequence, Mao et al. (1997) have shown that iridoviruses isolated from the same geographic region were identical or highly similar; meanwhile those from different regions were genetically more distant. Similar results were reported when 30 iridoviruses from different geographic sites were analyzed (Hyatt et al. 2000). Examination of the entire MCP sequence allowed the differentiation of Australian, European, and American ranavirus isolates (Marsh el al. 2002) A phylogeographic study of ranaviruses from salamander Ambystoma tigrinum in the USA was based a the comparison of 514 MCP nucleotides located at the 5′ end (Jancovich et al. 2005). Do et al. (2005) analyzed 13 iridoviruses isolated from fish in Korea using the complete MCP sequences and showed that the viruses belong to the some species, which is consistent with the introduction of a single infectious agent. The DNA polymerase of many large DNA viruses has been also used to evaluate virus relationships (Hanson et al. 2006).
The goals of the present study were to isolate the virus from sick Rana catesbeiana tadpoles and determine whether the virus had an etiological role in mass mortality episodes. To study the phylogenetic relationships with FV3 and other Ranavirus members, we analyzed the complete MCP coding region as well as partial sequences from the RNA polymerase DNA-dependent polymerase II (Pol II).
MATERIALS AND METHODS
Tadpole samples
Five hundred sick tadpoles in Gosner Stage 25 (Gosner 1960) were collected from 3 farms located in Goiás State, Brazil (Fig. 1) from 2003 to 2005, one-third being used for virus isolation, one-third for PCR, and the rest being necropsied and used for bacteriology, parasitology, and histopathology (Table 1). Continuous gravitational flow of surface water was maintained in all ponds where tadpoles received 45% crude protein powder feed from different sources. Samples were taken from tanks when acute mass mortality events were reported within the first 24 h of onset. Records of anamnestic and epizootiological data were collected as well as clinical evaluations of sick tadpoles. Dead animals were not sampled to avoid post-mortem changes that might result in the wrong microbiological or histopathological conclusions.
Fig. 1.
Location of frog farms sampled in Goiás State. Brazil: 1: Fujioka; 2: Laranjeiras; 3; Dos Reis
Table 1.
Source of tadpoles, location information, and number of tadpoles analyzed from each farm. See Fig. 1 for farm locations
| Farm name | Location |
Sampled tadpoles (n) | |
|---|---|---|---|
| County | GPS coordinates | ||
| Fujioka | Hidrolândia | 17° 02′ 17.03″S 49° 12′ 46.94″ W |
250 |
| Laranjeiras | Gameleira | 16° 20′ 28.75″S 48° 44′ 19.63″ W |
200 |
| Dos Reis | Gameleira | 16° 24′ 15.24″ S 48° 46′ 03.70″ W |
50 |
Samples for virus isolation
Tadpoles were preserved in 95% ethanol when fixed at the farm or frozen at −20°C when processed at the laboratory. For virus isolation, frozen tadpoles were macerated with the addition of sterile amphibian phosphate-buffered saline (APBS, 6.6 g 1−1 NaCl, 1.15 g 1−1 Na2HPO4, and 0.2 g 1−1 KH2PO4; pH 7.5). The suspension was collected and distributed in 2 ml sterile tubes, freeze-thawed 3 times at −80°C, and followed by centrifugation at 1000 × g for 10 min at 4°C. The supernatant containing virus was filtered with 0.45 μm Millipore filters, and 1.0 ml aliquots were stored at −80°C until used or immediately inoculated in 80% confluent Xenopus sp. kidney A6 cells (Rafferty 1969) in 25 cm2 bottles. After a 45 min incubation, 5 ml culture medium was added (Dulbecco’s modified Eagle medium [DMEM; Gibco] with addition of penicillin-streptomycin and amphotericin). Control bottles were inoculated wilh 1.0 ml APBS. Cells were incubated at 25 to 27°C in a dark chamber and examined twice daily during 1 wk to verify the cytopathic effect.
PCR
DNA was obtained using the saline extraction method (Miller et al 1988). Briefly, whole tadpoles were homogenized in a pestle and mortar in sterile conditions, and 50 mg samples were diluted in 500 μl lysis buffer (50 mM Tris-HCl, pH 8.0; 50 mM EDTA, pH 8.0; 1 % SDS, 50 mM NaCl) and 50 μl lysozyme (10 mg ml−1), then finger-vortexed and left overnight al 55°C. Tubes were centrifuged (Eppendort Microcentrifuge 5415 D) at 12000 × g for 15 min, and 400 μl supernatant was mixed with 240 μl 5 M NaCl, finger-vortexed for 15 s, and centrifuged for 30 min at 12000 × g. The supernatant (450 μl) was transferred to a new tube, to which 400 μl 100% ethanol at −20°C was added, and mixed by gently inverting the tubes for 30 s. Tubes were left for at least 2 h at −20°C or at 4°C overnight for DNA precipitation. For pelleting, tubes were centrifuged at 12000 × g for 30 min, washed twice with 70% ethanol, centrifuged at 5000 × g for 5 min, and dried at room temperature. DNA was resuspended in 100 μl TE buffer (10 mM Tris-HCl, pH 8.0; 1 mM EDTA) at 55°C in a dry air chamber for 2 h.
IE-ICP 18 was used as the screening tool for primary detection. Positive samples were then PCR-amplified using primers specific for MCP and Pol II genes (Table 2). Optimal melting temperature for these primers was determined by a thermal-gradient PCR, testing from 50 to 65°C. For each reaction, the mixture used was 2.5 μl 10× PCR buffer (20 mM Tris-HCl. pH 8.4; 50 mM KCl). 200 μM dNTP, 2.5 mM MgCl2, 2.5 μM of each primer. 1 U of Taq DNA polymerase, 5 μl of DNA template (100 ng μl−1), and distilled water to complete 50 μl. Amplification conditions were adapted for each set of primers according to the melting temperature and length of the amplified fragment. Briefly, an initial cycle at 90°C for 1 min was followed by 40 cycles at 94°C for 1 min, 60°C for 1 min, 72°C for 1 min, and a final extension step at 72°C for 5 min. For complete MCP primers, the extension time was 90 s, and for Pol II 30 s. Amplification products were run in 1 % agarose gel, immersed in TAE buffer (40 mM Tris-acetate, 1 mM EDTA) stained with 0.5 μg ml−1 ethidium bromide and visualized with UV transilluminator.
Table 2.
Primers used for PCR analysis: MCPcomp (MCP gene complete coding region); Pol II (RNA polymerase DNA dependent-Pol II); IE-ICP 18 (immediate early protein-ICP 18)
Sequencing and sequence analysis
The PCR fragments were sequenced using the Big Dye® Terminator (v. 3.1) kit in an ABI-3100 fragment analyzer (Applied Biosystems) following the manufacturer’s instructions. MCP sequences were analyzed from both ends, combined with 4 internal primers to overlap partial fragments and obtain the whole sequence (forward primers 5′-CAC CAG CGA TCT CAT CAA CC-3′ and 5′-CGC AGT CAA GGC CTT GAT CT-3′ [Hyatt et al. 2000], and reverse primers 5′-GTC TCT GGA GAA GAA GAA-3′ [MCP5; Mao et al. 1996] and 5′-GTA ATT GGA GCC GAC GGA AGG-3′ [designed by the authors for the present analysis]). Pol II products were sequenced using specific primers shown in Table 2. All sequences were analyzed at least 6 times from both ends.
The DNA and the deduced amino acid sequences were compared with sequences in the GenBank/EMBL databases using the basic local alignment search tool (BLAST; www.ncbi.nlm.nih.gov/BLAST/). Sequences were aligned using CLUSTAL W (Thompson et al. 1994), and the phylogenetic relationships among species were determined using the neighbor-joining method (Saitou & Nei 1987) and parsimony, and maximum likelihood using MEGA4 (Tamura et al. 2007), Paup* 4.0b10 (Swofford 2002), and ModelTest (Posada & Crandall 1998) software. The tree was rooted using the lymphocystis disease virus as the external group and its reliability was inferred using the Felsenstein (1985) bootstrap method with 10 000 replicates.
Bacteriology
Tadpoles were collected, immediately euthanized with an overdose of tricaine methane sulphonate (MS222, Finquel®, Argent Chemical Laboratories) and immersed for 30 s in 0.2% peracetic acid solution to reduce external contamination. The liver, kidneys, and ascitic exudate were obtained aseptically using sterilized surgical instruments. Samples were inoculated and incubated in 4 different broths: brain heart infusion, triplicase soy broth, glucose azide, and selenite cystine (DIFCO Labs), prepared according to the manufacturer’s indications. All samples were incubated at 30°C for 24 to 72 h.
Histopathology
Whole tadpoles were preserved in 10% buffered formaline (vol/vol) and processed according to routine techniques. Paraffin-embedded blocks were sectioned in 4 to 5 μm sections and stained with hematoxylin and eosin (H&E) for primary validation. In a second stage, selected samples were stained using MacCallum-Goodpasture for bacterial detection. Fite for mycobacteria, and PAS for fungus (Luna 1968).
Transmission electron microscopy (TEM)
Material for TEM was selected from lesions identified on histopathology slides. Once the lesion was precisely located in the paraffin-embedded block, a 1 mm3 tissue sample was extracted for analysis. The retrieved tissue was deparaffined and fixed in 2.5% glutaraldehyde with the addition of phosphate buffer (0.1 M, pH 7.0) and later processed for resin inclusion and positive staining with uranyl acetate and lead citrate. Samples were observed in a Philips EM 208 TEM.
Parasitology
Wet mounts of skin were observed under an optical microscope for the parasite survey.
RESULTS
Epizootiology and clinical signs
The disease occurred throughout the year in all farms in the present survey, although the incidence and prevalence were higher during the rainy season (October to April). Clinical signs were observed in tadpoles in Gosner Stage 25 (Gosner 1960) ranging from 15 to 28 d old. Mass mortalities affected the whole tank population within the first 24 h. In a short period, all tanks harboring tadpoles within the same age range were affected, with an overall mortality between 95 and 100%. Newly hatched tadpoles did not show any sign of disease, and those in pre-metamorphic stages were not affected.
Typically, diseased tadpoles stopped feeding and were lethargic. They lost their swimming ability and remained at the bottom of the tank with erratic movements Abdominal distention was a frequent finding, but some affected tadpoles evidenced pale color and cachexia (Fig. 2). Skin lesions like ulcers or petechiae were not observed.
Fig. 2.

External gross lesions in Rana calesbeiana tadpoles with Ranavirus infection. In the center is a normal tadpole, flanked by 2 ascitic tadpoles with ‘heart-shaped body’ (left) and 2 emaciated tadpoles showing ‘arrow-shaped body’ (right). Scale bar = 2 cm
Cell culture, PCR, and sequence analysis
Xenopus sp. A6 cells inoculated with virus suspension obtained from sick tadpoles showed signs of cytopathic effects after 48 h incubation. A6 cells damaged by the virus first rounded up, and later contracted and elongated, some showing a star shape, and their cytoplasm became very granular with signs of apoptosis. Then cells detached from the layer (Fig. 3).
Fig. 3.
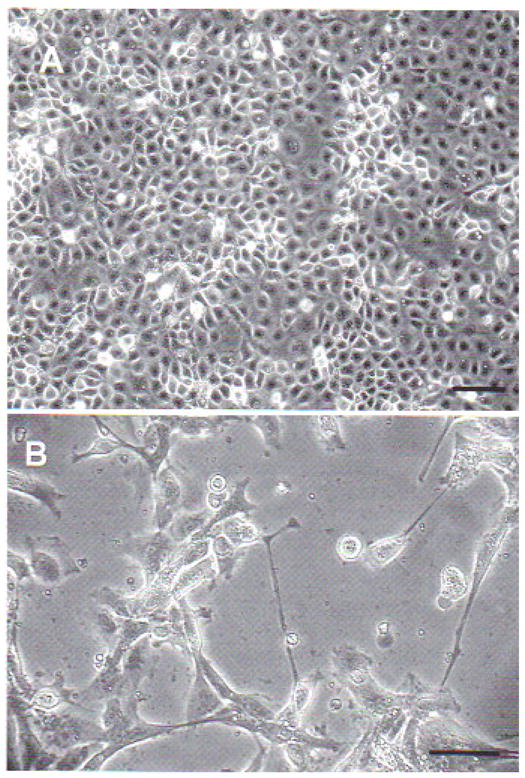
Xenopus sp. kidney A6 cells inoculated with viral suspension obtained from sick Rana catesbeiana tadpoles. (A) Mock-infected A6 cells; scale bar = 20 μm. (B) A6 cells with cytopathic effect 48 h after inoculation at 25 to 27°C; star-shaped and elongated cells are evidence of viral infection; scale bar = 10 μm
PCR on DNA extracted from sick tadpoles was positive for MCP and Pol II. The MCP gene primers yielded a specific fragment of about 1500 bp. Primers targeting the Pol II gene yielded a product of about 380 bp (Fig. 4).
Fig. 4.
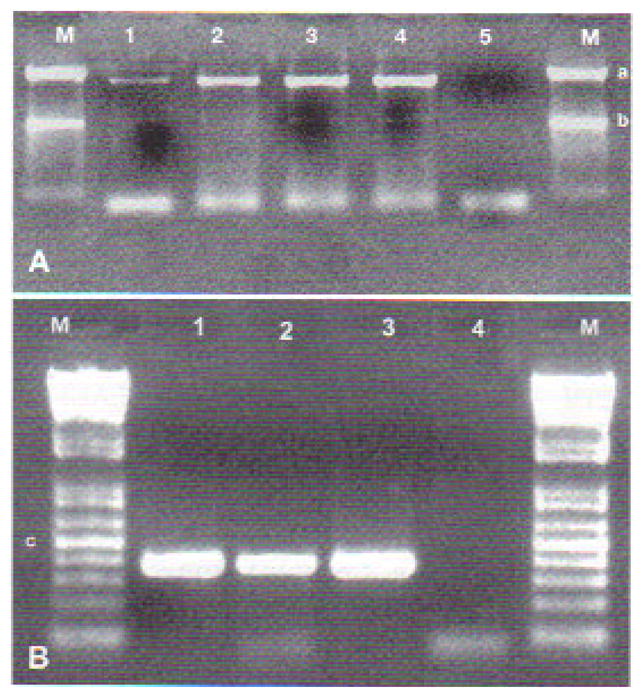
(A) Amplification products of the MCP gene from the 3 Brazilian Rana catesbeiana viral isolates (RCV-BR) (Lanes 1, 2, and 3) and frog virus 3 (Lane 4). Lane 5 is the negative control, and Lane M is the molecular marker (1 kb Plus DNA ladder; Invitrogen), where ‘a’ indicates 1500 bp and ‘b’ 1000 bp. (B) Amplification products of Pol II gene from the 3 RCV-BR isolates (Lanes 1, 2, and 3). Lane 4 is the negative control, and Lane M is the molecular marker (100 kb DNA ladder; Invitrogen), where ‘c’ indicates 500 bp. Purified virus genomic DNA was used as a template in PCR reactions described in ‘Materials and methods’. PCR products were separated on a 1 % agarose gel and visualized by staining with 0.5 μg ml1 ethidium bromide
To clarify the taxonomic position of our isolate, we cloned and sequenced the MCP and partial Pol II PCR fragments. Multiple alignments of MCP-deduced amino acid sequences showed the phylogenetic relationships of RCV-BR with FV3 and other ranaviruses (Fig. 5). High identities were obtained with published MCP sequences from anurans (Table 3). Multiple sequence alignments of the Pol II-deduced amino acid sequences showed 100% identity with FV3 (GenBank Accession No. AY548484), 98 % with tiger frog virus (GenBank Accession No. AF389451), and 96% with Regina ranavirus (GenBank Accession No. AF368230) and Ambystoma tigrinum stebbensi virus (GenBank Accession No. AY150217). Based on the alignments of MCP nucleotides, the neighbor-joining tree (Fig. 6) showed that selected ranaviruses and RCV-BR form a tight cluster of closely related viruses. Moreover, RCV-BR and FV3 are located in a single clade with high bootstrap values supporting this division. The same results were obtained for Pol II (data not shown).
Fig. 5.
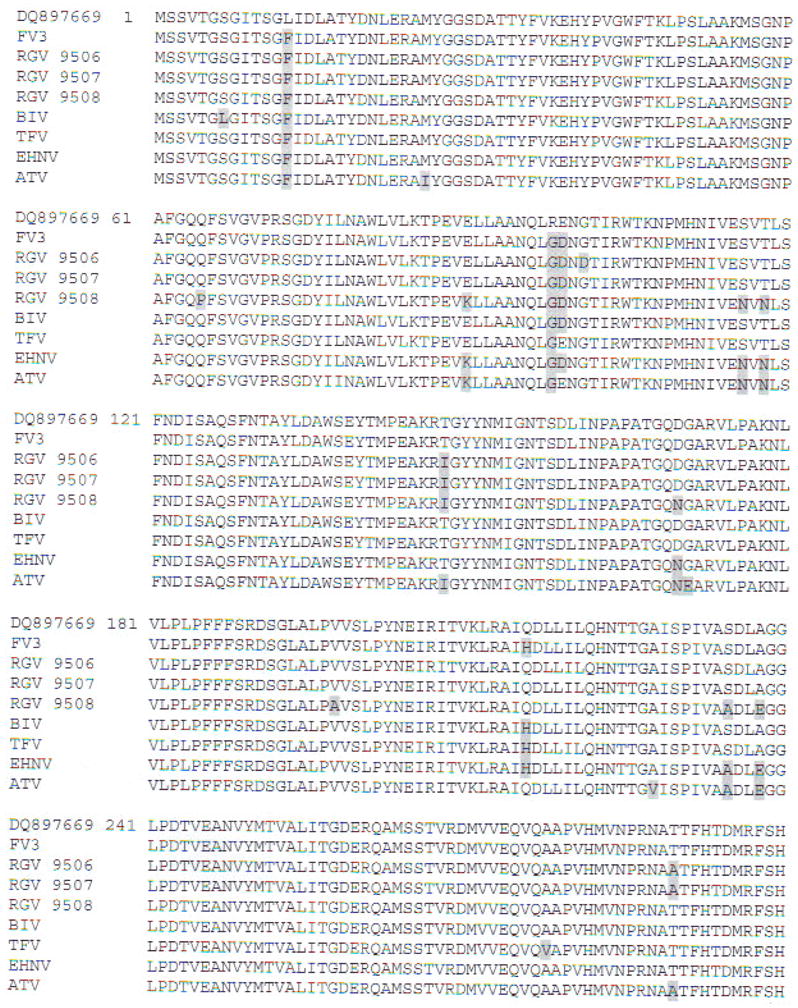

Multiple alignment of deduced amino acid sequence of MCP from the Brazilian Rana catesbeiana viral isolate (GenBank Accession No. DQ897669) along with other members of the genus Ranavirus: frog virus (FV3; GenBank Accession No. AY548484); Rana grylio virus (RGV 9806. 9807. 9808; Zhang et al. 2006); Bohle iridovirus (BIV; GenBank Accession No. AY 187046); tiger frog virus (TFV; GenBank Accession Nos. AY033630 and AF389451); epizootic haematopoietic necrosis virus (EHNV; GenBank Accession No. AY187045); and Ambystoma tigrinum virus (ATV; GenBank Accession No. AY50217). Amino acid residues differing from the Brazilian sequence are shaded
Table 3.
Identities obtained when the MCP nucleotide sequence from the Brazilian Rana catesbeiana viral isolate (GenBank Accession No. DQ897669) was aligned with published MCP sequences from anurans. ‘Source’ is GenBank Accession No. or literature reference
| Virus | % identity | Source |
|---|---|---|
| FV3 | 99.7 | AY548484 |
| RGV 9506 | 99 | Zhang et al. (2006) |
| RGV 9507 | 99 | Zhang et al. (2006) |
| Bohle iridovirus | 98 | AY187046 |
| Rana tigrina ranavirus | 98 | AY033630 |
| 98 | AP389451 | |
| RGV 9508 | 97 | Zhang et al. (2006) |
| Epizootic hematopoietic necrosis virus | 96 | AY187045 |
| Ambystoma tigrinum virus | 95 | AY150217 |
Fig. 6.
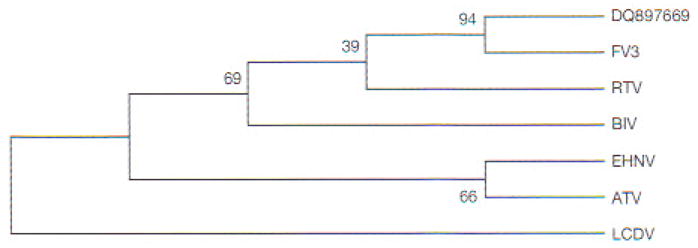
Phylogenetic analysis of MCP sequences. Neighbor-joining tree constructed using multiple alignment (and algorithms) presented in MEGA4 (Tamura et al. 2007). Degree of confidence for each branch point was determined by bootstrap analysis (10000 repetitions). For virus abbreviations, see Fig. 5. Lymphocystis disease virus (LCDV) used as external group from the genus Lymphocystisvirus
The RCV-BR MCP sequence was deposited to GenBank under Accession No. DQ897669.
Necropsy and histopathology
Autopsy of tadpoles with abdominal distension revealed abundant fluids filling the body cavity, with a pale-colored liver associated with congestive and slightly hemorrhagic organs. Cachectic tadpoles showed poor body conditions, including an absence of a fat body and pale coloration of the liver with congestive kidneys.
Histological analysis of infected tadpoles showed various degrees of tissue damage and cell necrosis over broad areas of tissue disruption. Lesions were prominent in the kidneys, liver, and spleen. The kidneys were the most affected organs, with large necrotic areas evidencing pyknosis and karyorrhexis associated with diffuse inflammatory infiltrate, mostly composed of mononuclear lymphocytes and eosinophils (Fig. 7a). The liver also showed necrosis with pyknosis and karyorrhexis, areas of steatosis, vacuolized hepatocytes, and mononuclear infiltrate (Fig. 7b). The spleen repeated the same pattern of necrosis and inflammatory infiltrate. Neither bacteria nor fungi were observed using specific histological detection techniques.
Fig. 7.
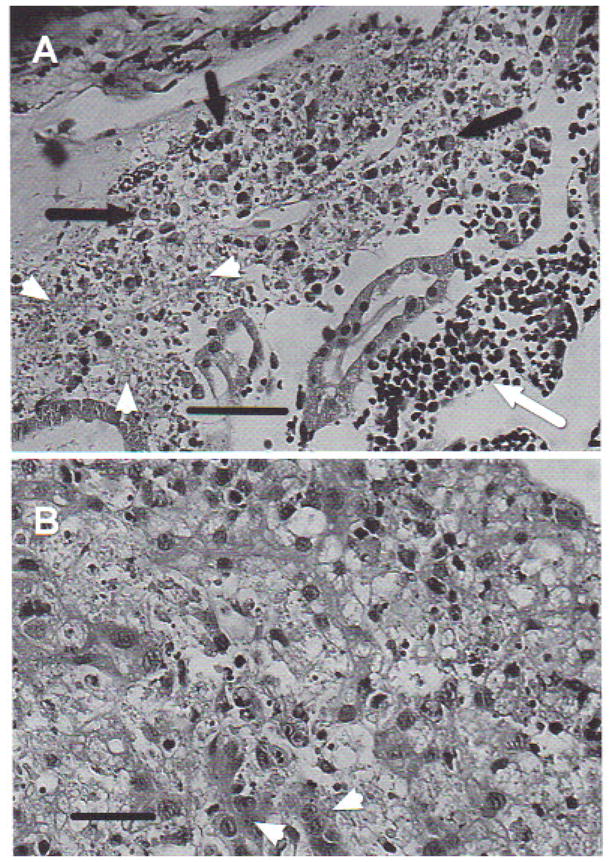
Rana catesbeiana tadpole. (A) Kidney showing lesions produced by Ranavirus. Complete loss of normal architecture. Cell necrosis with pyknosis and karyorrhexis (arrowheads), mononuclear lymphocyte infiltrate (white arrow), and eosinophilic infiltration (black arrows). H&E stain. Scale bar = 100 μm, (B) Liver with lesions produced by Ranavirus. Diffuse necrosis with pyknosis and karyorrhexis, areas of steatosis, vacuolized hepotocytes with loss of trabecular structure, and mononuclear infiltrate. Arrowheads indicate normal hepatocytes. H&E stain. Scale bar = 50 μm
TEM
Tissue samples of sick tadpoles obtained from formalin-fixed blocks and reprocessed for TEM analysis showed cell lyses, deformed nucleii with marginalized chromatin, absence of nuclear membrane, cytoplasm with empty or altered organelles, and mitochondria without crystals. Aggregates of iridovirus-like icosahedric particles in diverse assembly degrees were detected in kidney samples, confirming viral presence in areas of tissue damage (Fig. 8).
Fig. 8.
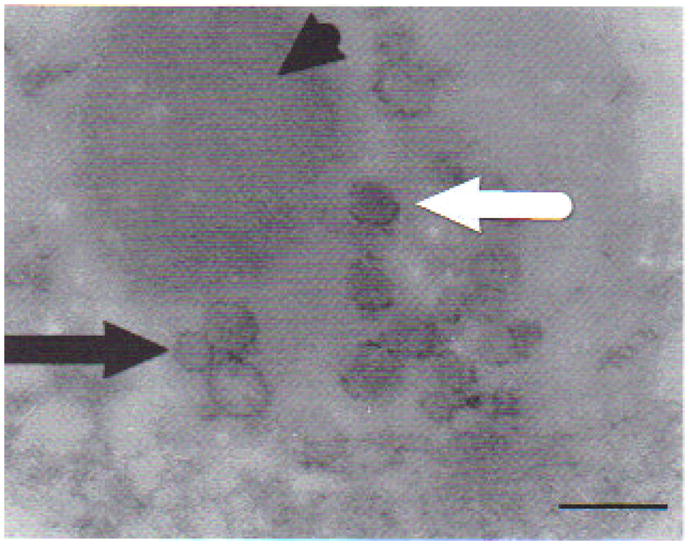
Transmission electron micrograph from kidney of affected Rana catesbeiana tadpole with signs of ascites. Samples were obtained from paraffin-embedded blocks after precisely locating lesions on histology slides. Complete (white arrow) and incomplete (black arrow) iridovirus-like particles. Viral particles seem to emerge from a finely granular, slightly electron-dense mass (arrowhead). Scale bar = 360 nm
Bacteriology and parasitology
Bacteriological analyses were negative for both tissue samples and ascitic exudates. Skin and gills samples showed several parasite species, of which Trichodina spp. and Oodinium spp. were the most common.
DISCUSSION
In the present study, we have identified a virus from the Ranavirus genus as the cause of mass mortality episodes in young Rana catesbeiana tadpoles in 3 Brazilian farms. Clinical and laboratory observations, including molecular characterization and TEM results, confirmed the viral etiology of these mass deaths. Histological examination of the tissue lesions indicated that bullfrog tadpoles died from viral injuries located mainly in the kidney, liver, and spleen. Bacteria were not associated with the disease, as tissue cultures and MacCallum-Goodpasture-stained histological slides were negative. Skin parasites were considered just an opportunistic invasion and not associated with clinical signs and lesions. Toxic causes, always suspected in acute syndromes, were discarded as each farm used different water sources and feed brands. The fact that different ponds were not affected simultaneously and that younger tadpoles remained healthy further ruled out this possibility.
The present evidence for a Ranavirus infection is consistent with the description of ‘abdominal distension disease’ in China, affecting farmed Rana grylio and R. tigrina rugulosa (Zhang et al. 2001, Weng et al. 2002).
Brazilian Ranavirus does not seem to be a great threat to Rana catesbeiana as farmed populations are well established, and disease outbreaks, despite their economic impact, were solved after biosecurity and good management practices were implemented. Environmental conditions within commercial frog farms should be the most important trigger for these outbreaks contributing to increased susceptibility to viral infections. This disease seems to be a typical epidemiological example of host–environment–pathogen interaction, where disease outbreaks and pathogen virulence enhancement are associated with environmental and management stressors. Similar considerations were quoted in reference to RGV pathogenesis in R. grylio with lethal syndrome (Zhang et al. 2001) and adult R. catesbeiana infections (Majji et al. 2006, Miller el al. 2007).
Ranviruses have been recognized as emerging pathogens, producing high mortalities in fish, reptiles, and amphibians; a minimally pathogenic virus in one species can cause serious diseases in others (Daszak et al. 1999. Chinchar 2002, Chinchar et al. 2005, Williams et al. 2005, Majji et al. 2006, Miller et al. 2007).
The high susceptibility of larval stages to ranaviruses has been established in Xenopus sp. (Gantress et al. 2003, Robert et al. 2005, Williams et al. 2005). This is consistent with the present results, as older tadpoles were not affected. However, others have reported that late-stage tadpoles and new metamorphs of other anuran species (e.g. Rana sylvatica and R. pipiens) appear to be more susceptible to Ranavirus infections (Green et al. 2002, Greer et al. 2005). This is not a contradiction, considering it may be an interplay between differential susceptibility of age classes, coupled with cycles of viral virulence and attenuation underlying this phenomenon, as well as the effect of Ranavirus infections often varying within host species (Daszak et al. 2003, Williams et al. 2005).
Wolf et al. (1968) isolated tadpole edema virus (TEV) from grossly edematous Rana catesbeiana and experimentally infected several amphibian species. Infected metamorphosing bullfrogs showed lethargy and edema prior to death. A generalized eosinophilia with marked infiltration in kidney tissues associated with necrosis and pyknotic nuclei was observed (Wolf et al. 1968). The similar increase in eosinophils found in the present study suggests that Ranavirus infection is associated with a pathophysiological host response involving an accumulation of eosinophils in infected tissues, at least in American bullfrogs. In mammals, eosinophils are pleiotropic multifunctional leukocytes involved in initiation and propagation of diverse inflammatory responses, as well as modulators of innate and adaptive immunity (Rothenberg & Hogan 2006). In addition to secreting various cytokines, they may participate in virus-induced immune responses by processing and presenting complex viral proteins and/or intact viral particles (Handzel et al. 1998). Whether eosinophils play a similar role in amphibian innate and adaptive immunity and how critical they are in amphibian anti-Ranavirus host response remains to be determined.
Sequence alignments of Pol II and MCP from RCV-BR showed 100 and 99.7% identity respectively with published FV3 sequences as well as high identities with other Ranavirus genus members. To determine the relationships among these viruses, we generated a neighbor-Joining tree (Saitou & Nei 1987) using the MCP sequence information in Fig. 5. The tree in Fig. 6 shows that all selected ranaviruses and RCV-BR form a tight cluster of closely related viruses. Moreover, RCV-BR and FV3 are located in a single clade, with high bootstrap values supporting this division. These results suggest that RCV-BR and FV3 are close relatives or perhaps strains of the same virus.
The highly conserved MCP is diverse enough to be used to distinguish among closely related isolates (Tidona et al. 1998). Do et al. (2005) analyzed 13 isolates of flounder (Paralichthys olivaceus) iridoviruses (FLIVs); MCP-deduced amino acid sequences of the 13 FLIVs showed 97 to 100% amino acid sequence identity to each other, suggesting that all 13 isolates have a single origin. Jancovich et al. (2005) concluded that 18 geographically widespread Ranavirus isolates in the USA corresponded to a single introduction, after determining genetic divergences of <1 % for MCP sequences.
Studies performed with Iridoviridae showed that viruses isolated from the same geographic region were genetically similar, whereas those from different areas were distinct (Mao et al. 1997, Hyatt et al. 2000, Marsh et al. 2002). FV3-like viruses were reported outside North America in Rana temporaria from the UK (Hyatt et al. 2000) and R. grylio viruses isolated in China (Zhang et al. 2001), as a consequence of amphibian translocations. TEV isolated from R. catesbeiana in North America was signaled as being endemic to this species (Wolf et al. 1968) and could be a strain of FV3 (Essani & Granoff 1989). The present results along with IE-ICP 18 identity reported by Galli et al. (2006) suggest that American bullfrogs may have carried the virus to Brazil, illustrating the key role of amphibian translocation in the world distribution of pathogens and particularly FV3. The minimum genetic differences between RCV-BR and FV3, located in 2 geographically distant places, could be explained by DNA viruses characteristically expressing mechanisms of co-divergence between host and virus, determined by vicariance, in association to persistent infections by sexual or vertical transmission (Holmes 2004).
Nevertheless, ranaviruses with high identities to FV3 within partial MCP sequences have been also found in Venezuela (Zupanovic et al. 1998) and Argentina (Fox et al. 2006), suggesting the possibility of on autochthonous species.
In agreement with Goldberg et al. (2003), it is necessary to continue studies on protein profiles using amplified fragment length polymorphism (AFLP) and to obtain the complete RCV-BR genome sequence to determine, with accuracy, similarities among these closely related viruses.
In conclusion, viruses of family Iridoviridae, genus Ranavirus are pathogenic to young Rana catesbeiana tadpoles, and likely the cause of mass mortality commonly observed on Brazilian frog farms. As for as we know, this is the first report of Ranavirus affecting aquatic organisms in Brazil. Given that ranaviruses are now ‘notifiable’ diseases according to the World Organization for Animal Health (OIE), this is of high relevance for frog farming and aquaculture industry and export in the USA. Although the Brazilian Ranavirus may become pathogenic for bullfrogs only under stressing conditions, it can be potentially virulent for other South American amphibians, fish, or reptiles. We believe that Ranavirus-like disease exemplifies the problems likely to arise with the intensive culturing of new species. More systematic animal husbandry including defined disease-control strategies will be required for long-term economically successful aquaculture industries. Further basic and applied research will be needed to answer questions raised in the present study, such as viral origin and the hazards associated with disease translocation and amphibian trade.
Acknowledgments
This work was performed with the financial support of Programa de Desarrollo Tecnológico-Dirección Nacional de Ciencia y Tecnología (Ministerio de Educación y Cultura, Uruguay) and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (Ministerio da Educação, Brazil), J.R. and H.M. were supported by NIH grant AI-2R24, AI059830-06. We thank S. Q. P. de Mesquita, U. N. Rauecker, and M. Hipólito for their excellent technical assistance. Fig. 1 was prepared by F. Mazzoni.
LITERATURE CITED
- Chinchar VG. Ranaviruses (family Iridoviridae); emerging cold-blooded killers. Arch Virol. 2002;147:447–470. doi: 10.1007/s007050200000. [DOI] [PubMed] [Google Scholar]
- Chinchar VG, Essbauer S, He JG, Hyatt AD, Miyazaki T, Seligy V, Williams T. Iridoviridae. In: Fauquet CM, Mayo MA, Manilott J, Desselberger U, Ball LA, editors. Virus taxonomy: 8th report of the International Committee on the Taxonomy of Viruses. Elsevier; London: 2005. pp. 163–175. [Google Scholar]
- Daszak P, Berger L, Cunningham AA, Hyatt AD, Green DE, Speare R. Emerging infectious diseases and amphibian population declines. Emerg Infect Dis. 1999;5:735–748. doi: 10.3201/eid0506.990601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daszak P, Cunningham AA, Hyatt AD. Infectious diseases and amphibian population declines. Divers Distrib. 2003;9:141–150. doi: 10.3201/eid0506.990601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do J, Cha S, Kim J, An E, et al. Phylogenetic analysis of the major capsid protein gene of iridovirus isolates from cultured flounders Paralichthys olivaceus in Korea. Dis Aquat Org. 2005;64:193–200. doi: 10.3354/dao064193. [DOI] [PubMed] [Google Scholar]
- Essani K, Granoff A. Amphibian and piscine iridoviruses proposal for nomenclature and taxonomy based on molecular and biological properties. Intervirology. 1989;30:187–193. doi: 10.1159/000150092. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Phylogenies and the comparative method. Am Nat. 1985;125:1–15. [Google Scholar]
- Fox SF, Greer AL, Torres-Cervantes R, Collins JP. First case of ranavirus-associated morbidity and mortality in natural populations of the South American frog Atelognathus patagonicus. Dis Aquat Org. 2006;72:87–92. doi: 10.3354/dao072087. [DOI] [PubMed] [Google Scholar]
- Galli L, Pereira A, Marquez A, Mazzoni R. Ranavirus detection by PCR in cultured ladpoles (Rana catesbeiana Shaw 1802) from South America. Aquaculture. 2006;257:78–82. [Google Scholar]
- Gantress J, Maniero G, Cohen N, Robert J. Development and characterization of a model system to study amphibian immune responses to iridoviruses. Virology. 2003;311:254–262. doi: 10.1016/s0042-6822(03)00151-x. [DOI] [PubMed] [Google Scholar]
- Goldberg T, Coleman D, Gram E, Inendino K, Philipp D. Strain variation in an emerging iridovirus of warm-water fishes. J Virol. 2003;77:8812–8818. doi: 10.1128/JVI.77.16.8812-8818.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goorha R, Murti KG. The genome of frog virus 3, an animal DNA virus, is circularly permuted and terminally redundant. Proc Natl Acad Sri USA. 1982;79:248–252. doi: 10.1073/pnas.79.2.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosner K. A simplified table for Staging anuran embryos and larva with notes on identification. Herpetologica. 1960;16:183–190. [Google Scholar]
- Green DE, Converse KA, Schrader AK. Epizootiology of sixty-four amphibian morbidity and mortality events in the USA, 1996–2001. Ann NY Acad Sci. 2002;969:323–339. doi: 10.1111/j.1749-6632.2002.tb04400.x. [DOI] [PubMed] [Google Scholar]
- Greer AL, Berrill M, Wilson PJ. Five amphibian mortality events associated with ranavirus infection in south central Ontario, Canada. Dis Aquat Org. 2005;67:9–14. doi: 10.3354/dao067009. [DOI] [PubMed] [Google Scholar]
- Handzel ZT, Busse WW, Sedgwick JB, Vrtis R, Lee WM, Kelly EAB, Gern JE. Eosinophils bind rhinovirus and activate virus-specific T cells. J Immunol. 1998;160:1279–1284. [PubMed] [Google Scholar]
- Hanson LA, Rudis MR, Vasquez-Lee M, Montgomery RD. A broadly applicable method to characterize large DNA viruses and adenoviruses based on the DNA polymerase gene. Virol J. 2006;3:28. doi: 10.1186/1743-422X-3-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He JG, Lu L, Deny M, He HH, et al. Sequence analysis of the complete genome of an iridovirus isolated from the tiger frog. Virology. 2002;292:185–197. doi: 10.1006/viro.2001.1245. [DOI] [PubMed] [Google Scholar]
- Hipólito M, Bach EE. Patologias em rã-touro (Rana catesbeiana, Shaw, 1802). Primeira revisão da bibliografia brasileira. Arq Inst Biol (Sao Paulo) 2002;69:113–120. [Google Scholar]
- Holmes EC. The phylogeography of human viruses. Mol Ecol. 2004;13:745–756. doi: 10.1046/j.1365-294x.2003.02051.x. [DOI] [PubMed] [Google Scholar]
- Houts GE, Gravell M, Granoff A. Electron microscopic observations on early events of frog virus 3 replication. Virology. 1974;58:589–594. doi: 10.1016/0042-6822(74)90093-2. [DOI] [PubMed] [Google Scholar]
- Hyatt AD, Gould AR, Zupanovic Z, Cunningham AA, et al. Comparative studies of piscine and amphibian iridoviruses. Arch Virol. 2000;145:301–331. doi: 10.1007/s007050050025. [DOI] [PubMed] [Google Scholar]
- Jancovich JK, Mao J, Chinchar VG, Wyatt C, et al. Genomic sequence of a ranavirus (family Iridoviridae) associated with salamander mortalities in North America. Virology. 2003;316:90–103. doi: 10.1016/j.virol.2003.08.001. [DOI] [PubMed] [Google Scholar]
- Jancovich JK, Davidson EW, Parameswaran N, Mao J, et al. Evidence for emergence of an amphibian iridoviral disease because of human-enhanced spread. Mol Ecol. 2005;14:213–224. doi: 10.1111/j.1365-294X.2004.02387.x. [DOI] [PubMed] [Google Scholar]
- Luna LG. Manual of histology staining methods of the Armed Forces Institute of Pathology. 3. McGraw-Hill; New York: 1968. [Google Scholar]
- Majji S, LaPatra S, Long SM, Sample R, Bryan L, Sinning A, Chinchar VG. Rana catesbeiana virus Z, (RCV-Z): a novel pathogenic ranavirus. Dis Aquat Org. 2006;73:1–11. doi: 10.3354/dao073001. [DOI] [PubMed] [Google Scholar]
- Mao J, Tham TN, Gentry GA, Aubertin A, Chinchar VG. Cloning, sequence analysis, and expression of the major capsid protein of the iridovirus frog virus 3. Virology. 1996;216:431–436. doi: 10.1006/viro.1996.0080. [DOI] [PubMed] [Google Scholar]
- Mao J, Hedrick RP, Chinchar VG. Molecular characterization, sequence analysis, and taxonomic position of newly isolated fish iridoviruses. Virology. 1997;229:212–220. doi: 10.1006/viro.1996.8435. [DOI] [PubMed] [Google Scholar]
- Marsh IB, Whittington BJ, O’Rourke B, Hyatt AD, Chisholm O. Rapid differentiation of Australian, European, and American ranaviruses based on variation in major capsid protein gene sequence. Mol Cell Probes. 2002;16:137–151. doi: 10.1006/mcpr.2001.0400. [DOI] [PubMed] [Google Scholar]
- Mathias M, Scott P. Série Agronegócios. SEBRAE/RJ; Rio de Janeiro: 2004. Potencial produtivo de criação de rãs no Estado do Rio de Janeiro. [Google Scholar]
- Mazzoni R. PhD thesis. Universidade Federal de Goiás; Goiânia: 2006. Infecções por Ranavirus e cocos Gram-positivos em girinos e rãs de criação (Rana catesbeiana Shaw, 1802) do estado de Goiás. [Google Scholar]
- Miller SA, Dykes DA, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DL, Rajeev S, Gray MJ, Baldwin CA. Frog virus 3 infection, cultured American bullfrogs. Emerg Infect Dis. 2007;13:342–343. doi: 10.3201/eid1302.061073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D, Crandall KA. ModelTest: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Rafferty KA. Mass culture of amphibian cells, methods and observations concerning stability of cell type. In: Mizell M, editor. Biology of amphibian tumors. Springer Verlag; New York: 1969. pp. 52–81. [Google Scholar]
- Robert J, Morales H, Buck W, Cohen A, Marr S, Gantress J. Adaptive immunity and histopathology in frog virus 3-infected Xenopus. Virology. 2005;332:667–675. doi: 10.1016/j.virol.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Rothenberg ME, Hogan SP. The eosinophil. Annu Rev Immunol. 2006;24:147–174. doi: 10.1146/annurev.immunol.24.021605.090720. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Swofford DL. Phylogenetic analysis using parsimony (* and other methods) Sinauer Associates; Sunderland, MA: 2002. PAUP*. [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Tan WGH, Barkman TJ, Chinchar VG, Essani K. Comparative genomic analyses of frog virus 3, type species of the genus Ranavirus (family Iridoviridae) Virology. 2004;323:70–84. doi: 10.1016/j.virol.2004.02.019. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTALW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tidona CA, Schnitzler P, Kehm R, Darai G. Is the major capsid protein of iridoviruses a suitable target for the study of viral evolution? Virus Genes. 1998;16:59–66. doi: 10.1023/a:1007949710031. [DOI] [PubMed] [Google Scholar]
- Webby R, Kalmarkoff J. Sequence comparison of the major capsid protein gene from 18 diverse iridoviruses. Arch Virol. 1998;143:1949–1966. doi: 10.1007/s007050050432. [DOI] [PubMed] [Google Scholar]
- Weng SP, He JG, Wang XH, Lu L, Deng M, Chan SM. Outbreaks of an iridovirus disease in cultured tiger frog, Rana tigrina rugulosa, in southern China. J Fish Dis. 2002;25:423–427. [Google Scholar]
- Williams T, Barbosa-Solomieu V, Chinchar VG. A decade of advances in iridovirus research. Adv Virus Res. 2005;65:173–248. doi: 10.1016/S0065-3527(05)65006-3. [DOI] [PubMed] [Google Scholar]
- Willis DB, Granoff A. Frog virus 3 DNA is heavily methylated at CpG sequences. Virology. 1980;107:250–257. doi: 10.1016/0042-6822(80)90290-1. [DOI] [PubMed] [Google Scholar]
- Wolf K, Bullock G, Dunbar C, Quimby M. Tadpole edema virus: a viscerotropic pathogen for anuran amphibians. J Infect Dis. 1968;118:253–262. doi: 10.1093/infdis/118.3.253. [DOI] [PubMed] [Google Scholar]
- Zhang QY, Xiao F, Li ZQ, Gui JF, Mao JH, Chinchar VG. Characterization of an iridovirus from the cultured pig frog Rana grylio with lethal syndrome. Dis Aquat Org. 2001;48:27–36. doi: 10.3354/dao048027. [DOI] [PubMed] [Google Scholar]
- Zhang QY, Zhe Z, Xiao F, Li ZQ, Gui JF. Molecular Characterization of three Rana grylio virus (RGV) isolates and Paralichthys olivaceus lymphocystis disease virus (LCDV-C) in iridoviruses. Aquaculture. 2006;251:1–10. [Google Scholar]
- Zupanovic Z, Musso C, López G, Loureiro CL, Hyatt AD, Hengstberger S, Robinson AJ. Isolation and characterization of iridoviruses from the giant toad Bufo marinus in Venezuela. Dis Aquat Org. 1998;33:1–9. doi: 10.3354/dao033001. [DOI] [PubMed] [Google Scholar]


