Abstract
Recent proteomics studies on human plasma high-density lipoprotein (HDL) have discovered up to 50 individual protein constituents. Many of these have known functions that vary surprisingly from the lipid transport roles commonly thought to mediate HDL’s ability to protect from coronary artery disease. Given newly discovered roles in inflammation, protease inhibition, complement regulation, and innate immunity, many have begun to view HDL as a broad collection of distinct particle subfamilies, each distinguished by unique protein compositions and functions. Herein we review recent applications of high-resolution proteomics to HDL and summarize evidence supporting the idea of HDL functional subspeciation. These studies have set the stage for a more complete understanding of the molecular basis of HDL functional heterogeneity and hold promise for the identification of new biomarkers that can predict disease or evaluate the success of clinical interventions.
Keywords: High density lipoprotein, Proteomics, Mass spectrometry, Lipoprotein, Apolipoprotein, Reverse cholesterol transport, Cardiovascular disease, Protein
Introduction
Epidemiologic studies dating back to the 1960s have shown that increased plasma high-density lipoprotein (HDL) cholesterol levels are a powerful negative risk factor for development of coronary artery disease (CAD) in a given population. HDL is a blood-borne assembly of amphipathic proteins (~50% by mass) that stabilize lipid emulsions composed of phospholipids (~25%), cholesterol (~4%), triglycerides (~3%), and cholesteryl esters (~12%). In addition to structural stability, these proteins (termed apolipoproteins) impart biological directionality to the lipid cargo by 1) targeting it to various tissues, 2) modifying its chemical form (ie, lipolysis or esterification), or 3) transferring it to other lipoproteins. Roughly 65% of HDL protein mass is comprised of apolipoprotein (apo)A-I with another 15% by apoA-II. The remainder includes around 50 proteins that are each too low in abundance to be present on all circulating HDL particles. The potential for differential accumulation of these proteins into certain subspecies likely drives the well-known polydispersity of HDL.
Such compositional variability suggests that HDL mediates numerous and highly diverse biological roles, an assertion borne out by hundreds of functional studies. Its most celebrated function is the shuttling of cholesterol from the periphery to the liver for catabolism/excretion during the process of reverse cholesterol transport (RCT). This is distinguished from the “forward” delivery of cholesterol from the liver to the periphery via the low-density lipoprotein (LDL) pathway. HDL and many of its apolipoproteins can promote the efflux of cholesterol from peripheral cells via a number of mechanisms [1] and can deliver cholesteryl esters to the liver in the process of selective uptake [2]. In vivo models of RCT show that genetic lowering of plasma HDL decreases the appearance of macrophage-derived cholesterol in the feces [3]. HDL has also been documented to prevent oxidative modification of LDL, which may reduce macrophage foam cell generation in the vessel wall—the hallmark of the developing atherosclerotic lesion [4]. Aside from lipid transport, HDL has anti-inflammatory traits. HDL can inhibit 1) the expression of cell adhesion molecules on endothelial cells, which sequester circulating monocytes during injury [5], and 2) the activity of macrophage chemotactic factor 1, which signals the infiltration of surface-adhered monocytes into the vessel wall [6]. In rabbits fitted with carotid periarterial collars, HDL administration resulted in 40% reductions in vascular cell adhesion molecule-1 expression and monocyte infiltration within 1 week [7]. There are many more examples of HDL mediation of vessel wall biology (for a review, see Rye et al. [8]). Intriguingly, HDL also appears to play important roles in host defense. It has been shown to be a major bacteriocidic factor in several species of fish [9]. In humans, it can deactivate particular oxidized phospholipids accumulating in macrophages that have been infected with Mycobacterium leprae, the organism that causes leprosy [10]. Recent work has also demonstrated that HDL can play a highly intriguing role in neutralizing the protozoan Trypanosoma brucei (discussed below).
Given the compositional heterogeneity of HDL on the one hand and its functional pleiotropy on the other, many are adopting the view that HDL refers to a broad collection of distinct particle subfamilies with unique protein compositions that perform distinct functions. In this context, a key challenge is to identify these subfamilies and determine their individual functions. This review focuses on recent progress in the separation and proteomic analyses of HDL, as well as evidence for its functional subspeciation. The reader should be aware of other excellent reviews and commentaries on related issues [11•, 12•, 13].
How is HDL and its Subfractions Defined?
In the basic science laboratory and the clinic, HDL is referred to by a complex and sometimes confusing set of definitions. Numerous modes of HDL separation from plasma have led to separate nomenclature systems that center on a specific physicochemical or immunological property. For example, HDL has been classically defined by its hydrated density (driven by the ratio of protein to lipid in the particles) as separated by ultracentrifugation. In humans, it is typically found as two major forms: HDL2 (density of 1.063–1.125 g/mL) and HDL3 (density of 1.125–1.210 g/mL), with diameters ranging from 70 to 120 Å. These have been further refined to five subspecies, HDL2(b, a) and HDL3(a, b, c), using tighter density cuts [14]. Given the method’s ease and wide use, the density-based nomenclature has been the most generally adopted. However, HDL has also been classified by major apolipoprotein content using immunoaffinity chromatography into apoA-I–containing particles that lack apoA-II (LpA-I) and those that contain both apoA-I and apoA-II (LpA-I/A-II) [15]. By contrast, agarose gel electrophoresis can classify HDL by charge density into α, pre-α, or pre-β forms, depending on the degree of negative charge [16]. In the clinic, human plasma is commonly treated with phospho-tungstic acid or heparin and magnesium chloride to precipitate the apoB-containing lipoproteins LDL and very low-density lipoprotein (VLDL) [17]. The precipitated lipoproteins are pelleted, and cholesterol in the supernate is referred to as HDL cholesterol (HDL-C). It is this HDL-C measurement that underlies the majority of epidemiological studies illustrating the inverse association with CAD. Other physical properties, such as molecular size (gel filtration chromatography) [18] and ionic character (ion exchange chromatography) [19], have been exploited for lipoprotein lipid analyses but have not yet been widely used for proteomics due to HDL coelution with unrelated high abundance plasma proteins.
Attempts to subclassify HDL have been driven by the promise that a better understanding of HDL subspeciation will yield a more detailed knowledge of its metabolism and perhaps predictive biomarkers for CAD. However, the functional basis for the high compositional heterogeneity of HDL has largely remained a mystery. One reason for this is that there is little concordance between the different HDL separation methodologies. For example, a sample of LpA-I as isolated by immunoaffinity chromatography contains particles of HDL2 and HDL3 density as well as α and pre-β electrophoretic species and vice versa. Thus, functional characterizations of HDL subspecies tend to be framed only within the context of the method used to isolate them. An HDL subfraction isolated by a particular method can be considered a true subfraction only in the physicochemical sense, not necessarily the functional sense. Another hurdle has been that functional conclusions about a given HDL subfraction can be compromised by any artifacts inherent to the method used to isolate it. It is increasingly clear that the high shear forces and ionic strengths experienced during potassium bromide (KBr) density ultracentrifugation strip off or redistribute certain proteins. For example, van’t Hooft and Havel [20] showed that ultracentrifugally isolated HDL interacted more avidly to the liver apoE receptor than gel-filtered HDL because of altered apoE content or depletion of other apolipoproteins. Another study showed that the use of deuterium oxide and sucrose in ultracentrifugation separations resulted in an altered HDL proteome compared with the classic salt-based method [21•]. Thus, KBr centrifugation may favor “core proteins” that survive the spins, but more transiently associated proteins—which may serve important biological functions—may be underestimated, found in nonphysiologic locations, or missed completely. Immunoaffinity isolation of lipoproteins holds the promise of more gentle separation conditions than ultracentrifugation. However, this approach has its own set of problems, chiefly the bias introduced by the specificity of the antibodies used in the separation. Given its abundance, many have been tempted to define HDL by the presence of apoA-I. Unfortunately, apoA-I is also present on LDL- and VLDL-sized particles [22], and lipidated particles that cofractionate with HDL have been shown to lack both apoA-I and apoA-II [23]. Thus, immunoprecipitation techniques that target a single protein may not cast a wide enough net for capturing all HDL particles. Finally, despite being epidemiologically predictive, the clinical HDL-C measurement fails to take into account the protein complement or any particle subspeciation within HDL. It is not clear exactly which HDL subspecies are captured by the precipitation techniques. For these reasons, prominent clinicians such as Movva and Rader [24] have argued that the HDL-C paradigm is “insufficient to capture the functional variation in HDL particles.”
Thus, from the viewpoint of understanding the function of individual particles within the HDL milieu, our current definitions of HDL may fall short. So what is the best way to define HDL? It may be that a more holistic view of HDL is warranted, one based on composition rather than method of separation. One universal constituent of all HDL subfractions is phospholipid. In cells, a critical role of the plasma membrane is to act as a two-dimensional solvent for the concentration of phospholipid-bound proteins and assembly into productive complexes. Perhaps phospholipids play an analogous role in circulation by acting as an organizing center for the assembly of extracellular lipid-associating proteins. When most small exchangeable amphipathic proteins interact with phospholipids, the resulting complexes usually fall into the size and density range of classically defined HDL [25]. Thus, HDL probably contains a host of protein/lipid complexes of defined composition that are as yet uncharacterized (and may not contain apoA-I at all) but may still carry out important biological functions. Given this, it may be useful to think of HDL in terms of “phospholipid-rich lipoproteins” (PL-rich LPs). This definition distinguishes from chylomicrons/ VLDL (triglyceride-rich lipoproteins) and LDL (cholesteryl ester-rich lipoproteins) and frees them from traditional apoA-I–centric or density-centric preconceptions that accompany the term “HDL.” Circulating phospholipid may be the platform upon which specific protein–protein or protein–lipid interactions drive the formation of distinct particles with distinct functions.
Applications of Modern Proteomics to HDL
Since HDL was first isolated by ultracentrifugation more than 60 years ago, hundreds of biochemical and immunological studies have been directed at characterizing its protein constituents. Prior to 2005, HDL was generally thought to contain proteins in four major functional groups: 1) proteins associated with lipid transport or lipoprotein integrity (ie, the “apos”: apoA-I, apoA-II, apoCs, apoD, apoF, apoM, apoE, etc.); 2) lipolytic enzymes such as lecithin cholesterol acyltransferase (LCAT), paraoxonase (PON), and platelet-activating factor aryl hydrolase; 3) lipid transfer proteins, including cholesteryl ester transfer protein (CETP) and phospholipid transfer protein (PLTP); and 4) acute phase response proteins such as serum amyloid A (SAA), clusterin, and apoA-IV. The presence of these proteins fit well into the general dogma of a primary HDL role in RCT.
However, this view has undergone significant revision in light of recent proteomics studies on HDL. With the advent of soft ionization techniques and the resulting leaps in mass spectrometry (MS) technology, modern proteomic approaches have become powerful tools for mapping complex protein mixtures. The advantage of proteomics over standard biochemical approaches lies in the ability to identify proteins without prior suspicion of their presence in HDL. Although strategies are almost as diverse as the laboratories performing them, the proteomic approaches applied to HDL can be classified into two general categories. The first uses gel electrophoresis to spread HDL proteins either by size only (1D) or by charge and then size (2D). The resulting gel spots are excised, digested with a protease such as trypsin, and identified by high-resolution tandem MS. In the second approach, sometimes referred to as a “shotgun” technique, tryptic peptides are first generated from HDL in solution. The peptides are then separated by high-performance liquid chromatography (commonly by reverse phase, but multidimensional separations are also used), subjected to electrospray ionization (ESI) as they elute from the column, and analyzed by tandem MS. A more detailed description of proteomic technologies can be found in an article by Canas et al. [26]. To date, there have been five major applications of modern proteomic approaches to human HDL. The findings are compared in Table 1.
Table 1.
HDL-associated proteins identified using modem proteomic strategies
| Study |
||||||
|---|---|---|---|---|---|---|
| Protein | Accession | 1 | 2 | 3 | 4 | 5 |
| Apo A-I | P02647 |
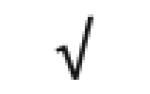
|
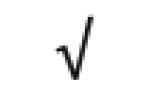
|
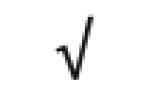
|
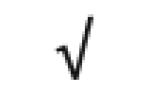
|
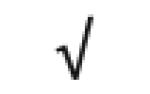
|
| Apo A-II | P02652 |
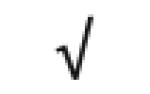
|
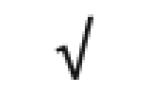
|
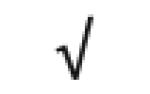
|
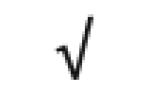
|
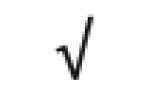
|
| Apo C-II | P02655 |
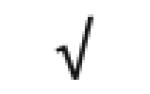
|
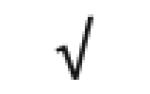
|
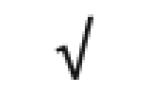
|
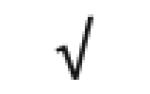
|
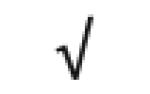
|
| Apo C-III | P02656 |
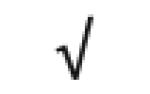
|
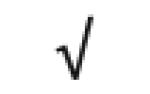
|
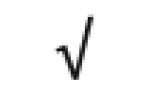
|
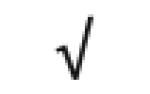
|
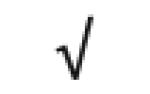
|
| Apo A-IV | P06727 |
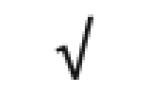
|
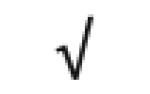
|
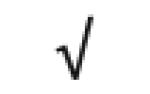
|
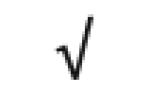
|
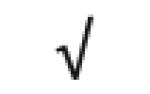
|
| Apo E | P02649 |
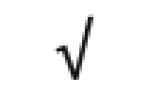
|
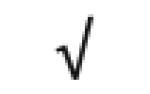
|
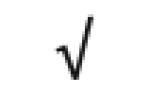
|
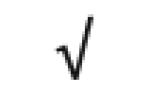
|
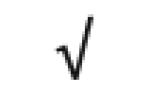
|
| Apo L-I | O14791 |
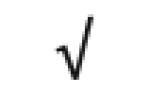
|
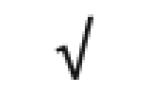
|
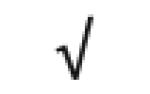
|
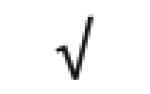
|
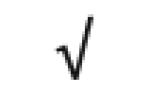
|
| SAA4 | P35542 |
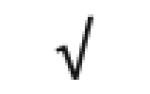
|
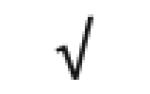
|
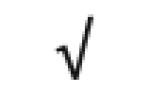
|
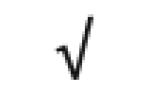
|
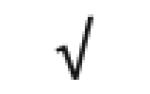
|
| SAA1&2 | P02735 |
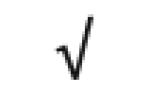
|
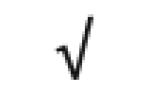
|
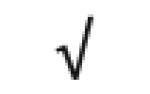
|
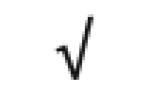
|
|
| Apo M | O95445 |
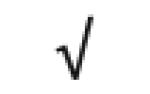
|
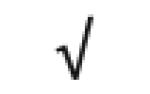
|
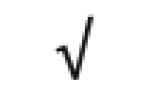
|
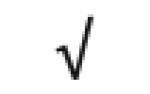
|
|
| Apo D | P05090 |
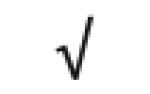
|
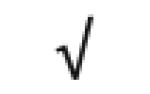
|
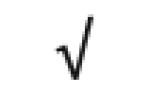
|
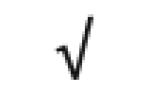
|
|
| Apo J | P10909 |
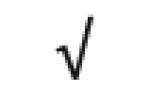
|
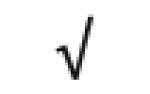
|
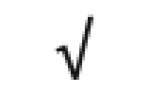
|
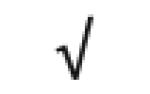
|
|
| Pon 1 | P27169 |
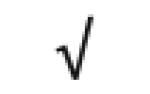
|
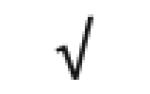
|
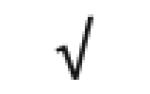
|
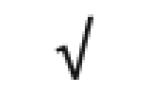
|
|
| HRP | P00739 |
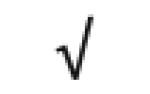
|
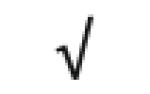
|
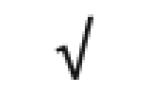
|
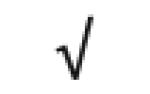
|
|
| Albumin | P02768 |
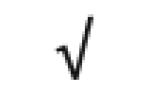
|
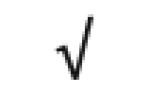
|
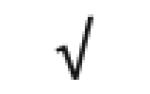
|
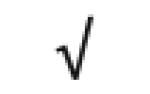
|
|
| Apo C-I | P02654 |
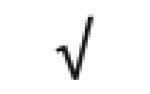
|
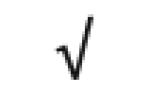
|
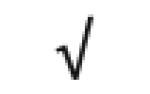
|
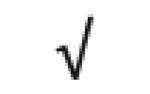
|
|
| α-1-antitrypsin | P01009 |
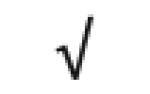
|
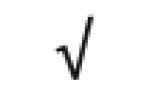
|
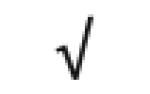
|
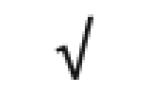
|
|
| Serotransferrin | P02787 |
|
|
|
||
| Transthyretin | P02766 |
|
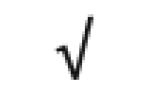
|
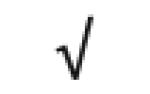
|
||
| Apo F | Q13790 |
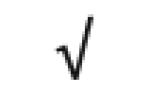
|
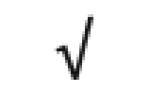
|
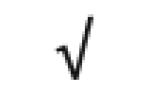
|
||
| α-1B-glycoprotein | P04217 |
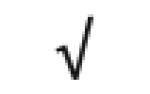
|
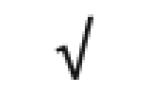
|
|||
| CETP | P11597 |
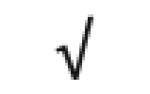
|
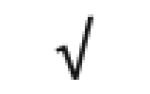
|
|||
| Complement C3 | P01024 |
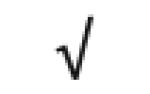
|
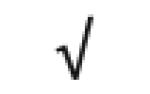
|
|||
| α1-acid glycoprotein 2 | P19652 |
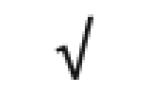
|
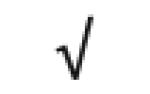
|
|||
| α2-HS-glycoprotein | P02765 |
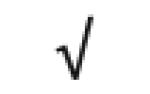
|
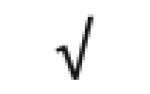
|
|||
| Vit-D Binding Protein | P02774 |
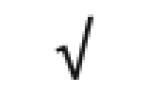
|
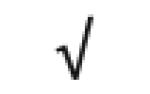
|
|||
| Apo C-IV | P55056 |
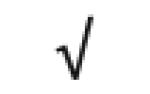
|
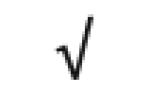
|
|||
| PLTP | P55058 |
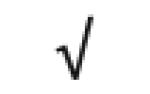
|
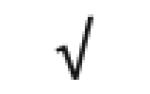
|
|||
| Pon3 | Q15166 |
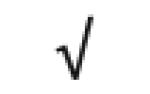
|
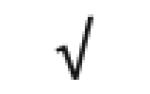
|
|||
| Fibrinogen | P02671 |
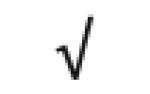
|
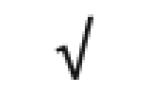
|
|||
| Apo B-100 | P04114 |
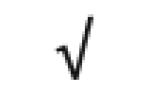
|
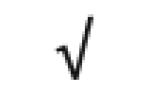
|
|||
| Apo H | P02749 |
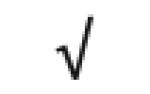
|
||||
| Complement C4A | P0C0L4 |
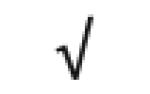
|
||||
| Complement C9 | P02748 |
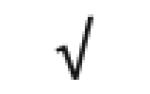
|
||||
| Vitronectin | P04004 |
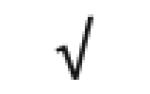
|
||||
| α-2 antiplasmin | P08697 |
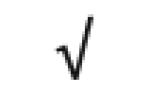
|
||||
| α-1-microglobulin | P02760 |
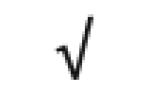
|
||||
| Inter α Trp. Inhib. | Q14624 |
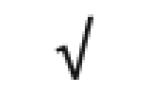
|
||||
| Angiotensin | P01019 |
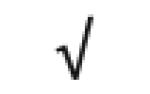
|
||||
| Kininogen-1 | P01042 |
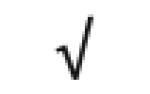
|
||||
| Hemopexin | P02790 |
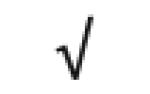
|
||||
| SERPINF1 | P36955 |
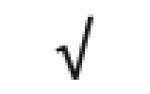
|
||||
| LCAT | P04180 |
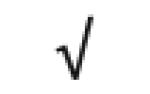
|
||||
| Complement C4B | P0C0L5 |
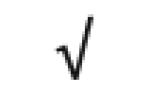
|
||||
| Ret. Bind. Prot. 4 | P02753 |
|
||||
| Prenylcystein oxidase | Q9UHG3 |
|
||||
| LPL | P06858 |
|
||||
| C-RP | P02741 |
|
||||
| Ceruloplasmin | P00450 |
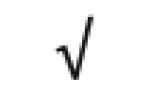
|
||||
| Serum Amyloid P | P02743 |
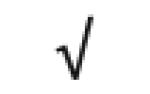
|
||||
| Complement Factor H | P08603 |
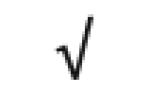
|
||||
| Kallikrein B1 | P03952 |
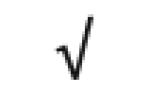
|
||||
| Factor VIII-assoc.prot | P00451 |
|
||||
| C1-INH | P05155 |
|
||||
| MSF-1 | P09603 |
|
||||
| Lymphocyte antigen | P06340 |
|
||||
| MGEA 5 | O60502 |
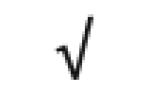
|
||||
| HLA-A protein | P04439 |
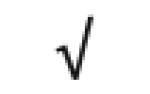
|
||||
| NOTCH 1 | P46531 |
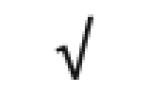
|
||||
| Siglec 5 | O15389 |
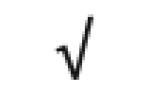
|
||||
| CLECSF 1 | Q9P126 |
|
||||
| Zinc Finger Protein IA-1 | Q01101 |
|
||||
| LTBP-1 | P22064 |
|
||||
| LTBP-2 | Q14767 |
|
||||
| Gas6 | Q61592 |
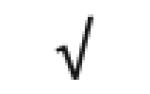
|
||||
| Ryanodine receptor 2 | Q92736 |
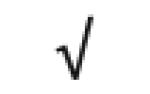
|
||||
| POU 5 domain product | Q8N7G0 |
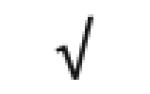
|
||||
| TFPI | P10646 |
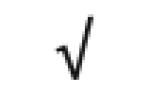
|
||||
| Unnamed protein product | gi10435007 |
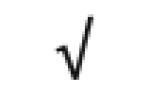
|
||||
| Unknown protein | gi12653035 |
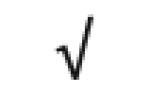
|
||||
| Unknown protein | gi12802992 |
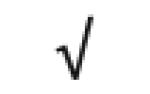
|
||||
| KIAA1095 | A8HTM6 |
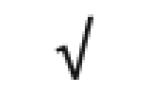
|
||||
| KIAA1730 protein | Q9C0D3 |
|
||||
| KIAA0675 gene product | Q86Y13 |
|
||||
| ZNF 356 | Q9ULV3 |
|
||||
| dj675G8.1 | gi 11137825 |
|
||||
| dj733D15.1 | Q31N78 |
|
||||
| TAT-interactive protein | gi 1427566 |
|
||||
| dj758N20.1 | gi 11493357 |
|
||||
| PTP | gi 13645209 |
|
||||
| IgG | P99007 |
|
||||
| dj1057B20.2 | Q9NQG5 |
|
||||
| Desmocollin | Q08554 |
|
||||
| Alpha-amylase salivary | P04745 |
|
||||
| Alpha-2-macroglobulin | P01023 |
|
||||
| IDH | P50213 |
|
||||
| ACTA | P03996 |
|
||||
| Galectin-7 (LEG7) | P47929 |
|
||||
| Platelet basic protein | P02775 |
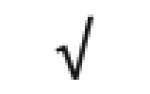
|
||||
| Prothrombin | P00734 |
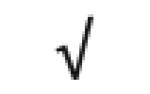
|
||||
| PAF-AH | P43034 |
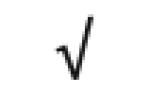
|
||||
| Apo(a) | P08519 |
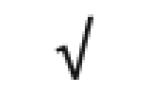
|
||||
| Totals: | 93 | 46 | 57 | 13 | 24 | 28 |
Dark gray: protein found in all 5 studies; Medium gray: in 4 of 5 studies; Light gray: in 3 of 5 studies Author (Reference), HDL fraction used; HDL isolation technique; MS technique
1: Vaisar (29), Total HDL or HDL3; UC; ESI-MS/MS 2: Rezaee (28), Total HDL; UC-IDE or 2DE or ICAT; MALDI or nLC-MS/MS 3: Karlsson (25), HDL2 and HDL3; UC-2DE; MALDI 4: Heller (27), HDL2 and HDL3; UC-2DE; MALDI 5: Davidson (31), HDL2b,a & HDL3a,b,c; UC; ESI-MS/MS
Abbreviations: ESI, electrospray ionization; MALDI, matrix assisted laser desorption ionization; MS, mass spectrometry; UC, ultracentrifugation; x-DE, x dimensional electrophoresis
Using a 2D electrophoresis approach (isoelectric focusing followed by sodium dodecyl sulfate polyacrylamide gel electrophoresis [SDS-PAGE]), Karlsson et al. [27] identified 13 proteins in pools of HDL2 and HDL3 separated by two-step discontinuous density ultracentrifugation from a pool of healthy donors. Of these, 11 were known HDL components by biochemical methods. One of the novel findings was α-1-antitrypsin (A1AT), a serine protease inhibitor (serpin) since confirmed by others. This protein is known to modulate activated neutrophils and thus supported a possible role for HDL in innate immunity [28]. In addition, this study highlighted a key advantage of the charge dimension of the electrophoretic approach in that multiple isoforms of apoA-I, apoA-II, apoC-III, apoE, apoM, SAA, and SAA-4 were identified that varied with respect to the presence of a propeptide, potential glycosylation, or sialylation. Another study in the same year utilized both the shotgun approach and a different 2D electrophoresis strategy (native separation followed by SDS-PAGE) to study density-isolated fractions roughly corresponding to HDL2 and HDL3 [29]. The study yielded 24 proteins including most found by Karlsson et al. [27], as well as A1AT. Serotransferin was postulated as a new HDL-associated protein, and α-2-macroglobulin, another protease inhibitor, was found in confirmation of previous biochemical reports.
Rezaee et al. [30] used a multipronged approach of 1D and 2D electrophoresis, shotgun proteomic methods, and immunologic assays to study centrifugally isolated total HDL from normal donors. This was the first study to employ an isotope-coded affinity tag method to identify lower abundance proteins that may be masked by the more common constituents. In this strategy, tryptic peptides containing cysteine residues were derivatized with a heavy or light isotopic tag. By selecting peptides with a threshold ratio of light to heavy label, the authors identified up to 40 cysteine-containing HDL proteins. Although many of these have yet to be confirmed, this study first identified key mediators of the complement system, C3, C1 inhibitor, and complement factor H, implicating HDL in hemostasis and innate immunity. Hortin et al. [31] examined ultracentrifugally isolated HDL from normal donors, but focused on low molecular weight peptides less than 4,000 kDa, and found many of the same proteins (or at least pieces of them). The finding of small peptides from fibrinogen suggested HDL may be a circulating reservoir for proteolytic fragments of major plasma proteins. This study pointed out a potential pitfall of bottom-up (ie, peptide-based) MS methods, in that peptide sequence identifications do not necessarily indicate the entire protein is present in HDL.
The most comprehensive proteomic analysis of HDL to date was performed by Vaisar et al. [32••]. Using centrifugally isolated total HDL or HDL3 from normal subjects, they used multidimensional protein identification technology and ESI-MS to identify 48 HDL proteins (35 previously known and 13 new identifications). Four of the new HDL associations play major roles in complement regulation, strengthening the argument for a role of HDL in innate immunity. Others showed a clear theme of protease inhibition. Most importantly, this study took HDL proteomics to the next level by comparing protein profiles of six normolipidemic control subjects with seven subjects with established CAD. Using a peptide counting strategy, it was found that HDL3 from CAD subjects was enriched in apoC-IV, PON1, complement C3, apoA-IV, and apoE. A follow-up study compared the HDL3 proteome of subjects with stenotic lesions verified by angiography before and 1 year after combination treatment with a statin and the HDL-raising drug niacin [33••]. The treatment was found to reduce apoE levels while increasing apoF and PLTP, partially remodeling the stenotic proteomic profile toward that of control subjects.
More recently, we applied a shotgun ESI-MS/MS approach to learn about the distribution of proteins across five different density subfractions (HDL2b,a and HDL3a,b,c) isolated by density ultracentrifugation [34••]. Using an abundance pattern analysis, we categorized 28 different proteins into five groups based on their distribution across the fractions. A correlational network suggested the existence of distinct protein clusters that may be suggestive of PL-rich LP subpopulations defined by specific protein interactions. Also, levels of apoL-I and PON were correlated with the capacity of HDL to protect against LDL oxidation.
Evidence for Protein Subspeciation and the Concept of Cooperative Protein Function in HDL
The proteins that comprise PL-rich LPs are called exchangeable apolipoproteins due to their ability to associate with and dissociate from lipoproteins as a function of their relative affinities for the particles versus a stable lipid-free form. Careful kinetic studies have documented the movement of apolipoproteins between VLDL and HDL [35]. Given this reversibility, one might be tempted to think of HDL as a transient ensemble of proteins randomly exchanging between lipid assemblies. Indeed, certain transfer proteins and enzymes likely ping-pong between different particles to perform their functions. Further, the recent X-ray crystal structure of CETP indicates that the boomerang-shaped protein acts as a mobile conduit that can move cholesteryl esters and triglycerides between HDL and triglyceride-rich LPs [36]. However, there is significant evidence that many apolipoproteins do in fact segregate into compositionally stable particles. Asztalos and Schaefer [37] have made extensive use of a native 2D gel electrophoresis system that separates human plasma first by charge and then by size. The gels are then probed with antibodies to visualize protein migration patterns. ApoA-I appears in up to 11 distinct spots that represent highly negatively charged species of various sizes (pre-α1, 2, 3), moderately negatively charged (α1, 2, 3), and less negatively charged species (pre-β1 a,b and pre-β2, a,b,c), all of various diameters [37]. When probing for other HDL-associated proteins, highly distinct patterns emerged [23]. ApoA-II associated with apoA-I in the α2 and α3 species, but not in the others. ApoE showed up on larger particles that also failed to overlap completely with apoA-I. They also found evidence for apoA-IV-only lipoproteins of similar diameter to apoA-I-containing particles. Further-more, our proteomic characterization of PL-rich LP density subspecies also showed clearly distinct abundance patterns for the different proteins [34••]. Some preferred small, dense particles, whereas others preferred large, light ones. These reports indicate that apolipoproteins are not randomly exchanging in plasma. The mechanisms driving such segregation are unknown but could be related to 1) affinity of a given protein to a given lipid composition or degree of surface curvature, or 2) specific protein–protein interactions on the particle surface that maintain protein segregation.
This latter idea of specific protein–protein interactions in PL-rich LPs opens the possibility for cooperative function. There is ample evidence that major activities of HDL rely on cooperative interactions between proteins cohabitating on a particle. The classic example is the relationship of apoA-I and LCAT. On its own, LCAT is relatively inefficient in mediating cholesterol esterification in lipoproteins. However, apoA-I acts as a cofactor to stimulate this activity by several orders of magnitude when present on the same particle [38]. ApoF, also known as lipid transport inhibitor protein, can inhibit the CETP-mediated exchange of cholesteryl esters between HDL and triglyceride-rich LPs, possibly by modulating the affinity of CETP for the HDL particle surface [39]. There is also growing evidence that one role of apoA-II may be to modulate apoA-I function. A study compared the hydrolysis rates of LpA-I, LpA-II, and LpA-I/A-II HDL by endothelial lipase, an important enzyme for the physiological regulation of HDL-C levels [40]. The lipid hydrolysis rate was highest in the mixed particles but lower in LpA-I and almost undetectable in LpA-II. That apoA-II facilitated lipid hydrolysis in mixed particles but not in LpA-II suggests it can affect apoA-I conformation to modulate endothelial lipase activation. However, the most striking example of on-particle protein cooperation relates to the role of HDL in innate immunity. In 1978, it was recognized that a dense fraction of HDL could mediate the lysis of T. brucei, a trypanosome that causes African sleeping sickness. This activity was referred to as trypanosome lytic factor (TLF) [41]. Immunoprecipitation studies demonstrated that TLF is a specific HDL subparticle that contains apoA-I, apoL-I, and haptoglobin-related protein (HRP). The current model for TLF lysis of T. brucei holds that the HDL particle is taken up by the trypanosome in a receptor-mediated pathway, possibly via the HRP moiety (for a review, see [42]). The complex is then targeted to the lysosome where apoL-I, via a colicin-like pore-forming domain, permeates the organelle to kill the organism. Interestingly, apoA-I may be required for the proper sequestration of apoL-I and HRP to form TLF. The unique composition of TLF is the strongest evidence yet that distinct particles within classically defined HDL exist and perform highly specialized functions that are quite distinct from traditional lipid transport roles [43]. Given the large number of proteins in play, it is easy to imagine many more PL-rich LPs yet to be discovered.
PL-Rich LP Subspeciation and its Impact on the Treatment of CAD
Statin therapies, although successful commercially, only reduce CAD risk by about one third [44]. Many have pointed to this as evidence for the need to raise HDL-C in combination with the statin-mediated lowering of LDL. Unfortunately, HDL is sometimes thought of in rather simplistic terms. Although its compositional heterogeneity is well cited, it tends to be discussed as a single entity (ie, HDL-C), most often tied to reverse cholesterol transport. Current pharmacological therapies such as niacin or those in development, including CETP inhibition and apoA-I transcription stimulation, aim to raise plasma HDL cholesterol in the generic sense without direct knowledge of the functionality (or lack thereof) of the elevated species. Although there is no question that high plasma levels of HDL are inversely correlated with CAD on a population basis, there are also many individuals with high HDL with CAD and vice versa. The implication of this is clear: not all HDLs are created equal. Among the numerous particle populations comprising HDL, some may be cardioprotective, some may not [45]. It is quite possible that raising HDL-C indiscriminately may increase the wrong particles at the expense of cardioprotective ones. We need a better understanding of the subparticle makeup of the fractions classically referred to as “HDL.” With such knowledge, perhaps more targeted therapies can be brought online to raise plasma levels of the cardioprotective PL-rich LPs. It is possible, maybe even likely, that a successful therapeutic strategy may not even raise HDL-C as it is measured in the clinic currently. Alternatively, small molecule therapies could be explored that mimic the cardioprotective effects of identified beneficial PL-rich subspecies. Given the tremendous resources that have been poured into nonspecific raising of plasma HDL-C, and the relatively meager successes to date, it seems prudent to invest more research into understanding HDL subspeciation.
Conclusions
The application of high-resolution MS-based proteomics has dramatically increased our understanding of HDL protein complexity. These findings leave little doubt that HDL is not only involved in lipid transport, but also proteinase inhibition, anti-inflammation, complement regulation, and innate immunity. Comparative proteomics studies offer the hope that new biomarkers can be identified to predict CAD. In addition to monitoring the ups and downs of particular proteins, we may find that the overall levels of a given protein may not differ between normal and CAD subjects, but that its distribution among PL-rich LPs or its associations with other proteins may change significantly. Realization of these goals will require solving technical problems, including increasing sample throughput to boost the number of subjects that can be efficiently studied and developing more sensitive and quantitative comparison strategies. Another challenge relates to the fact that, to date, all detailed proteomic characterizations have been performed on HDL isolated by ultracentrifugation. Given the potential for artifacts, these studies must be verified using alternative separations. Unfortunately, methods such as gel filtration, ion exchange, or native gel electrophoresis are compromised by the presence of high abundance contaminants that are not at issue in ultracentrifugation. New technologies for either lipoprotein isolation or improved MS dynamic range will be required to meet this important challenge.
Footnotes
Disclosure No potential conflicts of interest relevant to this article were reported.
Contributor Information
Scott Gordon, Center for Lipid and Arteriosclerosis Science, University of Cincinnati, 2120 East Galbraith Road, Cincinnati, OH 45237-0507, USA Gordonst@email.uc.edu.
Anita Durairaj, Center for Lipid and Arteriosclerosis Science, University of Cincinnati, 2120 East Galbraith Road, Cincinnati, OH 45237-0507, USA Duraira@email.uc.edu.
Jason L. Lu, Division of Biomedical Informatics, Cincinnati Children’s Hospital Research Foundation, 3333 Burnet Avenue, MLC 7024, Cincinnati, OH 45229-3039, USA long.lu@cchmc.org
W. Sean Davidson, Center for Lipid and Arteriosclerosis Science, University of Cincinnati, 2120 East Galbraith Road, Cincinnati, OH 45237-0507, USA.
References
Papers of particular interest, published recently, have been highlighted as:
• Of importance
•• Of major importance
- 1.Adorni MP, Zimetti F, Billheimer JT, et al. The roles of different pathways in the release of cholesterol from macrophages. J Lipid Res. 2007;48:2453–2462. doi: 10.1194/jlr.M700274-JLR200. [DOI] [PubMed] [Google Scholar]
- 2.Gu X, Trigatti B, Xu S, et al. The efficient cellular uptake of high density lipoprotein lipids via scavenger receptor class B type I requires not only receptor-mediated surface binding but also receptor-specific lipid transfer mediated by its extracellular domain. J Biol Chem. 1998;273:26338–26348. doi: 10.1074/jbc.273.41.26338. [DOI] [PubMed] [Google Scholar]
- 3.Moore RE, Navab M, Millar JS, et al. Increased atherosclerosis in mice lacking apolipoprotein A-I attributable to both impaired reverse cholesterol transport and increased inflammation. Circ Res. 2005;97:763–771. doi: 10.1161/01.RES.0000185320.82962.F7. [DOI] [PubMed] [Google Scholar]
- 4.Navab M, Hama SY, Anantharamaiah GM, et al. Normal high density lipoprotein inhibits three steps in the formation of mildly oxidized low density lipoprotein. Steps 2 and 3. J Lipid Res. 2000;41:1495–1508. [PubMed] [Google Scholar]
- 5.Barter PJ, Rye KA. High density lipoproteins and coronary heart disease. Atherosclerosis. 1996;121:1–12. doi: 10.1016/0021-9150(95)05675-0. [DOI] [PubMed] [Google Scholar]
- 6.Navab M, Imes SS, Hama SY, et al. Monocyte transmigration induced by modification of low density lipoprotein in cocultures of human aortic wall cells is due to induction of monocyte chemotactic protein 1 synthesis and is abolished by high density lipoprotein. J Clin Invest. 1991;88:2039–2046. doi: 10.1172/JCI115532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Puranik R, Bao S, Nobecourt E, et al. Low dose apolipoprotein A-I rescues carotid arteries from inflammation in vivo. Atherosclerosis. 2008;196:240–247. doi: 10.1016/j.atherosclerosis.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 8.Rye KA, Bursill CA, Lambert G, et al. The metabolism and anti-atherogenic properties of HDL. J Lipid Res. 2009;50(Suppl):S195–S200. doi: 10.1194/jlr.R800034-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Concha MI, Smith VJ, Castro K, et al. Apolipoproteins A-I and A-II are potentially important effectors of innate immunity in the teleost fish Cyprinus carpio. Eur J Biochem. 2004;271:2984–2990. doi: 10.1111/j.1432-1033.2004.04228.x. [DOI] [PubMed] [Google Scholar]
- 10.Cruz D, Watson AD, Miller CS, et al. Host-derived oxidized phospholipids and HDL regulate innate immunity in human leprosy. J Clin Invest. 2008;118:2917–2928. doi: 10.1172/JCI34189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11 •.Heinecke JW. The HDL proteome: a marker–and perhaps mediator–of coronary artery disease. J Lipid Res. 2009;50(Suppl):S167–S171. doi: 10.1194/jlr.R800097-JLR200. This excellent summary of HDL proteomics focuses on the work of the Heinecke laboratory.
- 12 •.Hoofnagle AN, Heinecke JW. Lipoproteomics: using mass spectrometry-based proteomics to explore the assembly, structure, and function of lipoproteins. J Lipid Res. 2009;50:1967–1975. doi: 10.1194/jlr.R900015-JLR200. This article presents another good review of HDL proteomics.
- 13.Reilly MP, Tall AR. HDL proteomics: pot of gold or Pandora’s box? J Clin Invest. 2007;117:595–598. doi: 10.1172/JCI31608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kontush A, Chantepie S, Chapman MJ. Small, dense HDL particles exert potent protection of atherogenic LDL against oxidative stress. Arterioscler Thromb Vasc Biol. 2003;23:1881–1888. doi: 10.1161/01.ATV.0000091338.93223.E8. [DOI] [PubMed] [Google Scholar]
- 15.Cheung MC, Albers JJ. Characterization of lipoprotein particles isolated by immunoaffinity chromatography. Particles containing A-I and A-II and particles containing A-I but no A-II. J Biol Chem. 1984;259:12201–12209. [PubMed] [Google Scholar]
- 16.Asztalos BF, Sloop CH, Wong L, Roheim PS. Two-dimensional electrophoresis of plasma lipoproteins: recognition of new apo A-I-containing subpopulations. Biochim Biophys Acta. 1993;1169:291–300. doi: 10.1016/0005-2760(93)90253-6. [DOI] [PubMed] [Google Scholar]
- 17.Warnick GR, Nauck M, Rifai N. Evolution of methods for measurement of HDL-cholesterol: from ultracentrifugation to homogeneous assays. Clin Chem. 2001;47:1579–1596. [PubMed] [Google Scholar]
- 18.Duong PT, Weibel GL, Lund-Katz S, et al. Characterization and properties of pre beta-HDL particles formed by ABCA1-mediated cellular lipid efflux to apoA-I. J Lipid Res. 2008;49:1006–1014. doi: 10.1194/jlr.M700506-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haginaka J, Yamaguchi Y, Kunitomo M. Anion-exchange high-performance liquid chromatography assays of plasma lipoproteins and modified low-density lipoproteins using a ProtEx-DEAE column. J Chromatogr B Biomed Sci Appl. 2001;751:161–167. doi: 10.1016/s0378-4347(00)00467-9. [DOI] [PubMed] [Google Scholar]
- 20.van’t Hooft F, Havel RJ. Metabolism of apolipoprotein E in plasma high density lipoproteins from normal and cholesterol-fed rats. J Biol Chem. 1982;257:10996–11001. [PubMed] [Google Scholar]
- 21 •.Stahlman M, Davidsson P, Kanmert I, et al. Proteomics and lipids of lipoproteins isolated at low salt concentrations in D2O/ sucrose or in KBr. J Lipid Res. 2008;49:481–490. doi: 10.1194/jlr.D700025-JLR200. This article presents a comparison of different methods for floating lipoproteins during density ultracentrifugation that shows the potential for artifacts in the KBr procedure.
- 22.Karlsson H, Leanderson P, Tagesson C, Lindahl M. Lipoproteomics I: mapping of proteins in low-density lipoprotein using two-dimensional gel electrophoresis and mass spectrometry. Proteomics. 2005;5:551–565. doi: 10.1002/pmic.200300938. [DOI] [PubMed] [Google Scholar]
- 23.Santos RD, Schaefer EJ, Asztalos BF, et al. Characterization of high density lipoprotein particles in familial apolipoprotein A-I deficiency. J Lipid Res. 2008;49:349–357. doi: 10.1194/jlr.M700362-JLR200. [DOI] [PubMed] [Google Scholar]
- 24.Movva R, Rader DJ. Laboratory assessment of HDL heterogeneity and function. Clin Chem. 2008;54:788–800. doi: 10.1373/clinchem.2007.101923. [DOI] [PubMed] [Google Scholar]
- 25.Davidson WS, Jonas A, Clayton DF, George JM. Stabilization of alpha-synuclein secondary structure upon binding to synthetic membranes. J Biol Chem. 1998;273:9443–9449. doi: 10.1074/jbc.273.16.9443. [DOI] [PubMed] [Google Scholar]
- 26.Canas B, Lopez-Ferrer D, Ramos-Fernandez A, et al. Mass spectrometry technologies for proteomics. Brief Funct Genomic Proteomic. 2006;4:295–320. doi: 10.1093/bfgp/eli002. [DOI] [PubMed] [Google Scholar]
- 27.Karlsson H, Leanderson P, Tagesson C, Lindahl M. Lipoproteomics II: mapping of proteins in high-density lipoprotein using two-dimensional gel electrophoresis and mass spectrometry. Proteomics. 2005;5:1431–1445. doi: 10.1002/pmic.200401010. [DOI] [PubMed] [Google Scholar]
- 28.Subramaniyam D, Glader P, von Wachenfeldt K, et al. C-36 peptide, a degradation product of alpha1-antitrypsin, modulates human monocyte activation through LPS signaling pathways. Int J Biochem Cell Biol. 2006;38:563–575. doi: 10.1016/j.biocel.2005.09.021. [DOI] [PubMed] [Google Scholar]
- 29.Heller M, Stalder D, Schlappritzi E, et al. A. Mass spectrometry-based analytical tools for the molecular protein characterization of human plasma lipoproteins. Proteomics. 2005;5:2619–2630. doi: 10.1002/pmic.200401233. [DOI] [PubMed] [Google Scholar]
- 30.Rezaee F, Casetta B, Levels JH, et al. Proteomic analysis of high-density lipoprotein. Proteomics. 2006;6:721–730. doi: 10.1002/pmic.200500191. [DOI] [PubMed] [Google Scholar]
- 31.Hortin GL, Shen RF, Martin BM, Remaley AT. Diverse range of small peptides associated with high-density lipoprotein. Biochem Biophys Res Commun. 2006;340:909–915. doi: 10.1016/j.bbrc.2005.12.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32 ••.Vaisar T, Pennathur S, Green PS, et al. Shotgun proteomics implicates protease inhibition and complement activation in the antiinflammatory properties of HDL. J Clin Invest. 2007;117:746–756. doi: 10.1172/JCI26206. Liquid chromatography ESI-MS/MS was used to identify proteins from ultracentrifugally isolated HDL of healthy control subjects and subjects with established CAD. A total of 48 proteins were identified. Using a novel peptide counting strategy, it was found that HDL3 from CAD patients was selectively enriched in apoE and other proteins that may play a role in macrophage biology, lipid metabolism, and the inflammatory response.
- 33 ••.Green PS, Vaisar T, Pennathur S, et al. Combined statin and niacin therapy remodels the high-density lipoprotein proteome. Circulation. 2008;118:1259–1267. doi: 10.1161/CIRCULATIONAHA.108.770669. This is the first study to utilize HDL proteomics as a measure of the efficacy of lipid-lowering therapies.
- 34 ••.Davidson WS, Silva RA, Chantepie S, et al. Proteomic analysis of defined HDL subpopulations reveals particle-specific protein clusters: relevance to antioxidative function. Arterioscler Thromb Vasc Biol. 2009;29:870–876. doi: 10.1161/ATVBAHA.109.186031. This study mapped the HDL proteome across five density subfractions indicating clearly distinct protein profiles. This study also associated specific apolipoproteins with antioxidative function.
- 35.Boyle KE, Phillips MC, Lund-Katz S. Kinetics and mechanism of exchange of apolipoprotein C-III molecules from very low density lipoprotein particles. Biochim Biophys Acta. 1999;1430:302–312. doi: 10.1016/s0167-4838(99)00009-6. [DOI] [PubMed] [Google Scholar]
- 36.Qiu X, Mistry A, Ammirati MJ, et al. Crystal structure of cholesteryl ester transfer protein reveals a long tunnel and four bound lipid molecules. Nat Struct Mol Biol. 2007;14:106–113. doi: 10.1038/nsmb1197. [DOI] [PubMed] [Google Scholar]
- 37.Asztalos BF, Schaefer EJ. High-density lipoprotein subpopulations in pathologic conditions. Am J Cardiol. 2003;91:12E–17E. doi: 10.1016/s0002-9149(02)03383-0. [DOI] [PubMed] [Google Scholar]
- 38.Jonas A. Lecithin-cholesterol acyltransferase in the metabolism of high-density lipoproteins. Biochim Biophys Acta. 1991;1084:205–220. doi: 10.1016/0005-2760(91)90062-m. [DOI] [PubMed] [Google Scholar]
- 39.Wang X, Driscoll DM, Morton RE. Molecular cloning and expression of lipid transfer inhibitor protein reveals its identity with apolipoprotein F. J Biol Chem. 1999;274:1814–1820. doi: 10.1074/jbc.274.3.1814. [DOI] [PubMed] [Google Scholar]
- 40.Jahangiri A, Rader DJ, Marchadier D, et al. Evidence that endothelial lipase remodels high density lipoproteins without mediating the dissociation of apolipoprotein A-I. J Lipid Res. 2005;46:896–903. doi: 10.1194/jlr.M400212-JLR200. [DOI] [PubMed] [Google Scholar]
- 41.Rifkin MR. Identification of the trypanocidal factor in normal human serum: high density lipoprotein. Proc Natl Acad Sci U S A. 1978;75:3450–3454. doi: 10.1073/pnas.75.7.3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nielsen MJ, Nielsen LB, Moestrup SK. High-density lipoprotein and innate immunity. Future Lipidol. 2006;1:729–734. [Google Scholar]
- 43.Shiflett AM, Bishop JR, Pahwa A, Hajduk SL. Human high density lipoproteins are platforms for the assembly of multi-component innate immune complexes. J Biol Chem. 2005;280:32578–32585. doi: 10.1074/jbc.M503510200. [DOI] [PubMed] [Google Scholar]
- 44.Maron DJ, Fazio S, Linton MF. Current perspectives on statins. Circulation. 2000;101:207–213. doi: 10.1161/01.cir.101.2.207. [DOI] [PubMed] [Google Scholar]
- 45.Ansell BJ, Navab M, Hama S, et al. Inflammatory/antiinflammatory properties of high-density lipoprotein distinguish patients from control subjects better than high-density lipoprotein cholesterol levels and are favorably affected by simvastatin treatment. Circulation. 2003;108:2751–2756. doi: 10.1161/01.CIR.0000103624.14436.4B. [DOI] [PubMed] [Google Scholar]


