Abstract
The amyloid-β 42 (Aβ42) peptide has been suggested to promote tau phosphorylation and toxicity in Alzheimer's disease (AD) pathogenesis; however, the underlying mechanisms are not fully understood. Using transgenic Drosophila expressing both human Aβ42 and tau, we show here that tau phosphorylation at Ser262 plays a critical role in Aβ42-induced tau toxicity. Co-expression of Aβ42 increased tau phosphorylation at AD-related sites including Ser262, and enhanced tau-induced neurodegeneration. In contrast, formation of either sarkosyl-insoluble tau or paired helical filaments was not induced by Aβ42. Co-expression of Aβ42 and tau carrying the non-phosphorylatable Ser262Ala mutation did not cause neurodegeneration, suggesting that the Ser262 phosphorylation site is required for the pathogenic interaction between Aβ42 and tau. We have recently reported that the DNA damage-activated Checkpoint kinase 2 (Chk2) phosphorylates tau at Ser262 and enhances tau toxicity in a transgenic Drosophila model. We detected that expression of Chk2, as well as a number of genes involved in DNA repair pathways, was increased in the Aβ42 fly brains. The induction of a DNA repair response is protective against Aβ42 toxicity, since blocking the function of the tumor suppressor p53, a key transcription factor for the induction of DNA repair genes, in neurons exacerbated Aβ42-induced neuronal dysfunction. Our results demonstrate that tau phosphorylation at Ser262 is crucial for Aβ42-induced tau toxicity in vivo, and suggest a new model of AD progression in which activation of DNA repair pathways is protective against Aβ42 toxicity but may trigger tau phosphorylation and toxicity in AD pathogenesis.
INTRODUCTION
Alzheimer's disease (AD) is a progressive neurodegenerative disease without effective therapies (1,2). Pathologically, AD is defined by an extensive loss of neurons and by formation of two characteristic protein deposits in the brain, extracellular amyloid plaques and intracellular neurofibrillary tangles [NFTs (1)]. The major components of amyloid plaques are the 40 or 42 amino acid amyloid-β peptides (Aβ40 or Aβ42) (3,4). Aβ peptides are derived from a type 1 transmembrane protein, the amyloid precursor protein (APP), by sequential cleavage by β- and γ-secretases (5). Molecular genetic studies of early-onset familial AD patients have identified causative mutations in APP, Presenilin 1 and Presenilin 2 (6), which increase Aβ42 production and/or Aβ aggregation (7,8). These results provide a strong causative link between Aβ42 and AD (7).
NFTs are intracellular protein inclusions composed of the hyperphosphorylated microtubule-associated protein tau (9–12). NFTs are detected in many neurodegenerative diseases (13), and multiple tau gene mutations and polymorphisms are associated with tauopathies, including hereditary frontotemporal dementia and parkinsonism linked to chromosome 17 (13). Tau mutations have not been associated with any known form of familial AD to date; however, tau haplotypes driving slightly higher tau expression increase the AD risk (14,15), suggesting that tau plays a role in the pathogenesis of AD as a modulator of disease progression.
An imbalance in phosphorylation and/or dephosphorylation of tau has been suggested to initiate the abnormal metabolism and toxicity of tau in AD (13,16,17). At least 30 putative Ser/Thr phosphorylation sites in tau are phosphorylated in NFTs (16). In vitro and in vivo studies have demonstrated that tau phosphorylation at some of the disease-associated sites plays critical roles in tau binding to microtubules (18–20) and tau fibril formation (21–24). Approximately half of AD-related sites are targets for serine/proline (SP) or threonine/proline (TP) kinases (25). In transgenic animal models, overexpression of kinases that phosphorylate tau at SP/TP sites, including GSK-3β and Cdk5 modify tau phosphorylation, NFT formation and tau toxicity (26–33). In addition, phosphorylation of tau at AD-related, non-SP/TP sites such as Ser262/356 increases tau phosphorylation at SP/TP sites and promotes tau toxicity (34–36).
Accumulating evidence suggests that Aβ and tau synergistically contribute to the pathogenesis of AD (17). Studies in human AD cases following Aβ immunization have shown decreases in amyloid burden and in phosphorylated tau in neurites surrounding the amyloid plaques (37,38). In transgenic mice overproducing human Aβ and tau proteins, Aβ facilitates the abnormal phosphorylation of tau at AD-related sites and enhances the formation of NFTs (39–41). Aβ immunization removes amyloid pathology as well as early stage tau lesions (42), and ameliorates cognitive decline in transgenic mice that form plaques and tangles (43–45). Knockdown of tau expression suppresses Aβ-induced neurotoxicity in cultured neurons (46,47), and lowering or eliminating endogenous tau expression in transgenic mice suppresses Aβ-induced behavioral deficits (48). In a Drosophila model expressing human Aβ42 and tau, Aβ42 synergistically enhances tau-induced neurodegeneration, and tau phosphorylation at AD-related SP/TP sites is important for Aβ42-induced tau toxicity (49). These reports suggest that Aβ lies upstream of aberrant phosphorylation and toxicity of tau in the pathogenesis of AD. However, the molecular mechanisms by which Aβ induces abnormal phosphorylation and toxicity of tau in vivo are not fully understood.
The transgenic Drosophila models of tauopathy, in which human tau is overexpressed, have been used as effective genetic model systems to reveal the mechanisms underlying human tau-induced neurodegeneration (33,34,49–57). Accumulation of disease-associated conformational changes and phospho-epitopes in tau has been detected in the fly brain and eye (34,50,53). NFT formation is not observed in fly neurons (50), indicating that tau toxicity is not conferred by large insoluble aggregates of tau in the Drosophila models. These results suggest that Drosophila models of tauopathies may recapitulate early pre-tangle events in tau-associated neurodegeneration (58).
In order to study the pathogenic interactions between Aβ42- and tau-induced toxicity in vivo, we use a transgenic Drosophila expressing human tau (50) in combination with a transgenic Drosophila model of human Aβ42 toxicity (59–61) (Please see the ‘Materials and Methods’ section for the details of the Aβ42 fly model). Double transgenic flies expressing human Aβ42 and tau enable the examination of the effect of Aβ42 on tau pathology and toxicity, in the absence of the effects of APP and other fragments of APP including C-terminal and N-terminal fragments and other Aβ species.
Here we show that tau phosphorylation at Ser262 plays a critical role in Aβ42-induced tau toxicity. We also demonstrate that expression of DNA repair genes, including DNA damage-activated Checkpoint kinase 2 (Chk2), is increased in Aβ42 fly neurons as a protective response against Aβ42 toxicity. Since Chk2 phosphorylates tau at Ser262 and enhances tau toxicity (36), our results suggest that the increased activity of the DNA repair pathways in response to Aβ42 may be one of the mechanisms that mediate Aβ42-induced tau phosphorylation and toxicity in vivo.
RESULTS
Human Aβ42 enhances human tau-induced toxicity in transgenic fly eyes and brains
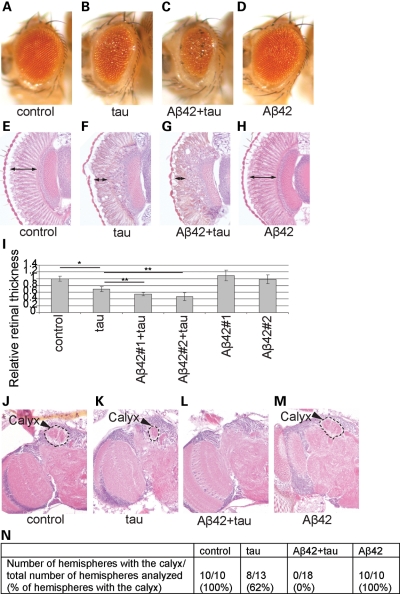
Expression of wild-type human tau (0N4R, see the ‘Materials and Methods’ section) in Drosophila eyes using the pan-retinal gmr-GAL4 driver causes eye degeneration characterized by small eye size, rough surface and reduced retinal thickness (33,50) (Fig. 1B, F and I). Co-expression of Aβ42 with tau significantly enhanced the reduction in the external size of eyes (Fig. 1C) as previously reported (49) and internal retina thickness (Fig. 1G and I), whereas Aβ42 expression alone did not significantly affect eye structures at this level of expression (Fig. 1D, H and I). Similar results were obtained from two independent Aβ42 transgenic fly lines (Aβ42#1 and Aβ42#2, Fig. 1I). These results indicate that Aβ42 expression exacerbates tau toxicity in vivo and that the fly eye can be used to investigate the molecular mechanisms of Aβ42-induced tau toxicity.
Figure 1.
Enhancement of human tau-induced neurodegeneration by Aβ42 in fly eyes and brains. (A–I) Enhancement of tau-induced retinal degeneration by Aβ42. External eyes (A–D) and internal retinal sections (E–H) from females (1 dae). Internal degeneration is apparent from the thickness of the retina, indicated by the double-headed arrows. (A and E) Eyes from control flies carrying the pan-retinal gmr-GAL4 driver only. (B and F) Expression of human tau in the eye reduces eye size and retinal thickness. (C and G) Eyes co-expressing human tau and Aβ42 have smaller eyes and thinner retinas than flies expressing tau alone. (D and H) Expression of Aβ42 alone does not change eye size or retinal thickness. (I) Retinal thickness was quantified and shown as a ratio relative to the control. Similar results were obtained from two independent Aβ42 transgenic fly lines (Aβ42#1 and Aβ42#2) [mean ± SD, n = 6–8, * and **P < 0.05 (Student's t-test)]. (J–N) Enhancement of human tau-induced defects in brain structures by co-expression of Aβ42. Sections containing the calyx neuropil of the mushroom body in brains of 10 dae male flies. (J) Control flies carrying the pan-neuronal elav-GAL4 driver only. The calyx is indicated by an arrowhead and hatched line. (K) Flies expressing tau have a smaller calyx or occasionally lack the calyx. (L) Flies co-expressing tau and Aβ42 do not have the calyx. (M) Flies expressing Aβ42 alone have a normal calyx structure. (N) The percentage of the hemispheres with the calyx neuropil in each genotype.
Expression of tau in neurons by the pan-neuronal elav-GAL4 driver caused an abnormality in fly brain structures. The mushroom body structures are paired structures, formed from approximately 2500 cells in each hemisphere, which play a central role in olfactory learning and memory in flies (Fig. 1J) (62). The calyx is a dendritic region in the mushroom body, and the calyx neuropil in flies expressing tau were smaller than those in controls (Fig. 1K) or were missing in some of the fly brains (Fig. 1N) (63). Co-expression of Aβ42 and tau caused a complete loss of calyx structures (Fig. 1L and N). In contrast, a normal calyx structure was observed in flies expressing Aβ42 alone (Fig. 1M and N). These results indicate that Aβ42 enhances tau-induced toxicity also in fly brain neurons.
Human Aβ42 increases human tau phosphorylation at Ser202, Thr231 and Ser262 in transgenic fly eyes and brains
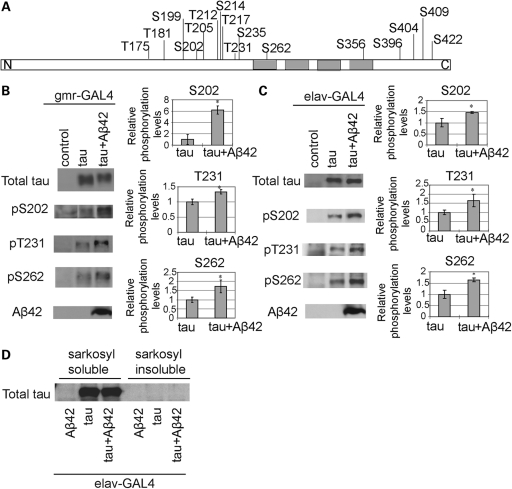
We examined whether enhancement of tau-induced toxicity by co-expression of Aβ42 in fly eyes and brains was accompanied by an increase in tau phosphorylation levels at AD-related sites. Tau phosphorylation at 16 AD-related sites (indicated in Fig. 2A), in the presence or absence of Aβ42 expression, was examined by western blotting using phospho-tau specific antibodies. This systematic analysis revealed that tau phosphorylation levels were significantly increased at Ser202, Thr231 and Ser262 when tau was co-expressed with Aβ42 in both eyes (using the gmr-GAL4 driver, Fig. 2B) and brains (using the pan-neuronal elav-GAL4 driver, Fig. 2C). These results were confirmed in three independent transgenic fly lines carrying Aβ42, and using two different antibodies to detect Ser202, Thr231 and Ser262 phosphorylation.
Figure 2.
Increase of human tau phosphorylation at Ser202, Thr231 and Ser262 by Aβ42 in fly eyes and brains. (A) Schematic representation of the structure of tau. Positions of the phosphorylation sites tested in this study are shown. Gray boxes, four repeats of the microtubule-binding domain. (B and C) Fly heads expressing human tau alone (tau) or with Aβ42 (tau + Aβ42) driven by the pan-retinal gmr-GAL4 at 1 dae (B) or pan-neuronal elav-GAL4 at 25 dae (C) were subjected to western blotting with anti-tau (total tau), anti-phospho-tau (pS202, pT231 and pS262) and anti-Aβ42 antibodies. Flies carrying the driver only were used as the negative control (control). The phosphorylation levels in the eye and brain of flies co-expressing tau and Aβ42 (tau + Aβ42) are shown as a ratio relative to that in flies expressing tau alone (tau). Representative blots are shown. Asterisks indicate significant differences from tau alone (tau) [n = 4 or 5, *P < 0.05 (Student's t-test)]. (D) Co-expression of Aβ42 did not increase sarkosyl-insoluble tau in the fly brain. Western blotting of sarkosyl-soluble and -insoluble fractions of head extracts from flies expressing tau alone (tau), or tau and Aβ42 (tau + Aβ42), driven by the pan-neuronal elav-GAL4 driver. Head extracts from flies expressing Aβ42 alone (Aβ42) was used as a negative control. Flies are at 35 dae.
To examine whether Aβ42 affects tau solubility in fly brains, fly brains expressing human tau in the presence or absence of Aβ42 were extracted with sarkosyl, and the sarkosyl-soluble and -insoluble fractions were subjected to western blotting with anti-human tau antibody. Expression of Aβ42 did not affect the distribution of tau in the sarkosyl-soluble and -insoluble fractions (Fig. 2D). Moreover, paired helical filaments were not detected in the sarkosyl-insoluble fractions from fly brains co-expressing Aβ42 and tau by transmission electron microscopy.
The Ser262 phosphorylation site of tau is critical for the pathogenic interaction between Aβ42 and tau in transgenic flies
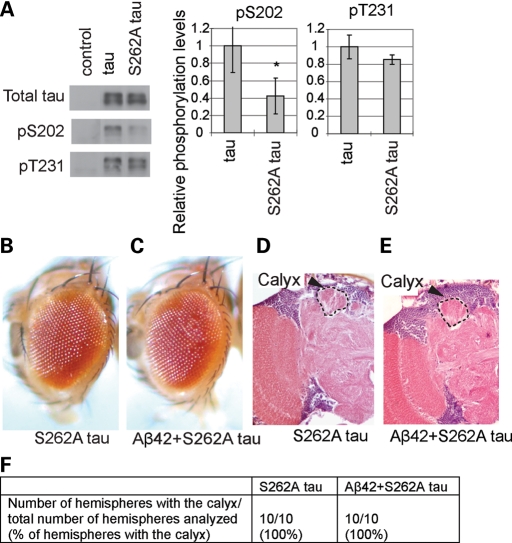
Tau phosphorylation at Ser262 has been shown to play a critical role in tau toxicity (34–36). To test whether the Ser262 phosphorylation site is required for the pathogenic interaction between Aβ42 and tau, we have established transgenic fly lines carrying human tau with an alanine mutation at the Ser262 site (S262A tau), which express comparable levels of S262A tau to wild-type human tau (Fig. 3A) (36). Consistent with the previous report using S262A/S356A tau (34), phosphorylation at Ser202 (AT8 epitope) was found to be significantly lower in S262A tau than in wild-type tau, whereas phosphorylation at Thr231 (AT180 epitope) was not significantly altered (Fig. 3A).
Figure 3.
The Ser262 phosphorylation site of tau is critical for the pathogenic interaction between Aβ42 and tau. (A) Effect of S262A mutation on tau phosphorylation at Ser202 and Thr231. Western blot of head lysates of 1 dae females with anti-tau, and anti-phospho-tau antibodies (pS202 and pT231). Control: flies carrying the pan-retinal gmr-Gal4 driver only. Phosphorylation of S262A tau at Ser202 or Thr231 is shown as a ratio relative to that of wild-type tau. Asterisks indicate significant differences from wild-type tau [n = 3 or 4, *P < 0.05 (Student's t-test)]. (B and C) Co-expression of Aβ42 and S262A tau using the pan-retinal gmr-GAL4 driver did not cause eye degeneration. External eyes of 1 dae females expressing S262A tau (B, S262A tau) or S262A tau with Aβ42 (C, Aβ42 + S262A tau). (D–F) Co-expression of Aβ42 and S262A tau using the pan-neuronal elav-GAL4 driver did not cause structural defects in the brains. Sections containing the calyx neuropil of the mushroom body (indicated by arrowheads and hatched line) in brains of 10 dae male flies. (D) Flies expressing S262A tau. (E) Flies co-expressing Aβ42 and S262A tau. (F) The percentage of the hemispheres with the calyx neuropil in each genotype.
The S262A single mutant dramatically suppressed tau toxicity in the retina as reported previously (Fig. 3B) (36), and also in the brain (Fig. 3D and F). Co-expression of Aβ42 and S262A tau using the pan-retinal gmr-GAL4 driver did not cause any reduction in eye size (Fig. 3C). In addition, co-expression of Aβ42 and S262A tau by the pan-neuronal elav-GAL4 driver did not cause structural defects in the mushroom body (Fig. 3E and F). These results indicate that the Ser262 site is critical for the pathogenic interaction between Aβ42 and tau.
Expression of Chk2, as well as a number of genes involved in DNA repair pathways, is increased by human Aβ42 expression in transgenic fly brains
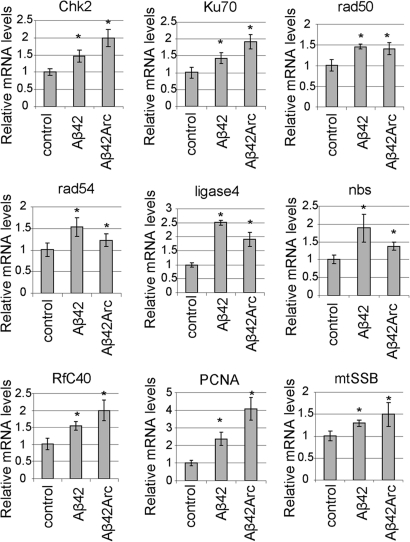
We have recently shown that the human DNA damage-activated Chk2 phosphorylate tau at Ser262 in vitro (36). Overexpression of Drosophila Chk2 enhances tau toxicity, and the Ser262 phosphorylation site is critical for the toxic interaction between Chk2 and tau in a transgenic fly model (36). Using quantitative real-time PCR, we found that mRNA levels of Chk2 were increased in the fly brain by the expression of human Aβ42 in neurons (Fig. 4). The increase in Chk2 mRNA levels was more prominent in the fly brains expressing Aβ42 with the familial Alzheimer's disease Arctic mutation (Aβ42Arc: E22G substitution), which shows more enhanced accumulation and toxicity than Aβ42 (60) (Fig. 4). These results suggest that the Aβ42-induced increase in Chk2 expression may underlie the increase in tau phosphorylation and the enhancement of tau toxicity.
Figure 4.
Aβ42-induced increases in expression of Chk2 and a number of DNA repair pathway genes in fly brains. mRNA levels in heads from flies (25 dae) expressing Aβ42 or Aβ42 with the familial Aβ42Arc driven by the pan-neuronal elav-GAL4 driver are shown as ratios relative to controls (flies carrying the elav-Gal4 driver only) [mean ± SD, n = 5, *P < 0.05 (Student's t-test)]. Relative mRNA levels of genes encoding Drosophila homologs of Chk2, and genes involved in DNA repair pathways (Ku70, Rad50, Rad54, Ligase 4, nbs, RfC40, PCNA and mtSSB), are increased in both Aβ42 and Aβ42Arc fly brains.
We examined whether a genetic reduction of Chk2 ameliorates the enhancement of tau toxicity caused by Aβ42. Because the homozygous null mutants of Chk2 are lethal (64), we tested the effect of a heterozygous loss-of-function mutation of Chk2 on tau-induced retinal degeneration in the presence or absence of Aβ42. A heterozygous loss-of-function mutation of Chk2 did not significantly suppress the enhancement of tau-induced retinal degeneration caused by Aβ42 (data not shown), suggesting that loss of one copy of Chk2 is not sufficient to reduce the enhancement of tau toxicity caused by Aβ42.
Why is Chk2 expression upregulated in the Aβ42 fly brain? Chk2 is a DNA damage transducer, which is activated in response to double-strand breaks in DNA (65,66). The non-homologous end-joining DNA repair pathway is the major repair pathway for DNA double-strand breaks in post-mitotic neurons (67). Genes involved in non-homologous end-joining DNA repair pathways (Ku70, rad50, rad54 and Ligase 4) and Nijmegen breakage syndrome (nbs), a component of the double-strand break sensor MRN complex, were upregulated in Aβ42 and Aβ42Arc fly brains (Fig. 4F). Increased expression of the replication factor C subunit 40 (RfC40) and proliferating cell nuclear antigen (PCNA), which is important for both DNA synthesis and DNA repair (68) and is abnormally re-expressed in human AD brains and animal models of AD (51,69,70), was also detected. In addition, expression of the Drosophila homologs of genes involved in direct repair (O-6-alkylguanine-DNA alkyltransferase), base excision repair (XRCC1) and mitochondrial single-strand break repair (mtSSB) was upregulated in both Aβ42 and Aβ42Arc flies (Fig. 4 and data not shown). These results suggest that an increase in expression of Chk2 is a part of the DNA repair response in Aβ42 flies.
Activation of DNA repair pathways is a protective response against Aβ42-induced toxicity
While genes involved in the DNA repair response are upregulated (Fig. 4), damaged DNA and apoptosis were not detected in the brains of flies expressing Aβ42, as indicated by TUNEL staining (71) and EM analysis (60). These results suggest that expression of DNA repair genes are induced as a protective response against Aβ42 toxicity. We tested this possibility by blocking the function of the tumor suppressor p53, which is a transcription factor that regulates DNA damage-induced transcription (72–74). Drosophila p53 regulates induction of pro-apoptotic genes and DNA repair genes, including components of the non-homologous end-joining repair pathway, after DNA damage (75). Two dominant negative forms of p53 have been used to disrupt p53 functions in Drosophila (76). DN-p53-259H carries a point mutation in the p53 DNA-binding domain, and DN-p53-Ct is a C-terminal p53 fragment. Both mutants form tetramers with endogenous p53, but fail to bind DNA and disrupt p53 functions (76).
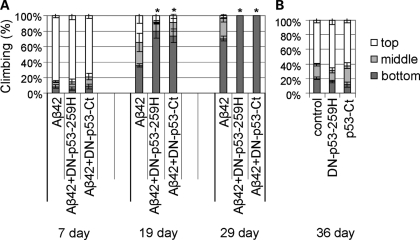
To test whether a reduction of p53 function would enhance Aβ42 toxicity, we examined the effect of neuronal expression of the dominant negative forms of p53 on Aβ42-induced locomotor defects. Aβ42 flies show age-dependent, progressive locomotor dysfunction starting around two weeks after eclosion, which can be detected by a climbing assay (59,60). In this assay, flies were placed in an empty plastic vial and tapped to the bottom. The number of flies at the top, middle or bottom of the vial was scored after 10 s. The neuronal expression of DN-p53-259H or DN-p53-Ct enhanced the locomotor defects induced by Aβ42 (Fig. 5A, 19 day and 29 day). In contrast, neuronal expression of DN-p53-259H or DN-p53-Ct alone did not cause locomotor defects at up to the age of 36 days after eclosion (Fig. 5B). These results indicate that the induction of DNA repair responses is protective against Aβ42 toxicity.
Figure 5.
Enhancement of Aβ42-induced locomotor defects by neuronal expression of dominant negative forms of p53. The average percentage of flies at the top (white), middle (light gray) or bottom (dark gray) of assay vials is shown (mean ± SD, n = 5). (A) The effect of overexpression of DN-p53-259H or DN-p53-Ct by the pan-neuronal elav-GAL4 on Aβ42-induced locomotor defects in flies at 7, 19 and 29 dae. Asterisks indicate the significant differences in the percentage of flies that stayed at the bottom [*P < 0.05 (Student's t-test)]. (B) In the absence of Aβ42, overexpression of DN-p53-259H or DN-p53-Ct does not cause any locomotor defects even at 36 dae.
DISCUSSION
Elucidation of the mechanisms by which Aβ42 induces abnormal phosphorylation and toxicity of tau is crucial to understanding the complex pathogenesis of AD. We have demonstrated here that, in transgenic Drosophila expressing both human Aβ42 and tau, Aβ42 increases tau phosphorylation at AD-related sites including Ser262 and enhances tau-induced neurodegeneration (Figs 1 and 2). Co-expression of Aβ42 and tau carrying the non-phosphorylatable Ser262Ala mutation did not cause neurodegeneration (Fig. 3), suggesting that the Ser262 phosphorylation site is required for the pathogenic interaction between Aβ42 and tau.
Tau phosphorylation at Ser262 is increased in pre-tangle neurons in AD (77,78). Increased tau phosphorylation at Ser262 is observed in cellular and animal models such as the cultured neurons treated with Aβ42 (79), brains of double transgenic mice expressing human APP and tau (80), and monkey cortex after injection of Aβ (81). Our results are consistent with these reports and suggest that the double transgenic fly model co-expressing human Aβ42 and tau recapitulates a pathological phosphorylation of tau induced by Aβ42 in mammalian neurons. In transgenic mice overproducing human Aβ and tau, Aβ enhances the formation of NFT (39–41). In contrast, in the double transgenic fly model, neither sarkosyl-insoluble tau nor PHF tau was detected (Fig. 2). These results suggest that large tau aggregates are not involved in the enhancement of Aβ42-induced tau toxicity in the transgenic fly model.
What are the mechanisms by which Aβ42 enhances tau toxicity through Ser262 phosphorylation? Ser262 is located in the microtubule-binding domain of tau, and phosphorylation at Ser262 reduces tau binding to microtubules (19), which may increase the chances of abnormal phosphorylation at other AD-related sites and, consequently, enhance tau toxicity (17,82,83). In the Drosophila model, tau phosphorylation at Ser262 triggers a temporally ordered series of phosphorylations at several proline-directed kinase target sites (SP/TP sites) and generates disease-associated phospho-epitopes (34). In the double transgenic fly model, we detected that phosphorylation of tau at two of the SP/TP sites, Ser202 and Thr231, was increased by Aβ42 (Fig. 2). Moreover, studies of transgenic flies co-expressing Aβ42 and tau have revealed that phosphorylation at AD-related SP/TP sites is involved in Aβ42-induced tau toxicity (49,84). These results suggest that the increase in tau phosphorylation at SP/TP sites followed by Ser262 phosphorylation may be one of the mechanisms underlying Aβ42-induced enhancement of tau toxicity. Interestingly, a recent study has shown that the introduction of the S262A/S356A mutation to tau can suppress the toxicity of tau hyperphosphorylated at SP/TP sites (35). This raises a possibility that tau phosphorylation at Ser262 affects tau toxicity not only by increasing tau phosphorylation at SP/TP sites but also controlling toxicity of tau phosphorylated at SP/TP sites.
Widespread single and double-strand DNA breaks have been detected in neurons in the brains of patients with AD and with mild cognitive impairment (85–98). More DNA damage was found in the aging hippocampus, one of the vulnerable regions of the brain in AD, than in the aging cerebellum (99). In postmortem brains from patients, the neurons that show NFT formation in AD are the same as those that show age-related accumulation of DNA damage (100). The Aβ42 peptide is known to cause oxidative stress (101,102), and damage to nucleic acids caused by reactive oxygen species includes base modifications such as 8-hydroxydeoxyguanosine, single-strand breaks and double-strand breaks if single-strand breaks are in close proximity (103). Expression of genes involved in DNA repair responses, including the DNA damage-activated Chk2, was increased in response to human Aβ42 expression in the fly brain (Fig. 4). Since Chk2 phosphorylates tau at Ser262 and enhances tau toxicity in a transgenic Drosophila model (36), these results suggest that increased expression of the DNA repair transducer Chk2 may be one of the mechanisms underlying Aβ42-induced phosphorylation and toxicity of tau in vivo.
While genes involved in the DNA repair response are upregulated (Fig. 4), damaged DNA and apoptosis were not detected in the brains of flies expressing Aβ42 (60,71). Furthermore, blocking the function of p53, which mediates expression of DNA repair genes, enhanced Aβ42-induced behavioral deficits (Fig. 5). These results suggest that the upregulation of genes involved in DNA repair pathways is protective against Aβ42 toxicity, but the increased activity of DNA damage-activated kinases such as Chk2 may cause tau phosphorylation and toxicity in AD progression.
In summary, this study has demonstrated that tau phosphorylation at Ser262 is critical for Aβ42-induced tau toxicity. Additionally, our results suggest that the activation of DNA damage-activated kinases by Aβ42 may be involved in the pathogenic interaction between Aβ42 and tau. Increases in DNA repair gene expression have been reported in aged brains (104) and in brains from Down's syndrome patients (105), and DNA repair efficiency is changed in AD brains (90,106–112). The DNA damage-activated Chk1 and Chk2 are expressed in post-mitotic neurons in the brain (113,114), and it will be important to investigate whether Chk1 and Chk2 are activated in AD brains.
MATERIALS AND METHODS
Transgenic fly models of Aβ42 toxicity
We have established the transgenic fly line carrying human Aβ42 and Aβ42 with Arctic mutation, which has been described previously in detail (59,60,115). Briefly, to produce human Aβ42 in the secretory pathway of fly neurons, the Aβ42 peptide sequence is directly fused to a secretion signal peptide at the N-terminus. Mass spectrometry analysis has revealed that the Aβ42 transgenic flies produce the intact human Aβ42 peptide in the fly brain (59,60), and immuno-electron microscopy has shown that the expressed Aβ42 is localized to the secretory pathways in neurons in the fly brain (60). These Aβ42 flies show late-onset, progressive short-term memory defects, locomotor dysfunctions, neurodegeneration and premature death, accompanied by the formation of Aβ42 deposits (59–61).
Fly stocks
The transgenic fly line carrying the human 0N4R tau, which has four tubulin-binding domains (R) at the C-terminal region and no N-terminal insert (N), was a kind gift from Dr Mel Feany (Harvard Medical School) (50). We have previously established the transgenic fly lines carrying S262A mutant tau (36). Other fly stocks were obtained from: Drs Wei Du (the University of Chicago) (Chk2[E51]) and the Bloomington Drosophila Stock Center (Indiana University) (UAS-DN-p53-Ct, UAS-DN-p53-259H, gmr-GAL4 and elav-GAL4). Crosses were maintained on standard cornmeal-based Drosophila medium at 25°C.
Histological analysis
To analyze internal eye structure, heads of female flies at 1 day-after-eclosion (dae) were fixed in Bouin's fixative (EMS) for 48 h at room temperature, incubated 24 h in 50 mm Tris/150 mm NaCl and embedded in paraffin. Serial sections (6 μm thickness) through the entire heads were prepared, stained with hematoxylin and eosin (Vector), and examined by bright-field microscopy. Images of the sections that include the retinal were captured, and retina thickness was measured using Image J. To analyze fly brain structures, paraffin sections of heads of 1 dae males were prepared as described previously (115). Heads from five to ten flies were analyzed for each genotype.
Western blotting
Total tau was probed with anti-tau monoclonal antibody (Tau46, Zymed), and phosphorylated tau was probed with phospho-tau specific antibodies against phospho-Thr175/181 (AT270, Pierce), phospho-Ser199 (Biosource), phospho-Ser202 (CP13, a kind gift from Dr Peter Davis, and Biosource), phospho-Thr205 (Biosource), phospho-Thr212 (Biosource), phospho-Ser214 (Biosource), phospho-Thr217 (Biosource), phospho-Thr231 (AT180, Thermo and Endogen), phospho-Ser235 (Biosource), phospho-Ser262 (Biosource and Calbiochem), phospho-Ser356 (Biosource), phospho-Ser396/404 (PHF1, a kind gift from Dr Peter Davis), phospho-Ser409 (Biosource) and phospho-Ser422 (Biosource). Fifteen fly heads for each genotype were collected at 1–3 dae and homogenized in SDS–Tris–Glycine sample buffer, separated by 10% Tris–Glycine gel and transferred to nitrocellulose membrane. The membranes were blocked with 5% milk (Nestle), blotted with the antibodies described above, incubated with appropriate secondary antibody and developed using ECL plus Western Blotting Detection Reagents (GE Healthcare). The signal intensity was quantified using ImageJ (NIH). Western blots were repeated a minimum of three times with different animals and representative blots are shown.
Extraction of sarkosyl-soluble tau
Sarcosyl-insoluble tau was prepared as described in (50,116,117). Briefly, fly heads were homogenized in 10 volumes buffer and centrifuged for 20 min at 15 000g. The supernatant was brought to 1% N-lauroylsarcosinate, incubated for 1 h at room temperature with shaking and then further centrifuged for 1 h at 100 000g. The resultant high-speed pellet was re-suspended at 10 μl per 50 mg of starting material. Tau levels in the sarcosyl-soluble and -insoluble fraction were analyzed by western blot. This sarcosyl-insoluble fraction was subjected to transmission electron microscopy. Flies were at 35 dae.
RNA extraction and quantitative real-time PCR analysis
For each sample, 30–40 flies were collected and frozen. Heads were mechanically isolated, and total RNA was extracted using TRIzol (Invitrogen) according to the manufacturer's protocol with an additional centrifugation step (11000g for 10 min) to remove cuticle membranes prior to the addition of chloroform. Total RNA was reverse-transcribed using Superscript II reverse transcriptase (Invitrogen), and the resulting cDNA was used as a template for PCR on a 7500 fast real-time PCR system (Applied Biosystems). The average threshold cycle value (Ct) was calculated from five replicates per sample. Expression of genes of interest was standardized relative to TBP. Relative expression values were determined by the deltaCt method according to quantitative PCR Analysis User Bulletin (Applied Biosystems). Primers were designed using Primer Express 3.0 (Applied Biosystems): Chk2 for 5′-AAACTGGGCTGCTGCTTCAC-3′, Chk2 rev 5′-GCGGAATGGTTTGCTGAAGA-3′, Ku70 for 5′-CAGCCGAATCTCATCAACGA-3′, Ku70 rev 5′-GTCCGGCAGCAGCTTTTCTA-3′, Rad50 for 5′-ATGTGAAGCCCTGAATTGCAT-3′, Rad50 rev 5′-ACCGACTTGCACTCCTCGTT-3′, Rad54 for 5′-AGAGGCCTGCCTGACAATATTC-3′, Rad54 rev 5′-GCGGCTTTCTGGTTTTTCCT-3′, Ligase 4 for 5′-GGCTGGCATAACCGCATCTA-3′, Ligase 4 rev 5′-CCGCCGTTTTGAAGAAACA-3′, nbs for 5′-TGCGCATGTGCAACTTAAACA-3′, nbs rev 5′-GCGCAACAATTCGCTTTTG-3′, RfC40 for 5′-GCGATGCTACGATTCACCAA-3′, RfC40 rev 5′-GTTGTTCAGTCCCTGTCGCATA-3′, PCNA for 5′-TTCAGCGAATCCGTTGTGATC-3′, PCNA rev 5′-CACCGGCTCCTGCATCTC-3′, mtSSB for 5′-CCGGTGGTCACCTTTTCG-3′, mtSSB rev 5′-GGTTCGCTGTCCCTTCTTCA-3′, TBP for 5′-GCGGCTGTGATTATGCGAAT-3′, TBP rev 5′-AGGGAAACCGAGCTTTTGGA-3′.
Climbing assay
The climbing assay was performed as previously described (60). Approximately 25 flies were placed in an empty plastic vial. The vial was gently tapped to knock the flies to the bottom, and the number of flies at the top, middle, or bottom of the vial was scored after 10 s. Experiments were repeated more than three times, and a representative result was shown.
FUNDING
This work was supported by start-up funds from the Farber Institute for Neurosciences, a pilot research grant from the Thomas Jefferson University, and in part by grants from the Gilbert Foundation/American Federation for Aging Research, the Alzheimer's Association (NIRG-08-91985) and the National Institutes of Health (R01AG032279-A1).
ACKNOWLEDGEMENTS
We thank Drs Peter Davis, Wei Du, Mel Feany, Yi Zhong and the Bloomington stock center for fly stocks and antibodies. We thank Dr Stephen Hearn for transmission electron microscopy analysis, Dr Miki Fujioka for her help with establishing the transgenic fly strains and Dr Nancy Bonini for technical advice on the histological analysis of Drosophila eyes. We thank Linda Granger, Christine Hostetter, Christopher Shenton and LiJuan Zhao for technical help.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Selkoe D.J. Alzheimer's disease: genes, proteins, and therapy. Physiol. Rev. 2001;81:741–766. doi: 10.1152/physrev.2001.81.2.741. [DOI] [PubMed] [Google Scholar]
- 2.Cummings J.L. Cognitive and behavioral heterogeneity in Alzheimer's disease: seeking the neurobiological basis. Neurobiol. Aging. 2000;21:845–861. doi: 10.1016/s0197-4580(00)00183-4. doi:10.1016/S0197-4580(00)00183-4. [DOI] [PubMed] [Google Scholar]
- 3.Glenner G.G., Wong C.W. Alzheimer's disease and Down's syndrome: sharing of a unique cerebrovascular amyloid fibril protein. Biochem. Biophys. Res. Commun. 1984;122:1131–1135. doi: 10.1016/0006-291x(84)91209-9. doi:10.1016/0006-291X(84)91209-9. [DOI] [PubMed] [Google Scholar]
- 4.Masters C.L., Simms G., Weinman N.A., Multhaup G., McDonald B.L., Beyreuther K. Amyloid plaque core protein in Alzheimer disease and Down syndrome. Proc. Natl. Acad. Sci. USA. 1985;82:4245–4249. doi: 10.1073/pnas.82.12.4245. doi:10.1073/pnas.82.12.4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sisodia S.S., St George-Hyslop P.H. Gamma-Secretase, Notch, Abeta and Alzheimer's disease: where do the presenilins fit in? Nat. Rev. Neurosci. 2002;3:281–290. doi: 10.1038/nrn785. doi:10.1038/nrn785. [DOI] [PubMed] [Google Scholar]
- 6.Bertram L., Tanzi R.E. The genetic epidemiology of neurodegenerative disease. J. Clin. Invest. 2005;115:1449–1457. doi: 10.1172/JCI24761. doi:10.1172/JCI24761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tanzi R.E., Bertram L. 20 years of the Alzheimer's disease amyloid hypothesis: a genetic perspective. Cell. 2005;120:545–555. doi: 10.1016/j.cell.2005.02.008. doi:10.1016/j.cell.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 8.Rovelet-Lecrux A., Hannequin D., Raux G., Le Meur N., Laquerriere A., Vital A., Dumanchin C., Feuillette S., Brice A., Vercelletto M., et al. APP locus duplication causes autosomal dominant early-onset Alzheimer disease with cerebral amyloid angiopathy. Nat. Genet. 2006;38:24–26. doi: 10.1038/ng1718. doi:10.1038/ng1718. [DOI] [PubMed] [Google Scholar]
- 9.Grundke-Iqbal I., Iqbal K., Quinlan M., Tung Y.C., Zaidi M.S., Wisniewski H.M. Microtubule-associated protein tau. A component of Alzheimer paired helical filaments. J. Biol. Chem. 1986;261:6084–6089. [PubMed] [Google Scholar]
- 10.Grundke-Iqbal I., Iqbal K., Tung Y.C., Quinlan M., Wisniewski H.M., Binder L.I. Abnormal phosphorylation of the microtubule-associated protein tau (tau) in Alzheimer cytoskeletal pathology. Proc. Natl. Acad. Sci. USA. 1986;83:4913–4917. doi: 10.1073/pnas.83.13.4913. doi:10.1073/pnas.83.13.4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kosik K.S., Joachim C.L., Selkoe D.J. Microtubule-associated protein tau (tau) is a major antigenic component of paired helical filaments in Alzheimer disease. Proc. Natl. Acad. Sci. USA. 1986;83:4044–4048. doi: 10.1073/pnas.83.11.4044. doi:10.1073/pnas.83.11.4044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wood J.G., Mirra S.S., Pollock N.J., Binder L.I. Neurofibrillary tangles of Alzheimer disease share antigenic determinants with the axonal microtubule-associated protein tau (tau) Proc. Natl. Acad. Sci. USA. 1986;83:4040–4043. doi: 10.1073/pnas.83.11.4040. doi:10.1073/pnas.83.11.4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee V.M., Goedert M., Trojanowski J.Q. Neurodegenerative tauopathies. Annu. Rev. Neurosci. 2001;24:1121–1159. doi: 10.1146/annurev.neuro.24.1.1121. doi:10.1146/annurev.neuro.24.1.1121. [DOI] [PubMed] [Google Scholar]
- 14.Myers A.J., Kaleem M., Marlowe L., Pittman A.M., Lees A.J., Fung H.C., Duckworth J., Leung D., Gibson A., Morris C.M., et al. The H1c haplotype at the MAPT locus is associated with Alzheimer's disease. Hum. Mol. Genet. 2005;14:2399–2404. doi: 10.1093/hmg/ddi241. doi:10.1093/hmg/ddi241. [DOI] [PubMed] [Google Scholar]
- 15.Kauwe J.S., Cruchaga C., Mayo K., Fenoglio C., Bertelsen S., Nowotny P., Galimberti D., Scarpini E., Morris J.C., Fagan A.M., et al. Variation in MAPT is associated with cerebrospinal fluid tau levels in the presence of amyloid-beta deposition. Proc. Natl. Acad. Sci. USA. 2008;105:8050–8054. doi: 10.1073/pnas.0801227105. doi:10.1073/pnas.0801227105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buee L., Bussiere T., Buee-Scherrer V., Delacourte A., Hof P.R. Tau protein isoforms, phosphorylation and role in neurodegenerative disorders. Brain. Res. Brain. Res. Rev. 2000;33:95–130. doi: 10.1016/s0165-0173(00)00019-9. doi:10.1016/S0165-0173(00)00019-9. [DOI] [PubMed] [Google Scholar]
- 17.Ballatore C., Lee V.M., Trojanowski J.Q. Tau-mediated neurodegeneration in Alzheimer's disease and related disorders. Nat. Rev. Neurosci. 2007;8:663–672. doi: 10.1038/nrn2194. doi:10.1038/nrn2194. [DOI] [PubMed] [Google Scholar]
- 18.Biernat J., Gustke N., Drewes G., Mandelkow E.M., Mandelkow E. Phosphorylation of Ser262 strongly reduces binding of tau to microtubules: distinction between PHF-like immunoreactivity and microtubule binding. Neuron. 1993;11:153–163. doi: 10.1016/0896-6273(93)90279-z. doi:10.1016/0896-6273(93)90279-Z. [DOI] [PubMed] [Google Scholar]
- 19.Sengupta A., Kabat J., Novak M., Wu Q., Grundke-Iqbal I., Iqbal K. Phosphorylation of tau at both Thr 231 and Ser 262 is required for maximal inhibition of its binding to microtubules. Arch. Biochem. Biophys. 1998;357:299–309. doi: 10.1006/abbi.1998.0813. doi:10.1006/abbi.1998.0813. [DOI] [PubMed] [Google Scholar]
- 20.Cho J.H., Johnson G.V. Glycogen synthase kinase 3beta phosphorylates tau at both primed and unprimed sites. Differential impact on microtubule binding. J. Biol. Chem. 2003;278:187–193. doi: 10.1074/jbc.M206236200. doi:10.1074/jbc.M206236200. [DOI] [PubMed] [Google Scholar]
- 21.Alonso A.C., Grundke-Iqbal I., Iqbal K. Alzheimer's disease hyperphosphorylated tau sequesters normal tau into tangles of filaments and disassembles microtubules. Nat. Med. 1996;2:783–787. doi: 10.1038/nm0796-783. doi:10.1038/nm0796-783. [DOI] [PubMed] [Google Scholar]
- 22.Necula M., Kuret J. Pseudophosphorylation and glycation of tau protein enhance but do not trigger fibrillization in vitro. J. Biol. Chem. 2004;279:49694–49703. doi: 10.1074/jbc.M405527200. doi:10.1074/jbc.M405527200. [DOI] [PubMed] [Google Scholar]
- 23.Alonso Adel C., Mederlyova A., Novak M., Grundke-Iqbal I., Iqbal K. Promotion of hyperphosphorylation by frontotemporal dementia tau mutations. J. Biol. Chem. 2004;279:34873–34881. doi: 10.1074/jbc.M405131200. [DOI] [PubMed] [Google Scholar]
- 24.Brunden K.R., Trojanowski J.Q., Lee V.M. Advances in tau-focused drug discovery for Alzheimer's disease and related tauopathies. Nat. Rev. Drug. Discov. 2009;8:783–793. doi: 10.1038/nrd2959. doi:10.1038/nrd2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gong C.X., Liu F., Grundke-Iqbal I., Iqbal K. Post-translational modifications of tau protein in Alzheimer's disease. J. Neural. Transm. 2005;112:813–838. doi: 10.1007/s00702-004-0221-0. doi:10.1007/s00702-004-0221-0. [DOI] [PubMed] [Google Scholar]
- 26.Spittaels K., Van den Haute C., Van Dorpe J., Geerts H., Mercken M., Bruynseels K., Lasrado R., Vandezande K., Laenen I., Boon T., et al. Glycogen synthase kinase-3beta phosphorylates protein tau and rescues the axonopathy in the central nervous system of human four-repeat tau transgenic mice. J. Biol. Chem. 2000;275:41340–41349. doi: 10.1074/jbc.M006219200. doi:10.1074/jbc.M006219200. [DOI] [PubMed] [Google Scholar]
- 27.Engel T., Lucas J.J., Gomez-Ramos P., Moran M.A., Avila J., Hernandez F. Cooexpression of FTDP-17 tau and GSK-3beta in transgenic mice induce tau polymerization and neurodegeneration. Neurobiol. Aging. 2006;27:1258–1268. doi: 10.1016/j.neurobiolaging.2005.06.010. doi:10.1016/j.neurobiolaging.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 28.Gomez-Sintes R., Hernandez F., Bortolozzi A., Artigas F., Avila J., Zaratin P., Gotteland J.P., Lucas J.J. Neuronal apoptosis and reversible motor deficit in dominant-negative GSK-3 conditional transgenic mice. EMBO J. 2007;26:2743–2754. doi: 10.1038/sj.emboj.7601725. doi:10.1038/sj.emboj.7601725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Noble W., Olm V., Takata K., Casey E., Mary O., Meyerson J., Gaynor K., LaFrancois J., Wang L., Kondo T., et al. Cdk5 is a key factor in tau aggregation and tangle formation in vivo. Neuron. 2003;38:555–565. doi: 10.1016/s0896-6273(03)00259-9. doi:10.1016/S0896-6273(03)00259-9. [DOI] [PubMed] [Google Scholar]
- 30.Plattner F., Angelo M., Giese K.P. The roles of cyclin-dependent kinase 5 and glycogen synthase kinase 3 in tau hyperphosphorylation. J. Biol. Chem. 2006;281:25457–25465. doi: 10.1074/jbc.M603469200. doi:10.1074/jbc.M603469200. [DOI] [PubMed] [Google Scholar]
- 31.Wen Y., Planel E., Herman M., Figueroa H.Y., Wang L., Liu L., Lau L.F., Yu W.H., Duff K.E. Interplay between cyclin-dependent kinase 5 and glycogen synthase kinase 3 beta mediated by neuregulin signaling leads to differential effects on tau phosphorylation and amyloid precursor protein processing. J. Neurosci. 2008;28:2624–2632. doi: 10.1523/JNEUROSCI.5245-07.2008. doi:10.1523/JNEUROSCI.5245-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gotz J., Gladbach A., Pennanen L., van Eersel J., Schild A., David D., Ittner L.M. Animal models reveal role for tau phosphorylation in human disease. Biochim. Biophys. Acta. 2009 doi: 10.1016/j.bbadis.2009.09.008. 10.1016/j.bbadis.2009.09.008. [DOI] [PubMed] [Google Scholar]
- 33.Jackson G.R., Wiedau-Pazos M., Sang T.K., Wagle N., Brown C.A., Massachi S., Geschwind D.H. Human wild-type tau interacts with wingless pathway components and produces neurofibrillary pathology in Drosophila. Neuron. 2002;34:509–519. doi: 10.1016/s0896-6273(02)00706-7. doi:10.1016/S0896-6273(02)00706-7. [DOI] [PubMed] [Google Scholar]
- 34.Nishimura I., Yang Y., Lu B. PAR-1 kinase plays an initiator role in a temporally ordered phosphorylation process that confers tau toxicity in Drosophila. Cell. 2004;116:671–682. doi: 10.1016/s0092-8674(04)00170-9. doi:10.1016/S0092-8674(04)00170-9. [DOI] [PubMed] [Google Scholar]
- 35.Chatterjee S., Sang T.K., Lawless G.M., Jackson G.R. Dissociation of tau toxicity and phosphorylation: role of GSK-3beta, MARK and Cdk5 in a Drosophila model. Hum. Mol. Genet. 2009;18:164–177. doi: 10.1093/hmg/ddn326. doi:10.1093/hmg/ddn326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Iijima-Ando K., Zhao L., Gatt A., Shenton C., Iijima K. A DNA damage-activated checkpoint kinase phosphorylates tau and enhances tau-induced neurodegeneration. Hum. Mol. Genet. 2010;19:1930–1938. doi: 10.1093/hmg/ddq068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nicoll J.A., Wilkinson D., Holmes C., Steart P., Markham H., Weller R.O. Neuropathology of human Alzheimer disease after immunization with amyloid-beta peptide: a case report. Nat. Med. 2003;9:448–452. doi: 10.1038/nm840. doi:10.1038/nm840. [DOI] [PubMed] [Google Scholar]
- 38.Ferrer I., Boada Rovira M., Sanchez Guerra M.L., Rey M.J., Costa-Jussa F. Neuropathology and pathogenesis of encephalitis following amyloid-beta immunization in Alzheimer's disease. Brain. Pathol. 2004;14:11–20. doi: 10.1111/j.1750-3639.2004.tb00493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lewis J., Dickson D.W., Lin W.L., Chisholm L., Corral A., Jones G., Yen S.H., Sahara N., Skipper L., Yager D., et al. Enhanced neurofibrillary degeneration in transgenic mice expressing mutant tau and APP. Science. 2001;293:1487–1491. doi: 10.1126/science.1058189. doi:10.1126/science.1058189. [DOI] [PubMed] [Google Scholar]
- 40.Gotz J., Chen F., van Dorpe J., Nitsch R.M. Formation of neurofibrillary tangles in P301l tau transgenic mice induced by Abeta 42 fibrils. Science. 2001;293:1491–1495. doi: 10.1126/science.1062097. doi:10.1126/science.1062097. [DOI] [PubMed] [Google Scholar]
- 41.Oddo S., Caccamo A., Shepherd J.D., Murphy M.P., Golde T.E., Kayed R., Metherate R., Mattson M.P., Akbari Y., LaFerla F.M. Triple-transgenic model of Alzheimer's disease with plaques and tangles: intracellular Abeta and synaptic dysfunction. Neuron. 2003;39:409–421. doi: 10.1016/s0896-6273(03)00434-3. doi:10.1016/S0896-6273(03)00434-3. [DOI] [PubMed] [Google Scholar]
- 42.Oddo S., Billings L., Kesslak J.P., Cribbs D.H., LaFerla F.M. Abeta immunotherapy leads to clearance of early, but not late, hyperphosphorylated tau aggregates via the proteasome. Neuron. 2004;43:321–332. doi: 10.1016/j.neuron.2004.07.003. doi:10.1016/j.neuron.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 43.Oddo S., Caccamo A., Smith I.F., Green K.N., LaFerla F.M. A dynamic relationship between intracellular and extracellular pools of Abeta. Am. J. Pathol. 2006;168:184–194. doi: 10.2353/ajpath.2006.050593. doi:10.2353/ajpath.2006.050593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oddo S., Caccamo A., Tran L., Lambert M.P., Glabe C.G., Klein W.L., LaFerla F.M. Temporal profile of amyloid-beta (Abeta) oligomerization in an in vivo model of Alzheimer disease: a link between Abeta and tau pathology. J. Biol. Chem. 2006;281:1599–1604. doi: 10.1074/jbc.M507892200. doi:10.1074/jbc.M507892200. [DOI] [PubMed] [Google Scholar]
- 45.Oddo S., Vasilevko V., Caccamo A., Kitazawa M., Cribbs D.H., LaFerla F.M. Reduction of soluble Abeta and tau, but not soluble Abeta alone, ameliorates cognitive decline in transgenic mice with plaques and tangles. J. Biol. Chem. 2006;281:39413–39423. doi: 10.1074/jbc.M608485200. doi:10.1074/jbc.M608485200. [DOI] [PubMed] [Google Scholar]
- 46.Rapoport M., Dawson H.N., Binder L.I., Vitek M.P., Ferreira A. Tau is essential to beta-amyloid-induced neurotoxicity. Proc. Natl. Acad. Sci. USA. 2002;99:6364–6369. doi: 10.1073/pnas.092136199. doi:10.1073/pnas.092136199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.King M.E., Kan H.M., Baas P.W., Erisir A., Glabe C.G., Bloom G.S. Tau-dependent microtubule disassembly initiated by prefibrillar beta-amyloid. J. Cell. Biol. 2006;175:541–546. doi: 10.1083/jcb.200605187. doi:10.1083/jcb.200605187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roberson E.D., Scearce-Levie K., Palop J.J., Yan F., Cheng I.H., Wu T., Gerstein H., Yu G.Q., Mucke L. Reducing endogenous tau ameliorates amyloid beta-induced deficits in an Alzheimer's disease mouse model. Science. 2007;316:750–754. doi: 10.1126/science.1141736. doi:10.1126/science.1141736. [DOI] [PubMed] [Google Scholar]
- 49.Fulga T.A., Elson-Schwab I., Khurana V., Steinhilb M.L., Spires T.L., Hyman B.T., Feany M.B. Abnormal bundling and accumulation of F-actin mediates tau-induced neuronal degeneration in vivo. Nat. Cell. Biol. 2007;9:139–148. doi: 10.1038/ncb1528. doi:10.1038/ncb1528. [DOI] [PubMed] [Google Scholar]
- 50.Wittmann C.W., Wszolek M.F., Shulman J.M., Salvaterra P.M., Lewis J., Hutton M., Feany M.B. Tauopathy in Drosophila: neurodegeneration without neurofibrillary tangles. Science. 2001;293:711–714. doi: 10.1126/science.1062382. doi:10.1126/science.1062382. [DOI] [PubMed] [Google Scholar]
- 51.Khurana V., Lu Y., Steinhilb M.L., Oldham S., Shulman J.M., Feany M.B. TOR-mediated cell-cycle activation causes neurodegeneration in a Drosophila tauopathy model. Curr. Biol. 2006;16:230–241. doi: 10.1016/j.cub.2005.12.042. doi:10.1016/j.cub.2005.12.042. [DOI] [PubMed] [Google Scholar]
- 52.Karsten S.L., Sang T.-K., Gehman L.T., Chatterjee S., Liu J., Lawless G.M., Sengupta S., Berry R.W., Pomakian J., Oh H.S., et al. A genomic screen for modifiers of tauopathy identifies puromycin-sensitive aminopeptidase as an inhibitor of tau-induced neurodegeneration. Neuron. 2006;51:549–560. doi: 10.1016/j.neuron.2006.07.019. doi:10.1016/j.neuron.2006.07.019. [DOI] [PubMed] [Google Scholar]
- 53.Jackson G.R., Wiedau-Pazos M., Sang T.-K., Wagle N., Brown C.A., Massachi S., Geschwind D.H. Human wild-type tau interacts with wingless pathway components and produces neurofibrillary pathology in Drosophila. Neuron. 2002;34:509–519. doi: 10.1016/s0896-6273(02)00706-7. doi:10.1016/S0896-6273(02)00706-7. [DOI] [PubMed] [Google Scholar]
- 54.Mershin A., Pavlopoulos E., Fitch O., Braden B.C., Nanopoulos D.V., Skoulakis E.M. Learning and memory deficits upon TAU accumulation in Drosophila mushroom body neurons. Learn. Mem. 2004;11:277–287. doi: 10.1101/lm.70804. doi:10.1101/lm.70804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chee F.C., Mudher A., Cuttle M.F., Newman T.A., MacKay D., Lovestone S., Shepherd D. Over-expression of tau results in defective synaptic transmission in Drosophila neuromuscular junctions. Neurobiol. Dis. 2005;20:918–928. doi: 10.1016/j.nbd.2005.05.029. doi:10.1016/j.nbd.2005.05.029. [DOI] [PubMed] [Google Scholar]
- 56.Mudher A., Shepherd D., Newman T.A., Mildren P., Jukes J.P., Squire A., Mears A., Drummond J.A., Berg S., MacKay D., et al. GSK-3beta inhibition reverses axonal transport defects and behavioural phenotypes in Drosophila. Mol. Psychiatry. 2004;9:522–530. doi: 10.1038/sj.mp.4001483. doi:10.1038/sj.mp.4001483. [DOI] [PubMed] [Google Scholar]
- 57.Iijima-Ando K., Iijima K. Transgenic Drosophila models of Alzheimer's disease and tauopathies. Brain Struct. Funct. 2009;214:245–262. doi: 10.1007/s00429-009-0234-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee V.M., Kenyon T.K., Trojanowski J.Q. Transgenic animal models of tauopathies. Biochim. Biophys. Acta. 2005;1739:251–259. doi: 10.1016/j.bbadis.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 59.Iijima K., Liu H.P., Chiang A.S., Hearn S.A., Konsolaki M., Zhong Y. Dissecting the pathological effects of human Abeta40 and Abeta42 in Drosophila: a potential model for Alzheimer's disease. Proc. Natl. Acad. Sci. USA. 2004;101:6623–6628. doi: 10.1073/pnas.0400895101. doi:10.1073/pnas.0400895101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Iijima K., Chiang H.C., Hearn S.A., Hakker I., Gatt A., Shenton C., Granger L., Leung A., Iijima-Ando K., Zhong Y. Abeta42 mutants with different aggregation profiles induce distinct pathologies in Drosophila. PLoS ONE. 2008;3:e1703. doi: 10.1371/journal.pone.0001703. doi:10.1371/journal.pone.0001703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Finelli A., Kelkar A., Song H.J., Yang H., Konsolaki M. A model for studying Alzheimer's Abeta42-induced toxicity in Drosophila melanogaster. Mol. Cell. Neurosci. 2004;26:365–375. doi: 10.1016/j.mcn.2004.03.001. doi:10.1016/j.mcn.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 62.Heisenberg M. Mushroom body memoir: from maps to models. Nat. Rev. Neurosci. 2003;4:266–275. doi: 10.1038/nrn1074. doi:10.1038/nrn1074. [DOI] [PubMed] [Google Scholar]
- 63.Kosmidis S., Grammenoudi S., Papanikolopoulou K., Skoulakis E.M. Differential effects of tau on the integrity and function of neurons essential for learning in Drosophila. J. Neurosci. 2010;30:464–477. doi: 10.1523/JNEUROSCI.1490-09.2010. doi:10.1523/JNEUROSCI.1490-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xu J., Xin S., Du W. Drosophila Chk2 is required for DNA damage-mediated cell cycle arrest and apoptosis. FEBS Lett. 2001;508:394–398. doi: 10.1016/s0014-5793(01)03103-9. doi:10.1016/S0014-5793(01)03103-9. [DOI] [PubMed] [Google Scholar]
- 65.Stracker T.H., Usui T., Petrini J.H. Taking the time to make important decisions: the checkpoint effector kinases Chk1 and Chk2 and the DNA damage response. DNA Repair (Amst) 2009;8:1047–1054. doi: 10.1016/j.dnarep.2009.04.012. doi:10.1016/j.dnarep.2009.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Reinhardt H.C., Yaffe M.B. Kinases that control the cell cycle in response to DNA damage: Chk1, Chk2 and MK2. Curr. Opin. Cell. Biol. 2009;21:245–255. doi: 10.1016/j.ceb.2009.01.018. doi:10.1016/j.ceb.2009.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lieber M.R. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. doi: 10.1146/annurev.biochem.052308.093131. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Essers J., Theil A.F., Baldeyron C., van Cappellen W.A., Houtsmuller A.B., Kanaar R., Vermeulen W. Nuclear dynamics of PCNA in DNA replication and repair. Mol. Cell. Biol. 2005;25:9350–9359. doi: 10.1128/MCB.25.21.9350-9359.2005. doi:10.1128/MCB.25.21.9350-9359.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Herrup K., Yang Y. Cell cycle regulation in the postmitotic neuron: oxymoron or new biology? Nat. Rev. Neurosci. 2007;8:368–378. doi: 10.1038/nrn2124. doi:10.1038/nrn2124. [DOI] [PubMed] [Google Scholar]
- 70.Busser J., Geldmacher D.S., Herrup K. Ectopic cell cycle proteins predict the sites of neuronal cell death in Alzheimer's disease brain. J. Neurosci. 1998;18:2801–2807. doi: 10.1523/JNEUROSCI.18-08-02801.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Iijima-Ando K., Hearn S.A., Shenton C., Gatt A., Zhao L., Iijima K. Mitochondrial mislocalization underlies Aβ42-induced neuronal dysfunction in a Drosophila model of Alzheimer's disease. PLoS ONE. 2009;4:e8310. doi: 10.1371/journal.pone.0008310. doi:10.1371/journal.pone.0008310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Amundson S.A., Bittner M., Meltzer P., Trent J., Fornace A.J., Jr Physiological function as regulation of large transcriptional programs: the cellular response to genotoxic stress. Comp. Biochem. Physiol. B. Biochem. Mol. Biol. 2001;129:703–710. doi: 10.1016/s1096-4959(01)00389-x. doi:10.1016/S1096-4959(01)00389-X. [DOI] [PubMed] [Google Scholar]
- 73.Flores E.R., Tsai K.Y., Crowley D., Sengupta S., Yang A., McKeon F., Jacks T. p63 and p73 are required for p53-dependent apoptosis in response to DNA damage. Nature. 2002;416:560–564. doi: 10.1038/416560a. doi:10.1038/416560a. [DOI] [PubMed] [Google Scholar]
- 74.Vousden K.H., Lu X. Live or let die: the cell's response to p53. Nat. Rev. Cancer. 2002;2:594–604. doi: 10.1038/nrc864. doi:10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- 75.Brodsky M.H., Weinert B.T., Tsang G., Rong Y.S., McGinnis N.M., Golic K.G., Rio D.C., Rubin G.M. Drosophila melanogaster MNK/Chk2 and p53 regulate multiple DNA repair and apoptotic pathways following DNA damage. Mol. Cell. Biol. 2004;24:1219–1231. doi: 10.1128/MCB.24.3.1219-1231.2004. doi:10.1128/MCB.24.3.1219-1231.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bauer J.H., Poon P.C., Glatt-Deeley H., Abrams J.M., Helfand S.L. Neuronal expression of p53 dominant-negative proteins in adult Drosophila melanogaster extends life span. Curr. Biol. 2005;15:2063–2068. doi: 10.1016/j.cub.2005.10.051. doi:10.1016/j.cub.2005.10.051. [DOI] [PubMed] [Google Scholar]
- 77.Augustinack J.C., Schneider A., Mandelkow E.M., Hyman B.T. Specific tau phosphorylation sites correlate with severity of neuronal cytopathology in Alzheimer's disease. Acta Neuropathol. 2002;103:26–35. doi: 10.1007/s004010100423. doi:10.1007/s004010100423. [DOI] [PubMed] [Google Scholar]
- 78.Lauckner J., Frey P., Geula C. Comparative distribution of tau phosphorylated at Ser262 in pre-tangles and tangles. Neurobiol. Aging. 2003;24:767–776. doi: 10.1016/s0197-4580(02)00228-2. doi:10.1016/S0197-4580(02)00228-2. [DOI] [PubMed] [Google Scholar]
- 79.Amadoro G., Corsetti V., Ciotti M.T., Florenzano F., Capsoni S., Amato G., Calissano P. Endogenous Abeta causes cell death via early tau hyperphosphorylation. Neurobiol. Aging. 2009 doi: 10.1016/j.neurobiolaging.2009.06.005. 10.1016/j.neurobiolaging.2009.06.005. [DOI] [PubMed] [Google Scholar]
- 80.Perez M., Ribe E., Rubio A., Lim F., Moran M.A., Ramos P.G., Ferrer I., Isla M.T., Avila J. Characterization of a double (amyloid precursor protein-tau) transgenic: tau phosphorylation and aggregation. Neuroscience. 2005;130:339–347. doi: 10.1016/j.neuroscience.2004.09.029. doi:10.1016/j.neuroscience.2004.09.029. [DOI] [PubMed] [Google Scholar]
- 81.Geula C., Wu C.K., Saroff D., Lorenzo A., Yuan M., Yankner B.A. Aging renders the brain vulnerable to amyloid beta-protein neurotoxicity. Nat. Med. 1998;4:827–831. doi: 10.1038/nm0798-827. [DOI] [PubMed] [Google Scholar]
- 82.Dickey C.A., Kamal A., Lundgren K., Klosak N., Bailey R.M., Dunmore J., Ash P., Shoraka S., Zlatkovic J., Eckman C.B., et al. The high-affinity HSP90-CHIP complex recognizes and selectively degrades phosphorylated tau client proteins. J. Clin. Invest. 2007;117:648–658. doi: 10.1172/JCI29715. doi:10.1172/JCI29715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dickey C.A., Dunmore J., Lu B., Wang J.W., Lee W.C., Kamal A., Burrows F., Eckman C., Hutton M., Petrucelli L. HSP induction mediates selective clearance of tau phosphorylated at proline-directed Ser/Thr sites but not KXGS (MARK) sites. FASEB J. 2006;20:753–755. doi: 10.1096/fj.05-5343fje. [DOI] [PubMed] [Google Scholar]
- 84.Folwell J., Cowan C.M., Ubhi K.K., Shiabh H., Newman T.A., Shepherd D., Mudher A. Abeta exacerbates the neuronal dysfunction caused by human tau expression in a Drosophila model of Alzheimer's disease. Exp. Neurol. 2009;223:401–409. doi: 10.1016/j.expneurol.2009.09.014. [DOI] [PubMed] [Google Scholar]
- 85.Smale G., Nichols N.R., Brady D.R., Finch C.E., Horton W.E., Jr Evidence for apoptotic cell death in Alzheimer's disease. Exp. Neurol. 1995;133:225–230. doi: 10.1006/exnr.1995.1025. doi:10.1006/exnr.1995.1025. [DOI] [PubMed] [Google Scholar]
- 86.Adamec E., Vonsattel J.P., Nixon R.A. DNA strand breaks in Alzheimer's disease. Brain Res. 1999;849:67–77. doi: 10.1016/s0006-8993(99)02004-1. doi:10.1016/S0006-8993(99)02004-1. [DOI] [PubMed] [Google Scholar]
- 87.Lassmann H., Bancher C., Breitschopf H., Wegiel J., Bobinski M., Jellinger K., Wisniewski H.M. Cell death in Alzheimer's disease evaluated by DNA fragmentation in situ. Acta Neuropathol. 1995;89:35–41. doi: 10.1007/BF00294257. doi:10.1007/BF00294257. [DOI] [PubMed] [Google Scholar]
- 88.Lucassen P.J., Chung W.C., Kamphorst W., Swaab D.F. DNA damage distribution in the human brain as shown by in situ end labeling; area-specific differences in aging and Alzheimer disease in the absence of apoptotic morphology. J. Neuropathol. Exp. Neurol. 1997;56:887–900. doi: 10.1097/00005072-199708000-00007. doi:10.1097/00005072-199708000-00007. [DOI] [PubMed] [Google Scholar]
- 89.Davydov V., Hansen L.A., Shackelford D.A. Is DNA repair compromised in Alzheimer's disease? Neurobiol. Aging. 2003;24:953–968. doi: 10.1016/s0197-4580(02)00229-4. doi:10.1016/S0197-4580(02)00229-4. [DOI] [PubMed] [Google Scholar]
- 90.Shao C., Xiong S., Li G.M., Gu L., Mao G., Markesbery W.R., Lovell M.A. Altered 8-oxoguanine glycosylase in mild cognitive impairment and late-stage Alzheimer's disease brain. Free Radic. Biol. Med. 2008;45:813–819. doi: 10.1016/j.freeradbiomed.2008.06.003. doi:10.1016/j.freeradbiomed.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Markesbery W.R., Kryscio R.J., Lovell M.A., Morrow J.D. Lipid peroxidation is an early event in the brain in amnestic mild cognitive impairment. Ann. Neurol. 2005;58:730–735. doi: 10.1002/ana.20629. doi:10.1002/ana.20629. [DOI] [PubMed] [Google Scholar]
- 92.Keller J.N., Schmitt F.A., Scheff S.W., Ding Q., Chen Q., Butterfield D.A., Markesbery W.R. Evidence of increased oxidative damage in subjects with mild cognitive impairment. Neurology. 2005;64:1152–1156. doi: 10.1212/01.WNL.0000156156.13641.BA. [DOI] [PubMed] [Google Scholar]
- 93.Ding Q., Markesbery W.R., Chen Q., Li F., Keller J.N. Ribosome dysfunction is an early event in Alzheimer's disease. J. Neurosci. 2005;25:9171–9175. doi: 10.1523/JNEUROSCI.3040-05.2005. doi:10.1523/JNEUROSCI.3040-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Butterfield D.A., Poon H.F., St Clair D., Keller J.N., Pierce W.M., Klein J.B., Markesbery W.R. Redox proteomics identification of oxidatively modified hippocampal proteins in mild cognitive impairment: insights into the development of Alzheimer's disease. Neurobiol. Dis. 2006;22:223–232. doi: 10.1016/j.nbd.2005.11.002. doi:10.1016/j.nbd.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 95.Wang J., Xiong S., Xie C., Markesbery W.R., Lovell M.A. Increased oxidative damage in nuclear and mitochondrial DNA in Alzheimer's disease. J. Neurochem. 2005;93:953–962. doi: 10.1111/j.1471-4159.2005.03053.x. doi:10.1111/j.1471-4159.2005.03053.x. [DOI] [PubMed] [Google Scholar]
- 96.Moreira P.I., Nunomura A., Nakamura M., Takeda A., Shenk J.C., Aliev G., Smith M.A., Perry G. Nucleic acid oxidation in Alzheimer disease. Free Radic. Biol. Med. 2008;44:1493–1505. doi: 10.1016/j.freeradbiomed.2008.01.002. doi:10.1016/j.freeradbiomed.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 97.Su J.H., Anderson A.J., Cummings B.J., Cotman C.W. Immunohistochemical evidence for apoptosis in Alzheimer's disease. Neuroreport. 1994;5:2529–2533. doi: 10.1097/00001756-199412000-00031. [DOI] [PubMed] [Google Scholar]
- 98.Stadelmann C., Bruck W., Bancher C., Jellinger K., Lassmann H. Alzheimer disease: DNA fragmentation indicates increased neuronal vulnerability, but not apoptosis. J. Neuropathol. Exp. Neurol. 1998;57:456–464. doi: 10.1097/00005072-199805000-00009. doi:10.1097/00005072-199805000-00009. [DOI] [PubMed] [Google Scholar]
- 99.Mandavilli B.S., Rao K.S. Neurons in the cerebral cortex are most susceptible to DNA-damage in aging rat brain. Biochem. Mol. Biol. Int. 1996;40:507–514. doi: 10.1080/15216549600201073. [DOI] [PubMed] [Google Scholar]
- 100.Ugolini G., Cattaneo A., Novak M. Co-localization of truncated tau and DNA fragmentation in Alzheimer's disease neurones. Neuroreport. 1997;8:3709–3712. doi: 10.1097/00001756-199712010-00010. [DOI] [PubMed] [Google Scholar]
- 101.Drake J., Link C.D., Butterfield D.A. Oxidative stress precedes fibrillar deposition of Alzheimer's disease amyloid beta-peptide (1–42) in a transgenic Caenorhabditis elegans model. Neurobiol. Aging. 2003;24:415–420. doi: 10.1016/s0197-4580(02)00225-7. doi:10.1016/S0197-4580(02)00225-7. [DOI] [PubMed] [Google Scholar]
- 102.Rival T., Page R.M., Chandraratna D.S., Sendall T.J., Ryder E., Liu B., Lewis H., Rosahl T., Hider R., Camargo L.M., et al. Fenton chemistry and oxidative stress mediate the toxicity of the beta-amyloid peptide in a Drosophila model of Alzheimer's disease. Eur. J. Neurosci. 2009;29:1335–1347. doi: 10.1111/j.1460-9568.2009.06701.x. doi:10.1111/j.1460-9568.2009.06701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Weterings E., Chen D.J. The endless tale of non-homologous end-joining. Cell Res. 2008;18:114–124. doi: 10.1038/cr.2008.3. doi:10.1038/cr.2008.3. [DOI] [PubMed] [Google Scholar]
- 104.Lu T., Pan Y., Kao S.Y., Li C., Kohane I., Chan J., Yankner B.A. Gene regulation and DNA damage in the ageing human brain. Nature. 2004;429:883–891. doi: 10.1038/nature02661. doi:10.1038/nature02661. [DOI] [PubMed] [Google Scholar]
- 105.Fang-Kircher S.G., Labudova O., Kitzmueller E., Rink H., Cairns N., Lubec G. Increased steady state mRNA levels of DNA-repair genes XRCC1, ERCC2 and ERCC3 in brain of patients with Down syndrome. Life Sci. 1999;64:1689–1699. doi: 10.1016/s0024-3205(99)00107-1. doi:10.1016/S0024-3205(99)00107-1. [DOI] [PubMed] [Google Scholar]
- 106.Parshad R.P., Sanford K.K., Price F.M., Melnick L.K., Nee L.E., Schapiro M.B., Tarone R.E., Robbins J.H. Fluorescent light-induced chromatid breaks distinguish Alzheimer disease cells from normal cells in tissue culture. Proc. Natl. Acad. Sci. USA. 1996;93:5146–5150. doi: 10.1073/pnas.93.10.5146. doi:10.1073/pnas.93.10.5146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jacobsen E., Beach T., Shen Y., Li R., Chang Y. Deficiency of the Mre11 DNA repair complex in Alzheimer's disease brains. Brain Res. Mol. Brain Res. 2004;128:1–7. doi: 10.1016/j.molbrainres.2004.05.023. doi:10.1016/j.molbrainres.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 108.Robbins J.H., Brumback R.A., Polinsky R.J., Wirtschafter J.D., Tarone R.E., Scudiero D.A., Otsuka F. Hypersensitivity to DNA-damaging agents in abiotrophies: a new explanation for degeneration of neurons, photoreceptors, and muscle in Alzheimer, Parkinson and Huntington diseases, retinitis pigmentosa, and Duchenne muscular dystrophy. Basic Life Sci. 1985;35:315–344. doi: 10.1007/978-1-4899-2218-2_20. [DOI] [PubMed] [Google Scholar]
- 109.Krishna T.H., Mahipal S., Sudhakar A., Sugimoto H., Kalluri R., Rao K.S. Reduced DNA gap repair in aging rat neuronal extracts and its restoration by DNA polymerase beta and DNA-ligase. J. Neurochem. 2005;92:818–823. doi: 10.1111/j.1471-4159.2004.02923.x. doi:10.1111/j.1471-4159.2004.02923.x. [DOI] [PubMed] [Google Scholar]
- 110.Weissman L., Jo D.G., Sorensen M.M., de Souza-Pinto N.C., Markesbery W.R., Mattson M.P., Bohr V.A. Defective DNA base excision repair in brain from individuals with Alzheimer's disease and amnestic mild cognitive impairment. Nucleic Acids Res. 2007;35:5545–5555. doi: 10.1093/nar/gkm605. doi:10.1093/nar/gkm605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Edwards J.A., Wang L.G., Setlow R.B., Kaminskas E. O6-methylguanine-DNA methyltransferase in lymphocytes of the elderly with and without Alzheimer's disease. Mutat Res. 1989;219:267–272. doi: 10.1016/0921-8734(89)90028-3. [DOI] [PubMed] [Google Scholar]
- 112.Kinsella T.J., Dobson P.P., Fornace A.J., Jr, Barrett S.F., Ganges M.B., Robbins J.H. Alzheimer's disease fibroblasts have normal repair of N-methyl-N′-nitro-N-nitrosoguanidine-induced DNA damage determined by the alkaline elution technique. Biochem. Biophys. Res. Commun. 1987;149:355–361. doi: 10.1016/0006-291x(87)90374-3. doi:10.1016/0006-291X(87)90374-3. [DOI] [PubMed] [Google Scholar]
- 113.Zhang Y., Qu D., Morris E.J., O'Hare M.J., Callaghan S.M., Slack R.S., Geller H.M., Park D.S. The Chk1/Cdc25A pathway as activators of the cell cycle in neuronal death induced by camptothecin. J. Neurosci. 2006;26:8819–8828. doi: 10.1523/JNEUROSCI.2593-06.2006. doi:10.1523/JNEUROSCI.2593-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lukas C., Bartkova J., Latella L., Falck J., Mailand N., Schroeder T., Sehested M., Lukas J., Bartek J. DNA damage-activated kinase Chk2 is independent of proliferation or differentiation yet correlates with tissue biology. Cancer Res. 2001;61:4990–4993. [PubMed] [Google Scholar]
- 115.Iijima-Ando K., Hearn S.A., Granger L., Shenton C., Gatt A., Chiang H.C., Hakker I., Zhong Y., Iijima K. Overexpression of neprilysin reduces Alzheimer amyloid-beta42 (Abeta42)-induced neuron loss and intraneuronal Abeta42 deposits but causes a reduction in cAMP-responsive element-binding protein-mediated transcription, age-dependent axon pathology, and premature death in Drosophila. J. Biol. Chem. 2008;283:19066–19076. doi: 10.1074/jbc.M710509200. doi:10.1074/jbc.M710509200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Greenberg S.G., Davies P. A preparation of Alzheimer paired helical filaments that displays distinct tau proteins by polyacrylamide gel electrophoresis. Proc. Natl. Acad. Sci. USA. 1990;87:5827–5831. doi: 10.1073/pnas.87.15.5827. doi:10.1073/pnas.87.15.5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Goedert M., Spillantini M.G., Cairns N.J., Crowther R.A. Tau proteins of Alzheimer paired helical filaments: abnormal phosphorylation of all six brain isoforms. Neuron. 1992;8:159–168. doi: 10.1016/0896-6273(92)90117-v. doi:10.1016/0896-6273(92)90117-V. [DOI] [PubMed] [Google Scholar]