Abstract
The goal of personalized medicine is to recommend drug treatment based on an individual’s genetic makeup. Pharmacogenomic studies utilize two main approaches: candidate gene and whole-genome. Both approaches analyze genetic variants such as single nucleotide polymorphisms (SNPs) to identify associations with drug response. In addition to DNA sequence variations, nongenetic but heritable epigenetic systems have also been implicated in regulating gene expression that could influence drug response. The International HapMap Project lymphoblastoid cell lines (LCLs) have been used to study genetic determinants responsible for expression variation and drug response. Recent studies have demonstrated that common genetic variants, including both SNPs and copy number variants (CNVs) account for a substantial fraction of natural variation in gene expression. Given the critical role played by DNA methylation in gene regulation and the fact that DNA methylation is currently the most studied epigenetic system, we suggest that profiling the variation in DNA methylation in the HapMap samples will provide new insights into the regulation of gene expression as well as the mechanisms of individual drug response at a new level of complexity. Epigenomics will substantially add to our knowledge of how genetics explains gene expression and pharmacogenomics.
Keywords: epigenetics, DNA methylation, gene expression, pharmacogenomics, HapMap, drug response
Introduction
Drug response is likely a multi-genic trait with numerous genes playing a role in drug response and toxicity. Scientists are beginning to appreciate the daunting level of complexity in the search for genes associated with drug response and/or toxicity. Predicting which patients will respond or experience an adverse effect would be highly beneficial to medicine. One reason is that adverse drug reactions (ADRs) have become one of the leading causes of hospitalization and death in the United States, accounting for more than 2.2 million serious cases and over 100,000 deaths a year estimated in a meta-analysis study (Lazarou et al 1998). Understandably, personalization of medical treatment would help to prevent this by identifying those individuals at greatest risk for toxicity prior to therapy (Ratain 2003, 2007).
There are several examples of the personalized medical approach, which include genotype-based selection of patients for effective therapy, to spare those who would not respond or would suffer undesirable side effects (Jain 2002). To fully understand the risk for a given patient, whole-genome data would be most comprehensive; however we are not yet at the point of translating that information into dosing recommendations (Ratain 2007). The challenge is that drug response is influenced by many different genetic and nongenetic factors and the extent to which each factor contributes to variation in response is not yet fully understood making the development of diagnostic tests that could predict an individual’s response to a particular drug difficult.
During the past decade or so, equipped with the human whole-genome sequences (Lander et al 2001; Venter et al 2001) and high throughput genotying technologies (eg, sequencing, microarrays) (Dearlove 2002; Twyman 2004; Romkes and Buch 2005), exciting progress has been made in understanding the contribution of human genetic variations, especially in the form of single nucleotide polymorphisms (SNPs), to how individuals respond to medications, either by changing the pharmacokinetics of drugs or by altering the cellular response to therapeutic agents (Roden et al 2006; Yong et al 2006; Giacomini et al 2007; Hartford and Dolan 2007; Vella and Camilleri 2008; Zhang et al 2008c). Examples include recent studies using a candidate gene approach (Liu et al 2007; Jarjanazi et al 2008; Rudin et al 2008) or a whole-genome analysis (Huang et al 2007a, 2007b; Zhang et al 2008b) to identify genetic variants that are correlated with cytotoxicity to anticancer agents. In addition to SNPs, other genetic elements such as DNA copy number variants (CNVs) have been implicated in drug response (Cappuzzo et al 2005; Greshock et al 2008). The association studies aim to link genetic variation, either in the form of SNPs or CNVs (both causal or as markers) to expression of targeted gene(s), which in turn may determine, in part, the variation in individual response to drugs. However, not only DNA sequence-based variations (SNPs and CNVs) can affect gene expression, other nongenetic factors such as environment/habitat (Idaghdour et al 2008) and epigenetic status (Zhang et al 2008d) can also regulate gene expression in vivo, suggesting the potential roles of these nongenetic factors in determining variation in drug response (Figure 1). Furthermore, environmental factors have been shown to affect epigenetic status (Wade and Archer 2006). For example, Kirk and colleagues (2008) reviewed the role of botanicals in epigenetic modulation. The effects of soy foods and exposure to genistein (a soy isoflavone) in breast cancer risk reduction have also been discussed (Warri et al 2008). Recent studies suggest that epigenetic regulation may in part mediate the complex gene-by-environment interactions that can lead to asthma (Miller and Ho 2008) and other diseases (Edwards and Myers 2007). Though three mechanisms, including CpG methylation, histone modification and RNA-associated silencing (Beck et al 1999; Holliday 2000; Fazzari and Greally 2004), have been defined as epigenetic modifiers, this review will focus on DNA methylation at CpG sites, which is currently the most well understood eukaryotic epigenetic modification, due to the availability of high throughput technology such as the Affymetrix ChIP-on-chip (Zhang et al 2006; Affymetrix 2007), the Illumina BeadArray platform (Bibikova et al 2006; Idaghdour et al 2008) and the Roche NimbleGen arrays (Weber et al 2005).
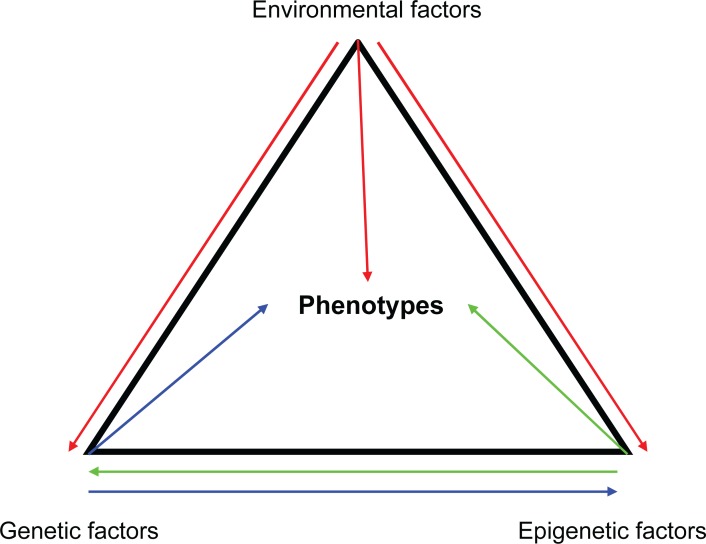
Figure 1.
Both genetic and nongenetic factors can affect phenotypes. Phenotypes (eg, gene expression, drug response) can be affected by genetic variations (eg, SNPs), environmental factors as well as epigenetic changes (eg, DNA methylation). The arrows indicate possible interactions between factors.
Methylation at CpG sites can be an accurate proxy for other epigenetic marks, such as histone methylation and acetylation (Lippman et al 2004; Zhang et al 2007b). DNA methylation can be either de novo (when CpG sites on both DNA strands are unmethylated) or maintenance (when CpG sites on one strand are methylated). Methylation of the C5 of cytosine residues in DNA has long been recognized as a fundamental epigenetic silencing mechanism (Holliday and Pugh 1975; Holliday 1989). 5-methylcytosine is also highly mutagenic, causing higher mutation rates at CpG sites for both transitions and transversions (Zhang et al 2007a). Given the critical role of DNA methylation in regulating gene expression, we suggest that genome-wide profiling of epigenetic variation (methylome) and association studies using these epigenomic data will provide new insights into gene regulation and individual drug response at a higher level of complexity relative to the current genetic variant-based pharmacogenomic studies. We suggest that the next wave of pharmacogenomic studies will have greater depth if both genetic variants and epigenetic variations are included in the analysis.
Gene expression, drug response, and DNA methylation
Gene expression acts as an intermediate phenotype situated between genetic (eg, sequence variation in DNA) and nongenetic factors (eg, environmental stimuli) and other more complex cellular, tissue, organ or whole-organism phenotypes (eg, drug response). Numerous studies have shown that gene expression can affect susceptibility to drug response. For instance, a widely appreciated example is the multi-drug resistance exhibited by tumor cells to anticancer agents through over-expression of MDR genes (Perez-Tomas 2006). Numerous recent studies have demonstrated the use of genomic data, particularly gene expression signatures, as clinical prognostic factors in cancer and other complex diseases (Garman et al 2007; Nevins and Potti 2007). These studies have demonstrated that gene expression signatures have the capability of predicting response to various commonly used and newly developing cancer therapeutics (Potti and Nevins 2008). For example, a recent study showed that incorporation of gene expression signatures into clinical risk stratification can refine prognosis in breast cancer (Acharya et al 2008).
On one hand, quantitative variation in gene-expression level (eg, mRNA transcript abundance) as a complex trait is heritable and has been mapped to the human genome as expression quantitative trait loci (eQTLs) in LCLs including the HapMap samples (Morley et al 2004; Spielman et al 2007; Stranger et al 2007b; Duan et al 2008; Zhang et al 2008a), which represent genomic regions marked by cis-acting or trans-acting SNPs for the genetic control of gene expression. Substantial contribution of DNA structural variation (eg, CNVs) (Redon et al 2006) to gene expression variation has also been demonstrated recently in the HapMap samples (Stranger et al 2007a), which include 210 individuals of major world populations in northern and western Europe (CEU, Caucasians from Utah, USA), Africa (YRI, Yoruba people from Ibadan, Nigeria) and Asia (JPT, Japanese from Tokyo, Japan; CHB, Han Chinese from Beijing, China) (Zhang et al 2008c). On the other hand, increased methylation in the promoter region of a gene has been shown to lead to reduced expression, whereas methylation in the transcribed region has a variable effect on gene expression (Jones 1999; Singal et al 2002). Thus DNA methylation patterns, together with SNPs or CNVs, likely play an important role in gene regulation, which in turn may affect drug response.
Integrating genetic variation, expression and sensitivity to drugs has been a focus of several recent studies. Both candidate gene approach (Liu et al 2007; Jarjanazi et al 2008; Rudin et al 2008) or whole-genome associations (Huang et al 2007a, 2007b; Zhang et al 2008b) have been employed to identify genetic variants that are associated with both drug-induced cytotoxicity and gene expression. These studies did not integrate promoter methylation; however epigenetic regulation of gene expression could explain much of the variation that is not due to SNPs or CNVs and improve our understanding of the role of gene expression in drug response.
Though the roles of DNA methylation in diseases such as cancer have been a topic of considerable interest in the past few years (Das and Singal 2004; Egger et al 2004; Shames et al 2007; Vucic et al 2008), the relationship between variation in DNA methylation status and gene expression has just begun to be studied. In a pilot study of the Human Epigenome Project (Bradbury 2003), DNA methylation profiling (∼1.9 million CpG sites) of human chromosomes 6, 20 and 22 from 12 different tissues was performed (Eckhardt et al 2006). A substantial fraction (∼17%) of genes were found differentially methylated in their 5’ UTRs and about one-third of the differentially methylated 5’ UTRs were inversely correlated with transcription even after controlling for factors reported to affect DNA methylation such as sex and age (Eckhardt et al 2006). In the past few years, the roles of DNA methylation through regulating gene expression in drug response have also been investigated by various laboratories (Gius et al 2005; Brown and Glasspool 2007; Strathdee 2007). One example is the DNA repair enzyme O6-methylguanine DNA methyltransferase (MGMT) efficiency of which has been associated with an increased susceptibility to alkylating agent toxicity (Kaina et al 1998; Gerson 2004; Rabik et al 2006). MGMT is transcriptionally silenced by promoter hypermethylation in several human cancers such as diffuse large B-cell lymphoma (Esteller et al 2002) as well as head and neck squamous cell carcinoma (Zuo et al 2004). In a retrospective cohort study, Esteller and colleagues (2002) demonstrated that MGMT promoter hypermethylation appeared to be a useful marker for predicting survival in patients with diffuse large B-cell lymphoma treated with multidrug regimens including cyclophosphamide. The demethylation of MGMT promoter was also shown to be involved in basic fibroblast growth factor–induced resistance against temozolomide in human melanoma cells (Fontijn et al 2007). More recently, Shen and colleagues (2007) assessed DNA methylation of 32 promoter-associated CpG islands in human cancer cell lines from the National Cancer Institute (NCI) drug-screening panel (NCI-60 panel). By correlating drug activity with DNA methylation, they identified a list of methylation markers that predicted sensitivity to chemotherapeutic drugs. For example, hypermethylation of the TP53 homologue TP73 and associated gene silencing was strongly correlated with sensitivity to alkylating agents. The methylation status of tumor suppressor genes (TIMP3, APC, and IGSF4) in the NCI-60 panel has also been found significantly correlated with the sensitivity to some antimetabolites such as fluorouracil (5-FU) (Sasaki et al 2008). In another study, Grovdal and colleagues (2007) showed a significant effect of methylation status of P15, CDH (E-cadherin) and HIC (hypermethylated in cancer 1) genes on the outcome of conventional chemotherapy in high-risk myelodysplastic syndromes (MDS) and acute myelogenous leukemia following MDS. Though the current studies using patients or patient-derived cell lines (eg, the NCI-60 panel) have shown the potential roles of DNA methylation of some candidate genes in drug response and cytotoxicity, the relationship between DNA methylation status of genes (not only candidate genes like tumor suppressors or MGMT) with drug resistance or sensitivity have not been comprehensively investigated.
Overall, these studies suggest that the variation in DNA methylation status can significantly contribute to the variation in gene expression, which in turn may affect drug response. Figure 2 shows a possible scenario for the relationship between DNA methylation and gene expression as well as drug response in a population. Similar to other genetic factors, cytosine methylation patterns are also heritable through cell division. Their preservation involves the “maintenance” methyltransferase DNMT1, which has specificity for hemi-methylated CpG dinucleotides; the enzyme thus can methylate CpGs in a newly synthesized DNA strand based on the presence of methylation in the CpG dinucleotide in the complementary template strand (Bernstein et al 2007). Therefore, this kind of epigenetic inheritance may allow cells of different phenotype (eg, drug response) but identical genotype to transmit their phenotype to their offspring. Although many genes have been identified that acquire methylation and whose expression is methylation-sensitive, little is known about how DNA methylation is controlled (Yoon et al 2002). Our current knowledge suggest DNA methylation may be regulated by different variants of DNA methyltransferase (Wang et al 2007) or RASGRF1 (a Ras protein-specific guanine nucleotide-releasing factor 1) (Yoon et al 2002). At present we have very poor understanding of the factors that contribute to interindividual epigenetic variation. The effects of DNA methylation status on gene regulation can be independent of those based on genetic variations (eg, SNPs), suggesting that the availability of a high-resolution DNA methylation resource could add another layer of information to our understanding of how genetic and epigenetic variation could be used in pharmacogenomic studies.
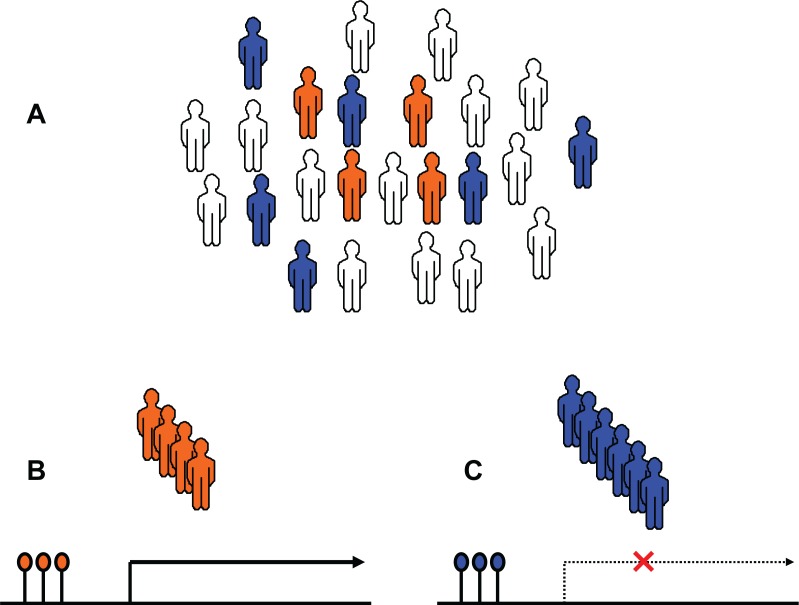
Figure 2.
DNA methylation status can affect variation in gene expression and drug response. A) For a particular drug response-related gene, there could be people with different epigenetic signatures within a population. Orange: sensitive people; Blue: nonrespondent people; white: all others. B) Sensitive people have an epigenetic signature that causes over-expression of the gene. C) In contrast, nonrespondent people have an epigenetic signature that inhibits gene expression.
Whole-genome approaches to profiling DNA methylation
Recent progress in biotechnology has provided tools to detect DNA methylation in the human genome, thus potentially allowing the study of the roles of DNA methylation in gene regulation. DNA methylation can be detected using bisulfite conversion, methylation-sensitive restriction enzyme (MSRE) digestion, methyl-binding proteins and anti-methylcytosine antibodies. Combining these techniques with DNA microarrays and high-throughput sequencing has made the mapping of DNA methylation feasible on a genome-wide scale (Zilberman and Henikoff 2007).
Bisulfite conversion is based on treatment of DNA with bisulfite, a chemical that results in the conversion of cytosine to uracil, but leaves 5-methylcytosine residues unaffected, thus introducing specific changes (C to T transitions) in the DNA sequence that depend on the methylation status of cytosine residues. The methylation status then can be deduced by techniques such as direct sequencing (Frommer et al 1992; Eckhardt et al 2006), pyrosequencing (Colella et al 2003) or microarray-based methods (Abecasis et al 2000), which can be used in a genome-wide analysis of DNA methylation. For the latter, microarrays can be designed using oligonucleotide pairs targeting CpG sites of interest, with one complementary to the unaltered methylated sequence, and the other to the C-to-T converted unmethylated sequence (Adorjan et al 2002). As the name implies, the method of MSRE digestion uses methylation specific restriction enzymes such as HpaII to cleave DNA at specific methylated-cytosine residues. Similarly, whole-genome oligonucleotide microarrays then can be used to differentiate the MSRE-digested products and background (eg, DNA fragments digested by nonmethylation-sensitive restriction enzymes such as MspI) (Zhang et al 2008d). In contrast, the methods based on methyl-binding proteins and anti-methylcytosine antibodies use specific methyl-group binding proteins or methylcytosine antibodies to enrich methylated DNA fragments through chromatin immunoprecipitation (ChIP) (Zhang et al 2006). Again, whole-genome oligonucleotide microarrays can be used to analyze the methylation status of the ChIP products.
In spite of some limitations, for example, the coverage of the MSRE digestion approach is limited to the CpG sites (Zhang et al 2008d); and incomplete conversion and degradation of DNA during bisulfite treatment could be confounding factors for the bisulfite conversion approach (Grunau et al 2001). These approaches have begun to allow genome-wide analyses of DNA methylation in humans (Eckhardt et al 2006). For example, in a previously mentioned study, using the bisulfite sequencing approach, the CpG methylation profiles of 43 samples derived from 12 different human tissues (eg, heart muscle, skeletal muscle) and various primary cell types (eg, dermal fibroblasts, CD4+ and CD8+ lymphocytes) were determined for chromosomes 6, 20, and 22 (Eckhardt et al 2006). Obviously, these whole-genome or epigenomic approaches provide promising tools to profile the variation in DNA methylation as well as to illustrate the relationship between DNA methylation and gene expression.
Conclusions and outlook
The realization of personalized medicine promises better treatment regimens for individuals based on the information contained in the human genome. To date, most pharmacogenomic studies have focused on the contribution of common genetic variants (SNPs and CNVs) to individual response to therapeutic treatments. However, nongenetic factors such as the epigenetic systems could also play an important role in gene regulation, which in turn could affect drug response. DNA methylation is currently the best-understood epigenetic system (Fazzari and Greally 2004). In the past few years, whole-genome DNA methylation profiling has become possible benefiting from the combination of methylation-specific and microarray-based techniques. Given the fact that common genetic variants only account for a fraction of the variation in drug response, other mechanisms such as DNA methylation status could help explain the remaining variation in drug response. We envision that in an ideal world, treatment decisions would be based on an individual’s whole-genome information which includes both genetic and nongenetic factors including DNA methylation status. A more comprehensive view of the genome that includes epigenetic information will also provide information on the use of agents aimed at pathways important in DNA methylation and gene regulation (Plimack et al 2007).
Research efforts such as the Human Epigenome Project, which aims to identify, catalog and interpret genome-wide DNA methylation profiles of all human genes in all major tissues (Bradbury 2003; Rakyan et al 2004; Eckhardt et al 2006), no doubt will provide new insights into the epigenetic components of the human genome and tissue specificity of epigenetic regulation of expression. However, integration of epigenomic profiling with resources such as those provided by the International HapMap Project (HapMap 2003, 2005), which has genotypic data and various publicly available phenotypes that include gene expression (see the Gene Expression Omnibus, http://www.ncbi.nlm.nih.gov/geo/), drug response (see http://www.PharmGKB.org) on a panel of LCLs derived from major world populations (Zhang et al 2008c), will greatly facilitate the identification of both genetic and nongenetic determinants responsible for the response to therapeutic treatments. The HapMap LCLs have been used to build cell-based models for pharmacogenomic discovery but currently lacks epigenomic information. This information could be integrated with publicly available genotypic, gene expression and drug sensitivity data on the same cell line (Zhang and Dolan 2008). Additional tissues including the liver or skin should be considered for pharmacogenomic discovery by integrating genomic, epigenomic, expression and drug metabolism data. These models could be of great value to the pharmacogenomic community.
Acknowledgments
Some of the research described in this article was funded through the Pharmacogenetics of Anticancer Agents Research (PAAR) Group (http://www.pharmacogenetics.org) by the NIH/NIGMS grant U01GM61393 and P50 CA125183. ME Dolan is on the Scientific Advisory Board to the NIGMS Human Genetic Cell Repository at the Coriell Institute for Medical Research, Camden, New Jersey, USA.
References
- Abecasis GR, Cardon LR, Cookson WO. A general test of association for quantitative traits in nuclear families. Am J Hum Genet. 2000;66:279–92. doi: 10.1086/302698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acharya CR, Hsu DS, Anders CK, et al. Gene expression signatures, clinicopathological features, and individualized therapy in breast cancer. JAMA. 2008;299:1574–87. doi: 10.1001/jama.299.13.1574. [DOI] [PubMed] [Google Scholar]
- Adorjan P, Distler J, Lipscher E, et al. Tumour class prediction and discovery by microarray-based DNA methylation analysis. Nucleic Acids Res. 2002;30:e21. doi: 10.1093/nar/30.5.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Affymetrix 2007. Application focus: ChIP-on-chip [online]. Accessed on Sept 18, 2008. URL: http://www.affymetrix.com.
- Beck S, Olek A, Walter J. From genomics to epigenomics: a loftier view of life. Nat Biotechnol. 1999;17:1144. doi: 10.1038/70651. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:669–81. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- Bibikova M, Lin Z, Zhou L, et al. High-throughput DNA methylation profiling using universal bead arrays. Genome Res. 2006;16:383–93. doi: 10.1101/gr.4410706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbury J. Human epigenome project – up and running. PLoS Biol. 2003;1:E82. doi: 10.1371/journal.pbio.0000082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown R, Glasspool R. Epigenetic modulation of resistance to chemotherapy? Ann Oncol. 2007;18:1429–30. doi: 10.1093/annonc/mdm415. [DOI] [PubMed] [Google Scholar]
- Cappuzzo F, Varella-Garcia M, Shigematsu H, et al. Increased HER2 gene copy number is associated with response to gefitinib therapy in epidermal growth factor receptor-positive non-small-cell lung cancer patients. J Clin Oncol. 2005;23:5007–18. doi: 10.1200/JCO.2005.09.111. [DOI] [PubMed] [Google Scholar]
- Colella S, Shen L, Baggerly KA, et al. Sensitive and quantitative universal pyrosequencing methylation analysis of CpG sites. Biotechniques. 2003;35:146–50. doi: 10.2144/03351md01. [DOI] [PubMed] [Google Scholar]
- Das PM, Singal R. DNA methylation and cancer. J Clin Oncol. 2004;22:4632–42. doi: 10.1200/JCO.2004.07.151. [DOI] [PubMed] [Google Scholar]
- Dearlove AM. High throughput genotyping technologies. Brief Funct Genomic Proteomic. 2002;1:139–50. doi: 10.1093/bfgp/1.2.139. [DOI] [PubMed] [Google Scholar]
- Duan S, Huang RS, Zhang W, et al. Genetic architecture of transcript-level variation in humans. Am J Hum Genet. 2008;82:1101–13. doi: 10.1016/j.ajhg.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckhardt F, Lewin J, Cortese R, et al. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat Genet. 2006;38:1378–85. doi: 10.1038/ng1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards TM, Myers JP. Environmental exposures and gene regulation in disease etiology. Environ Health Perspect. 2007;115:1264–70. doi: 10.1289/ehp.9951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger G, Liang G, Aparicio A, et al. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429:457–63. doi: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
- Esteller M, Gaidano G, Goodman SN, et al. Hypermethylation of the DNA repair gene O(6)-methylguanine DNA methyltransferase and survival of patients with diffuse large B-cell lymphoma. J Natl Cancer Inst. 2002;94:26–32. doi: 10.1093/jnci/94.1.26. [DOI] [PubMed] [Google Scholar]
- Fazzari MJ, Greally JM. Epigenomics: beyond CpG islands. Nat Rev Genet. 2004;5:446–55. doi: 10.1038/nrg1349. [DOI] [PubMed] [Google Scholar]
- Fontijn D, Adema AD, Bhakat KK, et al. O6-methylguanine-DNA-methyltransferase promoter demethylation is involved in basic fibroblast growth factor induced resistance against temozolomide in human melanoma cells. Mol Cancer Ther. 2007;6:2807–15. doi: 10.1158/1535-7163.MCT-07-0044. [DOI] [PubMed] [Google Scholar]
- Frommer M, McDonald LE, Millar DS, et al. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci U S A. 1992;89:1827–31. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garman KS, Nevins JR, Potti A. Genomic strategies for personalized cancer therapy. Hum Mol Genet. 2007;16(Spec No. 2):R226–32. doi: 10.1093/hmg/ddm184. [DOI] [PubMed] [Google Scholar]
- Gerson SL. MGMT: its role in cancer aetiology and cancer therapeutics. Nat Rev Cancer. 2004;4:296–307. doi: 10.1038/nrc1319. [DOI] [PubMed] [Google Scholar]
- Giacomini KM, Brett CM, Altman RB, et al. The pharmacogenetics research network: from SNP discovery to clinical drug response. Clin Pharmacol Ther. 2007;81:328–45. doi: 10.1038/sj.clpt.6100087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gius D, Bradbury CM, Sun L, et al. The epigenome as a molecular marker and target. Cancer. 2005;104:1789–93. doi: 10.1002/cncr.21395. [DOI] [PubMed] [Google Scholar]
- Greshock J, Cheng J, Rusnak D, et al. Genome-wide DNA copy number predictors of lapatinib sensitivity in tumor-derived cell lines. Mol Cancer Ther. 2008;7:935–43. doi: 10.1158/1535-7163.MCT-07-2072. [DOI] [PubMed] [Google Scholar]
- Grovdal M, Khan R, Aggerholm A, et al. Negative effect of DNA hypermethylation on the outcome of intensive chemotherapy in older patients with high-risk myelodysplastic syndromes and acute myeloid leukemia following myelodysplastic syndrome. Clin Cancer Res. 2007;13:7107–12. doi: 10.1158/1078-0432.CCR-07-1193. [DOI] [PubMed] [Google Scholar]
- Grunau C, Clark SJ, Rosenthal A. Bisulfite genomic sequencing: systematic investigation of critical experimental parameters. Nucleic Acids Res. 2001;29:E65–5. doi: 10.1093/nar/29.13.e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartford CM, Dolan ME. Identifying genetic variants that contribute to chemotherapy-induced cytotoxicity. Pharmacogenomics. 2007;8:1159–68. doi: 10.2217/14622416.8.9.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliday R, Pugh JE. DNA modification mechanisms and gene activity during development. Science. 1975;187:226–32. [PubMed] [Google Scholar]
- Holliday R. DNA methylation and epigenetic mechanisms. Cell Biophys. 1989;15:15–20. doi: 10.1007/BF02991575. [DOI] [PubMed] [Google Scholar]
- Holliday R. Epigenomics. Nat Biotechnol. 2000;18:243. doi: 10.1038/73588. [DOI] [PubMed] [Google Scholar]
- Huang RS, Duan S, Bleibel WK, et al. A genome-wide approach to identify genetic variants that contribute to etoposide-induced cytotoxicity. Proc Natl Acad Sci U S A. 2007a;104:9758–63. doi: 10.1073/pnas.0703736104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang RS, Duan S, Shukla SJ, et al. Identification of genetic variants contributing to cisplatin-induced cytotoxicity by use of a genomewide approach. Am J Hum Genet. 2007b;81:427–37. doi: 10.1086/519850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [HapMap] International HapMap Consortium The International HapMap Project. Nature. 2003;426:789–96. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- [HapMap] International HapMap Consortium A haplotype map of the human genome. Nature. 2005;437:1299–320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idaghdour Y, Storey JD, Jadallah SJ, et al. A genome-wide gene expression signature of environmental geography in leukocytes of Moroccan Amazighs. PLoS Genet. 2008;4:e1000052. doi: 10.1371/journal.pgen.1000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain KK. Personalized medicine. Curr Opin Mol Ther. 2002;4:548–58. [PubMed] [Google Scholar]
- Jarjanazi H, Kiefer J, Savas S, et al. Discovery of genetic profiles impacting response to chemotherapy: application to gemcitabine. Hum Mutat. 2008;29:461–7. doi: 10.1002/humu.20732. [DOI] [PubMed] [Google Scholar]
- Jones PA. The DNA methylation paradox. Trends Genet. 1999;15:34–7. doi: 10.1016/s0168-9525(98)01636-9. [DOI] [PubMed] [Google Scholar]
- Kaina B, Fritz G, Ochs K, et al. Transgenic systems in studies on genotoxicity of alkylating agents: critical lesions, thresholds and defense mechanisms. Mutat Res. 1998;405:179–91. doi: 10.1016/s0027-5107(98)00135-3. [DOI] [PubMed] [Google Scholar]
- Kirk H, Cefalu WT, Ribnicky D, et al. Botanicals as epigenetic modulators for mechanisms contributing to development of metabolic syndrome. Metabolism. 2008;57:S16–23. doi: 10.1016/j.metabol.2008.03.006. [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Lazarou J, Pomeranz BH, Corey PN. Incidence of adverse drug reactions in hospitalized patients: a meta-analysis of prospective studies. JAMA. 1998;279:1200–5. doi: 10.1001/jama.279.15.1200. [DOI] [PubMed] [Google Scholar]
- Liu W, Wu X, Zhang W, et al. Relationship of EGFR mutations, expression, amplification, and polymorphisms to epidermal growth factor receptor inhibitors in the NCI60 cell lines. Clin Cancer Res. 2007;13:6788–95. doi: 10.1158/1078-0432.CCR-07-0547. [DOI] [PubMed] [Google Scholar]
- Miller RL, Ho SM. Environmental epigenetics and asthma: current concepts and call for studies. Am J Respir Crit Care Med. 2008;177:567–73. doi: 10.1164/rccm.200710-1511PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morley M, Molony CM, Weber TM, et al. Genetic analysis of genome-wide variation in human gene expression. Nature. 2004;430:743–7. doi: 10.1038/nature02797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevins JR, Potti A. Mining gene expression profiles: expression signatures as cancer phenotypes. Nat Rev Genet. 2007;8:601–9. doi: 10.1038/nrg2137. [DOI] [PubMed] [Google Scholar]
- Perez-Tomas R. Multidrug resistance: retrospect and prospects in anti-cancer drug treatment. Curr Med Chem. 2006;13:1859–76. doi: 10.2174/092986706777585077. [DOI] [PubMed] [Google Scholar]
- Plimack ER, Stewart DJ, Issa JP. Combining epigenetic and cytotoxic therapy in the treatment of solid tumors. J Clin Oncol. 2007;25:4519–21. doi: 10.1200/JCO.2007.12.6029. [DOI] [PubMed] [Google Scholar]
- Potti A, Nevins JR. Utilization of genomic signatures to direct use of primary chemotherapy. Curr Opin Genet Dev. 2008;18:62–7. doi: 10.1016/j.gde.2008.01.018. [DOI] [PubMed] [Google Scholar]
- Rabik CA, Njoku MC, Dolan ME. Inactivation of O6-alkylguanine DNA alkyltransferase as a means to enhance chemotherapy. Cancer Treat Rev. 2006;32:261–76. doi: 10.1016/j.ctrv.2006.03.004. [DOI] [PubMed] [Google Scholar]
- Rakyan VK, Hildmann T, Novik KL, et al. DNA methylation profiling of the human major histocompatibility complex: a pilot study for the human epigenome project. PLoS Biol. 2004;2:e405. doi: 10.1371/journal.pbio.0020405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratain MJ. Finding the right dose. Clin Adv Hematol Oncol. 2003;1:517–8. 31. [PubMed] [Google Scholar]
- Ratain MJ. Personalized medicine: building the GPS to take us there. Clin Pharmacol Ther. 2007;81:321–2. doi: 10.1038/sj.clpt.6100092. [DOI] [PubMed] [Google Scholar]
- Redon R, Ishikawa S, Fitch KR, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–54. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roden DM, Altman RB, Benowitz NL, et al. Pharmacogenomics: challenges and opportunities. Ann Intern Med. 2006;145:749–57. doi: 10.7326/0003-4819-145-10-200611210-00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romkes M, Buch SC. Genotyping technologies: application to biotransformation enzyme genetic polymorphism screening. Methods Mol Biol. 2005;291:399–414. doi: 10.1385/1-59259-840-4:399. [DOI] [PubMed] [Google Scholar]
- Rudin CM, Liu W, Desai A, et al. Pharmacogenomic and pharmacokinetic determinants of erlotinib toxicity. J Clin Oncol. 2008;26:1119–27. doi: 10.1200/JCO.2007.13.1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki S, Kobunai T, Kitayama J, et al. DNA methylation and sensitivity to antimetabolites in cancer cell lines. Oncol Rep. 2008;19:407–12. [PubMed] [Google Scholar]
- Shames D, Minna J, Gazdar A. DNA methylation in health, disease, and cancer. Curr Mol Med. 2007;7:85–102. doi: 10.2174/156652407779940413. [DOI] [PubMed] [Google Scholar]
- Shen L, Kondo Y, Ahmed S, et al. Drug sensitivity prediction by CpG island methylation profile in the NCI-60 cancer cell line panel. Cancer Res. 2007;67:11335–43. doi: 10.1158/0008-5472.CAN-07-1502. [DOI] [PubMed] [Google Scholar]
- Singal R, Wang SZ, Sargent T, et al. Methylation of promoter proximal-transcribed sequences of an embryonic globin gene inhibits transcription in primary erythroid cells and promotes formation of a cell type-specific methyl cytosine binding complex. J Biol Chem. 2002;277:1897–905. doi: 10.1074/jbc.M105580200. [DOI] [PubMed] [Google Scholar]
- Spielman RS, Bastone LA, Burdick JT, et al. Common genetic variants account for differences in gene expression among ethnic groups. Nat Genet. 2007;39:226–31. doi: 10.1038/ng1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stranger BE, Forrest MS, Dunning M, et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007a;315:848–53. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stranger BE, Nica AC, Forrest MS, et al. Population genomics of human gene expression. Nat Genet. 2007b;39:1217–24. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strathdee G. Epigenetic markers and response to chemotherapy in cancer. Dis Markers. 2007;23:43–9. doi: 10.1155/2007/610815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twyman RM. SNP discovery and typing technologies for pharmacogenomics. Curr Top Med Chem. 2004;4:1423–31. doi: 10.2174/1568026043387656. [DOI] [PubMed] [Google Scholar]
- Vella A, Camilleri M. Pharmacogenetics: potential role in the treatment of diabetes and obesity. Expert Opin Pharmacother. 2008;9:1109–19. doi: 10.1517/14656566.9.7.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, et al. The sequence of the human genome. Science. 2001;291:1304–51. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Vucic EA, Brown CJ, Lam WL. Epigenetics of cancer progression. Pharmacogenomics. 2008;9:215–34. doi: 10.2217/14622416.9.2.215. [DOI] [PubMed] [Google Scholar]
- Wade PA, Archer TK. Epigenetics: environmental instructions for the genome. Environ Health Perspect. 2006;114:A140–1. doi: 10.1289/ehp.114-a140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Bhutani M, Pathak AK, et al. Delta DNMT3B variants regulate DNA methylation in a promoter-specific manner. Cancer Res. 2007;67:10647–52. doi: 10.1158/0008-5472.CAN-07-1337. [DOI] [PubMed] [Google Scholar]
- Warri A, Saarinen NM, Makela S, et al. The role of early life genistein exposures in modifying breast cancer risk. Br J Cancer. 2008;98:1485–93. doi: 10.1038/sj.bjc.6604321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M, Davies JJ, Wittig D, et al. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet. 2005;37:853–62. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- Yong WP, Innocenti F, Ratain MJ. The role of pharmacogenetics in cancer therapeutics. Br J Clin Pharmacol. 2006;62:35–46. doi: 10.1111/j.1365-2125.2006.02591.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon BJ, Herman H, Sikora A, et al. Regulation of DNA methylation of Rasgrf1. Nat Genet. 2002;30:92–6. doi: 10.1038/ng795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Bouffard GG, Wallace SS, et al. Estimation of DNA sequence context-dependent mutation rates using primate genomic sequences. J Mol Evol. 2007a;65:207–14. doi: 10.1007/s00239-007-9000-5. [DOI] [PubMed] [Google Scholar]
- Zhang W, Dolan ME. Ancestry-related differences in gene expression: findings may enhance understanding of health disparities between populations. Pharmacogenomics. 2008;9:489–92. doi: 10.2217/14622416.9.5.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Duan S, Kistner EO, et al. Evaluation of genetic variation contributing to differences in gene expression between populations. Am J Hum Genet. 2008a;82:631–40. doi: 10.1016/j.ajhg.2007.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Liu W, Poradosu E, et al. Genome-wide identification of genetic determinants for the cytotoxicity of perifosine. Hum Genomics. 2008b doi: 10.1186/1479-7364-3-1-53. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Ratain MJ, Dolan ME. The HapMap resource is providing new insights into ourselves and its application to pharmacogenomics. Bioinform Biol Insights. 2008c;2:15–23. doi: 10.4137/bbi.s455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Richards EJ, Borevitz JO. Genetic and epigenetic dissection of cis regulatory variation. Curr Opin Plant Biol. 2007b;10:142–8. doi: 10.1016/j.pbi.2007.02.002. [DOI] [PubMed] [Google Scholar]
- Zhang X, Shiu S, Cal A, et al. Global analysis of genetic, epigenetic and transcriptional polymorphisms in Arabidopsis thaliana using whole genome tiling arrays. PLoS Genet. 2008d;4:e1000032. doi: 10.1371/journal.pgen.1000032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Yazaki J, Sundaresan A, et al. Genome-wide high-resolution mapping and functional analysis of DNA methylation in arabidopsis. Cell. 2006;126:1189–201. doi: 10.1016/j.cell.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Zilberman D, Henikoff S. Genome-wide analysis of DNA methylation patterns. Development. 2007;134:3959–65. doi: 10.1242/dev.001131. [DOI] [PubMed] [Google Scholar]
- Zuo C, Ai L, Ratliff P, et al. O6-methylguanine-DNA methyltransferase gene: epigenetic silencing and prognostic value in head and neck squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev. 2004;13:967–75. [PubMed] [Google Scholar]