Abstract
Alternative splicing is a predominant form of gene regulation in higher eukaryotes. The evolution of alternative splicing provides an important mechanism for the acquisition of novel gene functions. In this work, we carried out a genome-wide phylogenetic survey of lineage-specific splicing patterns in the primate brain, via high-density exon junction array profiling of brain transcriptomes of humans, chimpanzees and rhesus macaques. We identified 509 genes showing splicing differences among these species. RT–PCR analysis of 40 exons confirmed the predicted splicing evolution of 33 exons. Of these 33 exons, outgroup analysis using rhesus macaques confirmed 13 exons with human-specific increase or decrease in transcript inclusion levels after humans diverged from chimpanzees. Some of the human-specific brain splicing patterns disrupt domains critical for protein–protein interactions, and some modulate translational efficiency of their host genes. Strikingly, for exons showing splicing differences across species, we observed a significant increase in the rate of silent substitutions within exons, coupled with accelerated sequence divergence in flanking introns. This indicates that evolution of cis-regulatory signals is a major contributor to the emergence of human-specific splicing patterns. In one gene (MAGOH), using minigene reporter assays, we demonstrated that the combination of two human-specific cis-sequence changes created its human-specific splicing pattern. Together, our data reveal widespread human-specific changes of alternative splicing in the brain and suggest an important role of splicing in the evolution of neuronal gene regulation and functions.
INTRODUCTION
Despite their close evolutionary relationships, humans and nonhuman primates have notable differences in phenotypic traits and susceptibility to various diseases (1). A long-standing challenge in evolutionary biology is to uncover genomic changes that account for the unique attributes of the human species. It has been proposed that evolution of gene regulation is a driving force for phenotypic divergence between species (2). Consistent with this theory, studies of human and nonhuman primate transcriptomes revealed widespread changes in steady-state transcript levels during recent human evolution (3–8).
In addition to transcriptional regulation, alternative splicing is another predominant mechanism of gene regulation in higher eukaryotes. Alternative splicing occurs among different tissues (9) or developmental states (10,11), during cellular responses to external cues (12,13) and in a wide range of human diseases (14,15). In humans, the vast majority of protein-coding genes are alternatively spliced (16–18). For example, deep RNA sequencing indicates that >90% of multi-exon human genes undergo alternative splicing (18,19). By producing multiple mRNA and protein products from a single gene, alternative splicing is capable of generating tremendous functional and regulatory diversity from a limited repertoire of protein-coding genes in eukaryotic genomes (20,21).
There are numerous examples of genes with species-specific exons or splicing patterns (22,23). Comparative analyses of cDNA and EST data indicate that many alternative splicing events are not conserved between species (24–27). In fact, during evolution, ancient exons undergo creation and loss of alternative splicing patterns (28–30), and new exons are frequently added to existing functional genes (23,24,31,32). Thus, evolution could use splicing to create species-specific post-transcriptional regulation of gene expression as well as novel protein function. One well-known example is the primate-specific exon in a human gene ADAR2 (adenosine de-aminase, RNA-specific, B1). This exon is created from an Alu retrotransposon during primate evolution (33). It inserts a new peptide segment into the catalytic domain of ADAR2, altering the catalytic activity of the protein product (34). Even between closely related species, such as humans and chimpanzees, differences in splicing exist (35,36). For example, using a custom Agilent microarray targeting approximately 1700 exon-skipping events and subsequent RT–PCR assays, we and colleagues identified 30 exons with different levels of transcript inclusion in corresponding tissues (frontal cortex or heart) of humans and chimpanzees (35). These data suggest that the splicing patterns of orthologous genes can diverge over relatively short evolutionary timescales [i.e. ∼5–7 million years that separate humans and chimpanzees (37)].
In this work, we conducted a systematic survey of splicing differences between human and nonhuman primate brains, particularly lineage-specific changes in splicing after humans diverged from chimpanzees. We carried out a high-throughput microarray analysis of alternative splicing in brain transcriptomes of human, chimpanzee and rhesus macaque, followed by extensive RT–PCR tests in a large panel of tissue samples from all three species. Our study revealed widespread changes of alternative splicing in the brain transcriptome during primate and human evolution.
RESULTS
Comparative analysis of alternative splicing between humans and nonhuman primates using a high-density exon junction array
To study alternative splicing in human and primate brains, we extracted total RNAs from the cerebellum of six chimpanzees, six rhesus macaques and two pooled human cerebellum samples each with 10 or more individual donors (see Materials and Methods). These RNA samples were processed and hybridized to the Affymetrix Human Exon Junction Array (HJAY) for analysis of alternative splicing patterns (38). The HJAY array is a next-generation Affymetrix exon array for genome-wide analysis of alternative splicing. It averages eight probes per probe set for 315 137 exons in the human genome, and also includes probe sets for 260 488 exon–exon junctions. The high probe density, inclusion of exon–exon junction probes and comprehensive coverage of known alternative splicing events make the HJAY array a powerful tool for analysis of alternative splicing (38–40). Our recent studies have shown that this new array detects changes in gene expression and alternative splicing at a very low false positive rate (38,41). Although originally designed for analysis of human genes, a large percentage of HJAY probes perfectly match orthologous transcripts from closely related nonhuman primates (41). Thus, we can use this array for comparative analysis of alternative splicing patterns by directly hybridizing RNA samples from closely related primate species to the HJAY array. For this work, we generated six HJAY profiles per species, with one replicate per sample for chimpanzee and rhesus cerebellums and three replicates per sample for the two human RNA pools.
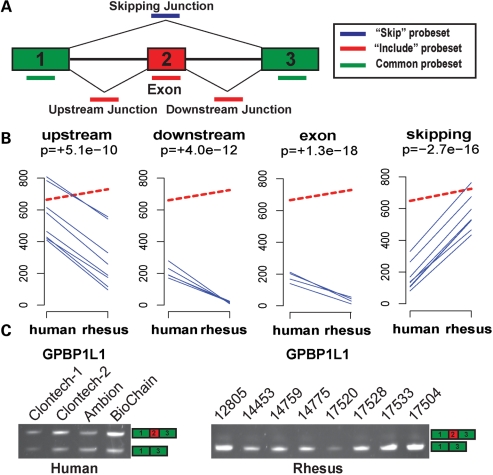
To identify alternative splicing differences of orthologous genes among these species, we compared HJAY profiles of human, chimpanzee and rhesus cerebellums. We used our published computational procedures for alternative splicing analysis of the HJAY array data (38,41–45), after filtering microarray probes with mismatches for orthologous chimpanzee/rhesus transcripts. The procedures of our HJAY array data analysis are summarized in Materials and Methods and described in detail in Supplementary Material, Methods. Briefly, we identified all HJAY exon probes and exon–exon junction probes that perfectly matched orthologous transcripts from chimpanzees or rhesus macaques, using the UCSC pairwise genome alignments of the human genome (hg18) to the genomes of chimpanzee (panTro2) or rhesus macaque (rheMac2) (46–47). Of the 20 815 mRNA/EST-supported human alternative splicing events interrogated by the HJAY array, 15 544 and 10 147 events had sufficient perfect-match probes for orthologous chimpanzee and rhesus transcript isoforms (see definition in Supplementary Material, Methods). For these events, we used our MADS+ (Microarray Analysis of Differential Splicing) algorithm (38,45) in pairwise comparisons of human versus chimpanzee or human versus rhesus array profiles to identify differential splicing events. Specifically, we restricted the MADS+ analysis to HJAY probes perfectly matching orthologous transcripts from multiple species. For every alternative splicing event analyzed, MADS+ calculated P-values of multiple probe sets targeting exons and exon–exon junctions (see Fig. 1A for the probe design for exon-skipping events), requiring opposite trends for probe sets targeting competing isoforms as evidence of differential alternative splicing (38). For example, as shown in Figure 1, the exon 2 of GPBP1L1 (GC-rich promoter binding protein 1-like 1) was predicted to be differentially spliced between human and rhesus cerebellums. From the HJAY array data of this exon, we observed significantly higher intensities of the exon inclusion probe sets (i.e. probe sets targeting the upstream exon–exon junction, downstream exon–exon junction and exon) and significantly lower intensities of the exon-skipping probe set in human samples compared with rhesus samples (Fig. 1B; solid blue lines). Meanwhile, the estimated gene expression level of GPBP1L1 was comparable in human and rhesus (Fig. 1B; dashed red lines). These data indicated that this exon was included at a higher level in the human cerebellum. Indeed, our subsequent RT–PCR validation confirmed that this exon was included in human transcripts at intermediate-to-high levels, but almost completely skipped in orthologous rhesus transcripts (Fig. 1C).
Figure 1.
Detection of a differentially spliced cassette exon between human and rhesus cerebellums by the HJAY array. (A) The HJAY probe design for cassette exons. Each probe set has eight probes. (B) The HJAY array data indicate significantly elevated exon inclusion of GPBP1L1 exon 2 in the human cerebellum compared with the rhesus cerebellum. In each panel, the dashed red line indicates the average gene expression levels of GPBP1L1 in six replicate samples from humans or rhesus macaques. The individual blue lines indicate the average background-corrected intensities of individual probes in a probe set. Only probes that perfectly match orthologous human and rhesus transcripts are shown. MADS+ P-value for differential splicing is shown below the name of the probe set. The ‘+’ and ‘−’ signs indicate the predicted direction of change in exon inclusion levels in the human cerebellum over the rhesus cerebellum. (C) The predicted splicing difference is confirmed by RT–PCR.
Using these computational procedures, our analysis of the HJAY profiles revealed widespread splicing differences between humans and closely related nonhuman primates. In total, we identified 336 alternative splicing differences between human and chimpanzee cerebellums and 415 differences between human and rhesus cerebellums. These events covered all types of alternative splicing patterns, with cassette exons (i.e. exon skipping) being the most prevalent type (Table 1). It should be noted that because of our filtering criteria, in particular the number of perfect-match probes for orthologous transcripts, a much smaller number of alternative splicing events can be analyzed for human–rhesus differences than for human–chimpanzee differences (1501 versus 2646, see Table 1). Despite this, we identified more alternative splicing events with human–rhesus differences (415) than with human–chimpanzee differences (336), representing 27.6% and 12.7% of all events analyzed between humans and rhesus macaques or between humans and chimpanzees, respectively. This was consistent with the phylogenetic relationship among these three species (37).
Table 1.
Summary of differential splicing events between human, chimpanzee and rhesus cerebellums identified by the HJAY array
| Cassette exons | Alternative 5′/3′ splice sites | Mutually exclusive exon usage | ||
|---|---|---|---|---|
| Human versus chimpanzee | Differential splicing eventsa | 281 | 29 | 26 |
| Splicing events analyzedb | 1985 | 323 | 338 | |
| Human versus rhesus | Differential splicing events | 340 | 51 | 24 |
| Splicing events analyzed | 1126 | 195 | 180 |
aIdentified by MADS+ (see Supplementary Material, Methods).
bThe total set of splicing events that passed the filters for gene expression level in the cerebellum, probe intensity and number of perfect-match probes for orthologous chimpanzee or rhesus transcripts (see Supplementary Material, Methods). These splicing events were analyzed by MADS+ for the detection of splicing differences between species.
The percentage of alternative splicing events with predicted human–chimpanzee differences in the cerebellum (12.7%) appeared to be higher than the percentage reported for two other tissues (frontal cortex and heart) (6–8%) in our earlier study (35). However, these statistics were not directly comparable, as distinct microarray platforms were used in these two studies. Specifically, in the present work, we used a high-density Affymetrix short oligonucleotide exon junction array, with approximately 32 probes for each exon-skipping event (38). In contrast, the earlier study used a custom Agilent long oligonucleotide array, with six probes designed to interrogate each exon-skipping event (35). Thus, the difference in the percentage of alternative splicing events with predicted human–chimpanzee differences could be largely attributed to the sensitivity of these two microarray platforms. We also noted that in the previous study using the Agilent custom array, we did not observe any significant difference in the rate of splicing evolution in frontal cortex and heart (35).
RT–PCR validation of alternative splicing evolution and human-specific splicing patterns in the brain
To confirm the identified alternative splicing differences between species, we selected 40 cassette exons for experimental validation, from 281 cassette exons predicted to have splicing differences between human and chimpanzee and 340 cassette exons predicted to have splicing differences between human and rhesus. All predicted events in the human versus chimpanzee comparison or the human versus rhesus comparison were ranked according to the overall level of statistical significance summarized from all available exon probes and exon–exon junction probes (see details in Supplementary Material, Methods). The 40 selected exons were analyzed by semi-quantitative RT–PCR in a large panel of cerebellum tissues from all three species. For a subset of these exons with subtle changes in inclusion levels, we also used a highly sensitive fluorescently labeled RT–PCR protocol, which allowed us to accurately quantify the levels of transcript inclusion in different species (see Materials and Methods). To distinguish bona fide inter-species splicing divergence from intra-species splicing variability, our RT–PCR analysis examined all individual samples used for generating the HJAY array profiles, as well as RNAs from additional human/chimpanzee/rhesus tissue samples (see Supplementary Material, Table S1). Of the 40 exons tested, 33 showed splicing differences between species as predicted by the HJAY array, yielding a validation rate of 83%. For most of the RT–PCR confirmed events, the splicing patterns of orthologous exons were consistent within species, while different between species. For example, the exon 2 in the 5′-UTR of GPBP1L1 was predicted according to the HJAY array data to be differentially spliced between humans and rhesus macaques (Fig. 1B). This exon was completely skipped in all eight rhesus cerebellum samples from different animals. In contrast, we observed a consistent pattern of intermediate-to-high levels of exon inclusion in four human cerebellum samples from single or multiple donors, and in eight chimpanzee cerebellum samples from different animals (Fig. 2A).
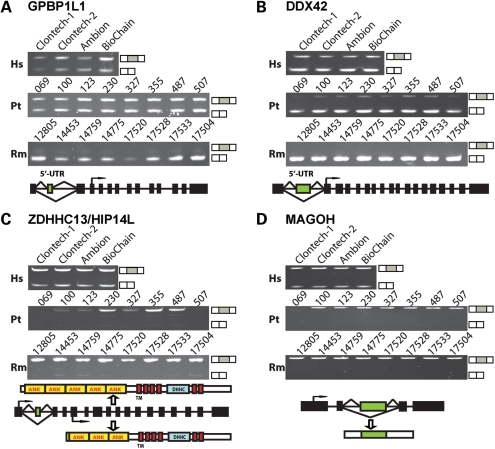
Figure 2.
Examples of exons with splicing differences between human and chimpanzee/rhesus cerebellums. (A) RT–PCR analysis of the exon 2 of GPBP1L1 in human (Hs), chimpanzee (Pt) and rhesus macaque (Rm) samples. (B) RT–PCR analysis of the exon 2 of DDX42 in human (Hs), chimpanzee (Pt) and rhesus macaque (Rm) samples. (C) RT–PCR analysis of the exon 2 of HIP14L (also referred to as ZDHHC13) in human (Hs), chimpanzee (Pt) and rhesus macaque (Rm) samples. ANK, ankyrin repeat; TM, transmembrane domain; DHHC (Asp–His–His–Cys), cysteine-rich domain. (D) RT–PCR analysis of the exon 3 of MAGOH in human (Hs), chimpanzee (Pt) and rhesus macaque (Rm) samples. In each figure, the exon with differential splicing between species is indicated as the green box in the gene structure diagram. Translation start site is indicated by the forward arrow. The sizes of exons and introns are not drawn according to the scale.
It should be emphasized that in selecting candidate exons for RT–PCR analysis, we included exons with various levels of statistical significance and did not cherry-pick candidates from the top of our predicted events to achieve a high validation rate. In fact, a primary selection criterion for experimental validation was to facilitate RT–PCR primer design and data interpretation, i.e. the selected cassette exons did not have alternative splice site usage at the upstream and downstream exon–intron boundaries and were flanked by constitutively spliced exons based on the UCSC Genome Browser annotation. All cassette exons analyzed by the HJAY array in the human versus chimpanzee or the human versus rhesus comparison were ranked by a single combined P-value for the overall statistical significance of inter-species splicing difference, which was summarized from individual P-values of all available perfect-match probes targeting orthologous exons and exon–exon junctions as in Shen et al. (38) and Xing et al. (45). Among the 281 predicted cassette exon events in the human versus chimpanzee comparison, the median rank of RT–PCR confirmed events was 65 with an inter-quartile range (IQR) of 9 to 152. The lowest rank of validated events was 239. In the human versus rhesus comparison, among the 340 predicted cassette exon events, the median rank of RT–PCR confirmed events was 80 with an IQR of 43 to 169 and the lowest rank of 320. Thus, based on our validation rate (83%), we expect that the vast majority of our HJAY-predicted events represent real splicing differences between humans and closely related nonhuman primates. The list of validated events and their RT–PCR gel pictures are provided in Supplementary Material, Table S2. The rankings of RT–PCR-validated events among all HJAY-predicted events are provided in Supplementary Material, Table S3. The complete lists of HJAY-predicted cassette exon events are provided in Supplementary Material, Tables S4 and S5.
Our RT–PCR analysis also revealed a number of exons with human-specific changes in splicing compared with both chimpanzee and rhesus orthologous exons. In this phylogenetic analysis, the observed splicing pattern in the rhesus cerebellum was used as the outgroup to infer the direction of splicing changes of the human exon after humans diverged from chimpanzees. For example, in DDX42 (DEAD box protein 42), the exon 2 in its 5′-UTR had different levels of exon inclusion in human, chimpanzee and rhesus cerebellums (Fig. 2B). Based on RT–PCR analysis, this exon was completely skipped in the rhesus transcripts, weakly included in the chimpanzee transcripts and included at an intermediate level in the human transcripts. This suggests that the splicing activity of DDX42 in the cerebellum was gradually strengthened in the lineage leading to humans. Together, of these 33 exons with RT–PCR-validated splicing differences among species, 13 exons (39%) had consistently higher or lower levels of transcript inclusion in the human cerebellum compared with both chimpanzee and rhesus cerebellums, suggesting recent increases or decreases in splicing activities after humans diverged from chimpanzees. These genes covered a broad range of functional categories (Table 2). Extrapolating from the total number of events identified by the HJAY analysis (Table 1), the overall validation rate in our RT–PCR analysis (83%) and the percentage of exons with human-specific splicing changes among all exons with validated splicing differences (39%), we estimated that more than 100 alternative splicing events in our entire data set underwent human-specific splicing changes. These splicing events represent a group of unique transcriptome signatures in the human brain that can distinguish humans from other closely related primate species.
Table 2.
Exons with human-specific changes in transcript inclusion levels compared with chimpanzee and rhesus cerebellums
| Gene symbol | Exon location (hg18) | Phylogenetic patterns of exon inclusion levels | Impact on mRNA/protein | Gene function |
|---|---|---|---|---|
| MAGOH | chr1:53471801-53471912 | Human (medium-major) ≪ chimpanzee (constitutive)/rhesus (constitutive) | Coding | RNA and protein binding, NMD |
| PIGX | chr3:197937922-197938039 | Human (minor-medium) ≫ chimpanzee (no inclusion)/rhesus (no inclusion) | Coding, exon inclusion introduces premature termination codon | Component of glycosylphosphatidylinositol-mannosyltransferase I |
| ZDHHC13/HIP14L | chr11:19121105-19121246 | Human (medium) ≪ chimpanzee (almost constitutive)/rhesus (almost constitutive) | Coding, exon 2 skipping causes usage of a downstream ATG start site on exon 5 | Palmitoyl transferase, Mg2+ transport |
| LPHN3/CIRL3 | chr4:62461031-62461070 | Human (medium) ≪ chimpanzee (major)/rhesus (major) | Coding | G-protein coupled receptor, cell adhesion and signal transduction |
| NUPL1 | chr13:24781384-24781420 | Human (major-constitutive) ≫ chimpanzee (medium)/rhesus (medium-major) | Coding | Component of the nuclear pore complex (NPC); nucleocytoplasmic transporter activity |
| DDX42 | chr17:59206199-59206270 | Human (minor-medium) > chimpanzee (minor) > rhesus (no inclusion) | 5′-UTR | ATP-dependent helicase activity, protein displacement and RNA annealing |
| CAMTA1 | chr1:7738284-7738315 | Human (minor-medium) < chimpanzee (medium) < rhesus (medium-major) | Coding, alternative C-terminus | Calmodulin binding, transcription regulator |
| GSTM3 | chr1:110083977-110084053 | Human (major) < chimpanzee (constitutive)/rhesus (constitutive) | Coding, alternative N-terminus | Glutathione S-transferase |
| ACSL3 | chr2:223473635-223473742 | Human (medium) > chimpanzee (minor)/rhesus (minor) | 5′-UTR | Acetate–CoA ligase |
| GLS | chr2:191527725-191527789 | Human (major) < chimpanzee (major)/rhesus (major) (differences confirmed by FAM-labeled RT–PCR) | Coding, alternative C-terminus | Glutaminase, glutamine catabolic process |
| PTPRZ1 | chr7:121474963-121475080 | Human (minor) > chimpanzee (no inclusion)/rhesus (no inclusion) | Coding | Transmembrane receptor protein tyrosine phosphatase |
| KCNJ3/GIRK1 | chr2:155274360-155274577 | Human (medium) < chimpanzee (medium-major)/rhesus (major) | Coding, truncated protein | G-protein activated inward-rectifier type potassium channel |
| NAV2 | chr11:19862415-19862484 | Human (medium) > chimpanzee (minor-medium)/rhesus (minor-medium) | Coding | ATP-binding helicase |
Previous studies have suggested a difference in the rate of protein sequence evolution between the human and chimpanzee lineages (48–50). Using our RT–PCR data of 33 exons (Supplementary Material, Table S2), we compared the numbers of exons with human-specific or chimpanzee-specific splicing changes in the brain. To enable an unbiased comparison, we focused on 16 exons predicted by the HJAY array to have splicing differences between humans and chimpanzees and subsequently validated by RT–PCR. Of these 16 exons, 2 exons did not have RT–PCR data in rhesus macaques due to the difficulty of primer design. For the remaining 14 exons, using the rhesus macaque as the outgroup, we identified 9 exons with human-specific splicing changes and 5 exons with chimpanzee-specific splicing changes. However, we caution that any conclusion on the rate difference of splicing changes in the human and chimpanzee lineages would require a much more extensive set of experimentally validated splicing differences between all three species.
Functional and regulatory implications of alternative splicing evolution in the brain
On all genes with identified splicing differences between human, chimpanzee and rhesus cerebellums, Gene Ontology analysis using the DAVID tool (51) found four strongly enriched GO terms: ‘cytoskeleton organization and biogenesis’ (P = 0.007), ‘RNA processing’ (P = 0.012), ‘cell–cell adhesion’ (P = 0.025) and ‘neurological system process’ (P = 0.028) (see the DAVID analysis procedure in Materials and Methods). A previous study of natural selection on protein-coding regions of human genes found cytoskeletal proteins to be under strong negative selection pressure during human evolution (52). Our result thus suggests an intriguing scenario that genes encoding cytoskeletal proteins could experience contrasting modes of selection pressure at the RNA level and at the protein level during recent human evolution.
Next, we investigated potential evidence of positive selection for exons with human-specific splicing changes. Because of the small size of individual exons, methods to detect positive selection using divergence data between species do not have enough power (31). On the other hand, a variety of approaches have been developed to detect signatures of recent positive selection using SNP data (53,54). We compiled the results from 13 genome- or chromosome-wide scans for positive selection in the human genome (55–67). Of the 13 exons with RT–PCR-validated human-specific splicing patterns (Table 2), two exons (in NUPL1 and PTPRZ1) were located within the positively selected genomic regions identified by Tang et al. (65) and Huttley et al. (55), respectively. Notably, Tang et al. (65) used an approach based on contrasting the extended haplotype homozygosity profiles between populations. Huttley et al. (55) was among the earliest scans for positive selection signals using long LD blocks. It is important to note that these SNP-based approaches are only suitable for detecting signals of positive selection during very recent human evolution (i.e. ≤250 kya) (53–54), while the acquisition of a human-specific splicing pattern and the selection pressure acting on such an event could potentially occur any time during the 5–7 million years after humans diverged from chimpanzees.
Interestingly, of the 13 genes showing human-specific changes in brain splicing patterns after humans diverged from chimpanzees, several are implicated in the etiology of human diseases, in particular neurological diseases, including Huntington's disease [HIP14L (68)], Alzheimer's disease [GSTM3 (69)] and schizophrenia [PTPRZ1 (70)] (Table 2). For example, HIP14L (Huntingtin-interacting protein 14-related; also referred to as ZDHHC13) encodes a neuronal-specific palmitoyl acyltransferase that interacts with and palmitoylates Huntingtin, a gene when mutated causes Huntington's disease (68). Our HJAY array and RT–PCR analysis found a substantially elevated level of HIP14L exon 2 skipping in the human cerebellum (Fig. 2C). This exon-skipping event affects the N-terminus of the encoded HIP14L protein product. While the mRNA isoform including exon 2 is translated from the first exon of the HIP14L mRNA, the skipping of exon 2 causes the utilization of a downstream ATG start codon located in exon 5. This truncates two N-terminal Ankyrin repeats, a critical structural motif mediating protein–protein interactions (71) (Fig. 2C).
Another example of human-specific splicing changes with interesting functional implications is the alternative splicing of MAGOH (Mago-nashi homolog, proliferation-associated). MAGOH encodes a key component of the exon junction complex (EJC) and is involved in mRNA nonsense-mediated decay (NMD) (72). The human-specific evolution of MAGOH transcripts resulted in pronounced skipping of exon 3 in the human cerebellum, while the orthologous exon was 100% included in chimpanzee and rhesus transcripts (Fig. 2D). Importantly, exon 3 encodes a peptide segment critical for the interaction between MAGOH and the ribosome-associated protein PYM (72). In a previous study, site-directed mutagenesis within exon 3 abolishes MAGOH-PYM interaction, leading to impaired EJC removal and recycling (72). Although the exact functional impact of the exon 3 skipping isoform remains to be determined experimentally, this human-specific splicing event could provide a novel regulatory mechanism to modulate the MAGOH-PYM interaction, thus affecting the NMD pathway in a human-specific manner.
Of the 13 exons whose human-specific changes in splicing activities are confirmed by RT–PCR (Table 2), 11 are located within the protein-coding regions. Alternative splicing of these exons results in either in-frame insertion/deletion of a peptide segment (e.g. MAGOH), alterations of the N-terminus or the C-terminus of the protein product (e.g. HIP14L) or introduction of a premature termination codon and possibly mRNA nonsense-mediated decay (e.g. PIGX). The remaining two exons (DDX42, ACSL3) are located in the 5′-UTR. It is well known that the 5′-UTRs of mRNAs contain regulatory signals for modulating mRNA stability and protein translation (73), and alternative splicing within 5′-UTRs can affect translational efficiency (74,75). Thus, human-specific splicing changes within the 5′-UTR may influence post-transcriptional regulation of gene expression. We also note that the 5′-UTR appears to be a frequent spot for the creation of new exons, based on previous studies on exonization of transposable elements (TEs) during primate evolution (31,76,77).
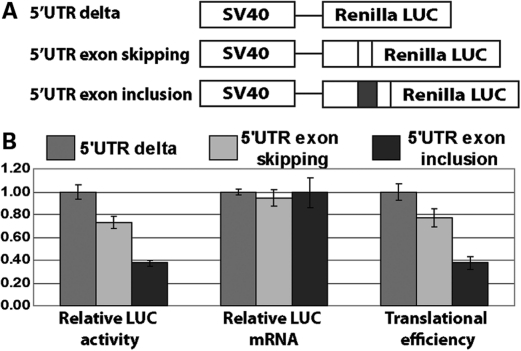
To further confirm the regulatory impact of human-specific splicing changes in the 5′-UTR, we tested the exon 2 in the 5′-UTR of DDX42 which had a much higher transcript inclusion level in humans compared with chimpanzees and rhesus macaques (Fig. 2B). We performed 5′-UTR luciferase reporter assays (78,79) to assess whether the inclusion of this exon affected translational efficiency of the DDX42 mRNA. Briefly, 5′-UTR isoforms containing or skipping the exon 2 of DDX42 were cloned into the psiCHECK2 luciferase reporter system (Promega). For each 5′-UTR isoform, the resulting reporter construct expressed both the firefly luciferase and the Renilla luciferase fused downstream of the cloned 5′-UTR isoform (Fig. 3A). After transfection into HeLa cells, we measured luciferase activities and mRNA levels. For each 5′-UTR construct, the Renilla luciferase activity and mRNA level were normalized to the firefly luciferase, and the translational efficiency was estimated using the fold change of luciferase activity normalized to mRNA concentration (78,79) (see details in Materials and Methods). We observed a 2-fold reduction in the estimated translational efficiency when the exon 2 of DDX42 was inserted to its 5′-UTR (Fig. 3B). The mRNA concentration remained unchanged (Fig. 3B). Thus, the human-specific increase in the transcript inclusion level of DDX42 exon 2 provides a regulatory mechanism for reducing protein production at the post-transcriptional level without changing the protein-coding sequence.
Figure 3.
Modulation of translational efficiency by exon 2 in DDX42 5′-UTR with a human-specific splicing change. (A) Schematic diagrams of the DDX42 5′-UTR luciferase reporter constructs. 5′-UTR delta construct: psiCHECK2 vector (Promega) expressing Renilla luciferase under the control of the SV40 promoter and firefly luciferase under the control of the HSV-TK promoter (not shown). 5′-UTR exon-skipping construct: Renilla luciferase downstream of the DDX42 5′-UTR isoform, which skips exon 2. 5′-UTR exon inclusion construct: Renilla luciferase downstream of the DDX42 5′-UTR isoform, which includes exon 2 (black exon). (B) Inclusion of exon 2 into DDX42 5′-UTR results in a 2-fold reduction in translational efficiency according to the luciferase reporter assay. Relative luciferase (LUC) activity: shown as the ratios of Renilla and firefly luciferase activity. The luciferase activity ratio of the control construct 5′-UTR delta (psiCHECK2) was designated as 1 and the values of 5′-UTR exon-skipping/inclusion constructs were normalized accordingly. Relative luciferase (LUC) mRNA levels: shown as the ratios of Renilla and firefly mRNA concentration. The mRNA ratio of the control construct 5′-UTR delta (psiCHECK2) was designated as 1 and the values of 5′-UTR exon-skipping/inclusion constructs were normalized accordingly. Translational efficiency: shown as fold change of luciferase activity normalized to the mRNA level. The fold change of luciferase activity over the mRNA level of the control construct 5′-UTR delta (psiCHECK2) was designated as 1 and the values of 5′-UTR exon-skipping/inclusion constructs were normalized accordingly. In all plots, the mean represents average value from six independent experiments, and the error bar indicates standard error of the mean.
Correlation between splicing evolution and sequence divergence of exons and flanking introns
The large number of identified differential splicing events between human and nonhuman primate brains also allowed us to investigate mechanisms important for the evolution of splicing. It is well-known that cis-elements within exons and flanking introns, such as splice sites and splicing enhancer/silencer elements, play critical roles in the regulation of splicing (80). To assess how evolution of exonic and intronic sequences influenced the evolution of splicing, we analyzed the nucleotide substitution patterns of cassette exons with HJAY-predicted splicing differences among species. As the control, we also analyzed a separate group of cassette exons that passed the same probe-filtering procedure but were not found to have splicing differences among species by the HJAY analysis. Strikingly, for exons showing splicing differences among species, we observed a significant increase in the rate of silent substitutions within exons, coupled with accelerated sequence evolution in flanking introns. Within the region spanning from the 100 nt upstream intron to the 100 nt downstream intron, cassette exons with identified splicing changes between human and chimpanzee cerebellums had an average rate of nucleotide differences of 1.83% (including substitutions and indels), compared with 1.69% for exons without splicing changes (P = 9.4e − 4, one-sided Fisher's exact test; Table 3). We also observed this pattern in the human versus rhesus comparison (P = 6.0e − 4). The same trend was reproducible when we analyzed the 100 nt upstream or 100 nt downstream intronic regions separately (Table 3). For exonic regions, it is well known that the pattern of nucleotide sequence conservation is influenced by selective constraints at both the protein level and the RNA level (22,81,82). To remove the potential confounding effect of protein-level selection pressure, we calculated the synonymous substitution rate (Ks rate) between species (see Materials and Methods). As synonymous substitutions do not alter protein products, the Ks rate has been proposed as an informative measure for exonic selection pressure at the level of RNA splicing (22,82–85). We found a significantly increased Ks rate in exons showing splicing differences between species, compared with exons without splicing differences (Table 4). For example, for cassette exons with identified splicing differences between humans and chimpanzees, the overall Ks rate in exons was 0.0129. In contrast, for cassette exons without identified splicing differences between these two species, the overall Ks rate was significantly lower (0.0089, P = 2.5e − 3). The same trend was reproducible in the human versus rhesus comparison (Table 4).
Table 3.
Exons with identified splicing changes between species have a higher rate of nucleotide differences between orthologous exons and flanking introns
| Human versus | Region | Splicing change | Conserved nucleotide | Number of nucleotide differencesa (%) | Fisher's exact test P-valueb |
|---|---|---|---|---|---|
| Chimpanzee | Upstream intron (100 nt) + exon + downstream intron (100 nt) | Yes | 87 418 | 1633 (1.83) | 9.4e − 4 |
| No | 537 142 | 9213 (1.69) | |||
| Upstream intron (100 nt) | Yes | 27 295 | 780 (2.78) | 1.2e − 6 | |
| No | 163 126 | 3925 (2.35) | |||
| Downstream intron (100 nt) | Yes | 27 350 | 646 (2.31) | 1.4e − 2 | |
| No | 163 260 | 3499 (2.10) | |||
| Rhesus | Upstream intron (100 nt) + exon + downstream intron (100 nt) | Yes | 107 735 | 5327 (4.71) | 6.0e − 4 |
| No | 229 724 | 10 742 (4.47) | |||
| Upstream intron (100 nt) | Yes | 31 906 | 2100 (6.18) | 3.1e − 3 | |
| No | 72 329 | 4415 (5.75) | |||
| Downstream intron (100 nt) | Yes | 31 826 | 2217 (6.51) | 1.6e − 4 | |
| No | 72 144 | 4561 (5.95) |
aNucleotide differences include substitutions, insertions and deletions.
bP-value from one-sided Fisher's exact test on the rate of between-species nucleotide differences of cassette exons with identified splicing changes versus cassette exons without splicing changes.
Table 4.
Exons with identified splicing changes between species have a higher synonymous substitution rate (Ks rate) between orthologous exons
| Human versus | Splicing change | Number of synonymous substitutions | Number of synonymous sites | Ks | Fisher's exact test P-value (one-sided) |
|---|---|---|---|---|---|
| Chimpanzee | Yes | 78 | 6040 | 0.0129 | 2.5e − 3 |
| No | 343 | 38 477 | 0.0089 | ||
| Rhesus | Yes | 277 | 7722 | 0.0359 | 5.3e − 6 |
| No | 428 | 16 797 | 0.0255 |
Together, these results indicate that the evolution of cis-regulatory sequences is a major contributor to the emergence of human-specific splicing patterns. It must be emphasized that these patterns are not artifacts due to sequence mismatches that affect hybridization of the HJAY probes to chimpanzee/rhesus transcripts, as our array analysis is entirely based on probes that perfectly match orthologous transcripts from multiple species.
Next, we investigated various types of cis-changes that could contribute to the evolution of splicing. First, small-scale genomic structural changes, including exon duplication and insertion of TEs, have been associated with the creation of new exons during mammalian evolution (23,31). However, as our study focused on exons whose genomic sequences were conserved between humans and nonhuman primates, this mechanism was not an important contributor to the inter-species splicing differences identified in this work. For example, of the 33 exons with RT–PCR-validated splicing differences, only three exons overlapped with TEs, and all three overlapped with TEs in all three species. Therefore, the splicing differences did not result from insertions of TEs during recent human evolution. Next, we investigated evolutionary changes at the 5′ and 3′ splice sites, which were essential signals for exon recognition. For every exon analyzed by the HJAY array, we scored its 5′ and 3′ splice sites in human, chimpanzee and rhesus genomes using MAXENT (86). In the human versus chimpanzee comparison, 16 (5.8%) exons with detected splicing differences had a difference in the 5′ or 3′ splice site scores between humans and chimpanzees of at least 2, compared with 49 (3.0%) exons without detected splicing differences between these two species (P = 0.03, two-sided Fisher's exact test). However, this trend was not reproduced in the human versus rhesus comparison. Twenty-five (7.5%) exons with detected splicing differences had a difference in the 5′ or 3′ splice site scores between humans and rhesus macaques of at least 2, compared with 56 (7.4%) exons without detected splicing differences between these two species. This was not unexpected, as the longer evolutionary distance between humans and rhesus macaques allowed for compensatory cis-changes to occur, which could buffer the changes in the splice site strength. Together, these results suggest that the evolution of the splice sites contributed to inter-species divergence of splicing, although this mechanism could only explain a small percentage of all inter-species differences reported. Finally, we investigated the creation and loss of exonic splicing regulatory elements. On a list of exonic splicing enhancers (ESEs) and silencers (ESSs) collected by Burge and colleagues (87,88), we did not observe any overall correlation between splicing differences among species and evolutionary changes that created or disrupted known ESEs and ESSs. Although this could be due to the lack of statistical power, it is more likely that this observation reflects our limited understanding of the tissue-specific ‘splicing code’ in higher eukaryotes (35,89). Indeed, the precise splicing outcomes of individual exons are controlled by a complex array of exonic and intronic regulatory signals (80). Nucleotide changes at any position within the exon or surrounding introns could have the possibility to cause the evolution of splicing patterns. Moreover, these ESEs and ESSs were identified and validated in the HeLa or HEK293 cell lines (87,88), while there are substantial differences in splicing regulation in the brain (90). Additionally, this analysis considered all ESEs or all ESSs as a whole, while the change to a single ESE or ESS may contribute to the evolution of splicing in individual exons.
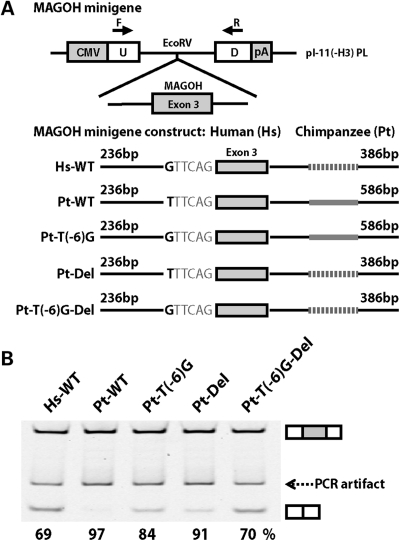
To further elucidate the molecular mechanisms that could create human-specific splicing patterns, we selected the exon 3 of MAGOH for a detailed case study. This exon was 100% included in chimpanzee and rhesus transcripts, but had a substantial level of exon skipping in the human transcript (Fig. 2D). By comparing the exonic and flanking intronic sequences of this exon in human, chimpanzee and rhesus genomes, we identified two human-specific changes in the flanking intronic sequences. We found a human-specific T-to-G substitution at the sixth intronic nucleotide of the upstream intron–exon boundary, which reduced the score of the 3′ (acceptor) splice site from 8.26 in chimpanzees to 6.13 in humans. Additionally, we identified a human-specific deletion of a 200 bp segment in the downstream intronic region (Fig. 4A). To assess which cis-sequence change(s) contributed to the human-specific evolution of MAGOH exon 3 splicing, we tested the effects of these cis-changes using minigene splicing reporter assays. We cloned the exon and its flanking intronic regions into the minigene reporter pI-11-H3 [see reference (91) and Materials and Methods] and made two minigene constructs corresponding to the wild-type human and chimpanzee genomic sequences (Hs-WT and Pt-WT, see Fig. 4A). The splicing difference between the human and chimpanzee wild-type minigene constructs was consistent with the difference of endogenous splicing patterns in human and chimpanzee tissues. The chimpanzee minigene construct had an exon inclusion level of 97%, while the human minigene construct had a much lower exon inclusion level of 69% (Fig. 4B). We then conducted site-directed mutagenesis of the wild-type chimpanzee minigene construct to introduce the human-specific cis-change(s). The T-to-G change within the 3′ splice site reduced the exon inclusion level of the mutant chimpanzee construct (Pt-T(-6)G) to 84%. However, its inclusion level was still higher than that of the wild-type human construct (Hs-WT, 69%). The deletion of the 200 bp downstream intronic segment reduced the exon inclusion level of the mutant chimpanzee construct (Pt-Del) to 91%. Strikingly, when both cis-changes were introduced simultaneously, the inclusion level of the mutant chimpanzee construct (Pt-T(-6)G-Del) was 70%, almost the same as the inclusion level of the wild-type human minigene construct (69%). These data suggest that both human-specific cis-sequence changes contributed to the evolution of MAGOH exon 3 splicing. This example highlights the complexity of molecular events causing splicing differences between species. Additionally, our sequence and minigene analysis of MAGOH illustrates a general strategy to pinpoint the causal cis-regulatory change(s) responsible for lineage-specific splicing patterns.
Figure 4.
Minigene analysis of the human-specific splicing change of MAGOH exon 3. (A) Schematic diagrams of the pI-11-H3 minigene splicing reporter and the wild-type/mutant minigene constructs of MAGOH exon 3. The human-specific T-to-G substitution at the sixth intronic nucleotide of the upstream intron–exon boundary and the human-specific deletion of a 200 bp segment in the downstream intronic region are indicated. (B) Minigene assays of wild-type and mutant MAGOH exon 3 constructs suggest that both human-specific cis-sequence changes contributed to the evolution of splicing. In the gel picture, the number below each lane represents the percent exon inclusion level estimated by fluorescently labeled RT–PCR.
Impact of alternative splicing evolution on the brain transcriptome
An intriguing topic of investigation is whether our identified splicing differences preferentially influence the transcriptome of the brain (cerebellum) rather than other tissues. In this study, we chose to analyze a large panel of available human/primate samples from the cerebellum. It must be noted that the regulation of gene expression and alternative splicing could be either ubiquitous (92) or tissue specific (3,18,93). Thus, two interesting questions arise. First, do the 509 genes showing splicing differences between human and chimpanzee/rhesus cerebellums tend to exhibit brain-specific expression? Second, what fraction of our reported splicing evolution events are ubiquitous in multiple tissues or reflect changes in tissue-specific patterns of splicing?
To address the first question, we analyzed an RNA-Seq data set of nine human tissues, including cerebellum and eight other tissues: adipose, lymph node, heart, muscle, liver, breast, testes and colon (18). For every human gene, we estimated its overall gene expression levels in these nine tissues by calculating the RPKM value [reads per kilobase of exon model per million mapped reads (18,94)] within its constitutive exons. Not surprisingly, we found that the 509 genes with identified splicing differences exhibited strong brain-specificity in their gene expression patterns. The median RPKM value of these 509 genes was significantly higher in the cerebellum than in seven other tissues (see Supplementary Material, Fig. S1). The only exception was in the testes, a tissue known to have similar expression profiles as the brain (95,96). Moreover, of the 13 genes showing human-specific changes in brain splicing patterns (Table 2), the expression levels of 12 genes in the cerebellum were higher than their median expression levels in eight tissues (excluding the testes). Ten genes were expressed at the highest level in the cerebellum. These results indicate that our identified splicing evolution events affect genes preferentially expressed in the brain.
The second question is whether our identified splicing evolution events among human and chimpanzee/rhesus cerebellums are ubiquitous in multiple tissues or reflect changes in tissue-specific patterns of splicing. To distinguish these scenarios, from the 13 exons displaying human-specific changes in brain splicing patterns (Table 2), we selected 10 exons whose inter-species splicing differences in the cerebellum were readily identified by semi-quantitative RT–PCR assays, and examined the splicing patterns of these 10 exons in three additional tissues (kidney, liver and muscle) of all three species. Based on the observed inter/intra-species splicing differences in multiple species, these 10 exons could be classified into three major categories. For one exon (PTPRZ1), the evolution of splicing occurred in a gene whose expression was strongly restricted to the cerebellum (Supplementary Material, Fig. S2A). In seven genes (CAMTA1, DDX42, GSTM3, LPHN3, NUPL1, PIGX, ZDHHC13), the splicing of the exon showed tissue specificity within species, and the evolution of splicing in the cerebellum was different from the patterns observed in some other tissues (Supplementary Material, Fig. S2B–H). For example, in PIGX, although the exon was almost completely skipped in all chimpanzee and rhesus tissues, we found varying degrees of human-specific increase in its exon inclusion level in the four tissues. The inclusion level of this exon was the highest in the cerebellum, followed by muscle, kidney and liver (Supplementary Material, Fig. S2G). In the remaining two genes (ACSL3, MAGOH), the splicing pattern of the exon appeared to be identical among all tested tissues within each individual species. There was no detectable difference in the extent of splicing evolution among different tissues (Supplementary Material, Fig. S2I and J).
Together, these experiments provide examples of both tissue-specific and ubiquitous evolution of alternative splicing. It must be noted that even alternative splicing events that ubiquitously affect a wide range of tissues of broadly expressed genes could have significant functional consequences in the brain/nervous system. Two well-known examples are the disease-associated aberrant alternative splicing of SMN in patients with SMA (spinal muscular atrophy) (97) and of MAPT in patients with FTDP-17 (frontotemporal dementia and parkinsonism linked to chromosome 17) (98,99). In both cases, the disease gene is broadly expressed, and the disease mutation causes splicing defects in many tissues. However, the pathological phenotypes of these aberrant alternative splicing events are strongly restricted to neuronal cells (15,100). Thus, it is entirely possible that a splicing evolution event is shared by a broad range of tissues but still has a significant functional impact in the brain.
DISCUSSION
This study represents a genome-scale phylogenetic survey of alternative splicing evolution in humans and two closely related nonhuman primate species. Using a new high-density exon junction array with a high accuracy for alternative splicing analysis (38–40), we identified 509 genes with splicing differences in the cerebellums of humans and chimpanzees/rhesus macaques. Semi-quantitative and quantitative RT–PCR analyses of a large panel of cerebellum tissues provided strong experimental evidence for a number of genes with human-specific splicing changes after humans diverged from chimpanzees (Table 2). Of these genes, several were previously implicated in the etiology of human neurological diseases. These data are consistent with the view that alternative splicing provides an important mechanism for the creation of evolutionary novelty and species-specific traits (22,23,35). Based on published literature and the analysis of resulting protein isoforms, the identified human-specific splicing changes in several genes (HIP14L, MAGOH) may have a significant functional impact, by removing protein domains/segments that modulate key protein–protein interactions. We also experimentally demonstrated the regulatory impact of a human-specific splicing pattern within the 5′-UTR of DDX42 (Fig. 3). Together, these data suggest an important role of splicing in the evolution of neuronal gene regulation and functions and provide a number of intriguing candidates for detailed functional studies.
An important issue in comparative analysis of alternative splicing is whether the identified inter-species splicing divergence could in fact be attributed to artifacts of intra-species splicing variability due to genetic or environmental factors (3,101–103). This is of particular concern to the analysis of primate tissues, due to the scarcity of these samples and the difficulty in matching age, gender and health conditions of corresponding tissues from multiple species. In our study, two independent lines of evidence suggest that the vast majority of our identified events represent bona fide inter-species divergence of splicing. First, in the HJAY array profiling and subsequent RT–PCR experiments, we examined cerebellum tissues from a large number of individuals from all three species. For most of the RT–PCR-confirmed splicing differences between human and chimpanzee/rhesus cerebellums, we observed consistent splicing patterns within species (see Fig. 2 for examples). Second, we found a strong correlation between the evolution of splicing and the sequence divergence of exons and flanking intronic regions. For exons showing splicing differences across species, we observed a significant increase in the rate of silent substitutions within exons, coupled with accelerated sequence divergence in flanking introns. This was consistent with the role of cis-sequence signals in the regulation of splicing (15,80). This pattern would not be expected if our identified splicing differences between species were significantly contaminated by artifacts due to intra-species splicing variability.
A related question is whether genes showing splicing differences among human, chimpanzee and rhesus cerebellums display sex-biased splicing. Recent studies of primate and rodent tissues have identified evolutionarily conserved sex-biased splicing events (36,104). However, whether the evolutionary divergence of splicing patterns could be sex-specific has not been investigated before. For the 33 exons with RT–PCR data in our work, we compared their splicing patterns in samples from male and female donors. We found only one exon (in FACE2) whose exon inclusion level appeared to be slightly higher in female chimpanzees than in male chimpanzees. However, even in this case, the difference in splicing between male and female chimpanzees was very minor compared with the inter-species difference, which was shared by all single-donor male and female samples from humans and chimpanzees (Supplementary Material, Table S2). It must be emphasized that the goal of this study is to identify robust inter-species differences of splicing, regardless of other factors (including sex) that could contribute to intra-species splicing variability. Therefore, we designed our HJAY array experiments to profile multi-donor human RNAs mixed with male and female samples, as well as single-donor chimpanzee/rhesus samples of both sexes. It is possible that certain splicing events could evolve differently in males and females. However, to identify sex-specific evolution of splicing and possibly compare the rate of such events in males and females, it is necessary to perform genomic profiling and extensive RT–PCR analysis of a much larger panel of samples from both sexes of all three species.
Although we have already identified over 500 genes with differential splicing between human and chimpanzee/rhesus cerebellums, we expect that they constitute only a subset of all splicing differences among these tissues. Despite the high validation rate for the new HJAY array in detecting differential splicing events (38), our approach has several sources of false negatives. First, array-based analysis of alternative splicing is best at the detection of known alternative splicing events. Novel alternative splicing events that are not interrogated by the initial array design will be missed by this analysis. Second, as the HJAY array is designed from human exon annotations, it cannot identify lineage-specific exon loss in humans or lineage-specific exon creation in chimpanzees or rhesus macaques. Third, in order to accurately assess alternative splicing patterns in nonhuman primate tissues, it is necessary to restrict the analysis of HJAY profiles to probes that perfectly match orthologous transcripts from multiple species (41,105,106). Thus, exons with a high rate of sequence divergence between humans and nonhuman primates, which could be hotspots for splicing evolution (35), are less likely to have sufficient perfect-match probes allowing comparative analysis by the HJAY array. These limitations in the array-based approach may be addressed by other genomic technologies for high-throughput splicing analysis. For example, RNA-Seq has emerged as a powerful tool for transcriptome profiling (18,19,107,108). RNA-Seq can detect novel transcripts, exons and splice junctions independent of prior annotations, which is an attractive feature for comparing transcriptomes of closely related species. However, for RNA-Seq to obtain quantitative sampling of alternative splicing events in the entire transcriptome, the sequencing has to go extremely deep. Differential splicing events of genes with intermediate or low expression levels could be missed by RNA-Seq (107). As a result, currently RNA-Seq analysis of alternative splicing is strongly biased towards events in highly expressed genes. For example, in our ongoing study of the epithelia-specific splicing regulator ESRP1/2 (38,91,109), using 76 bp RNA-Seq with over 60 million reads per sample, we can detect only <30% of known ESRP-dependent splicing events previously identified by the HJAY array and confirmed by RT–PCR (Xing Y and Carstens RP, unpublished data). Thus, we anticipate that studies using RNA-Seq may generate a complementary list of splicing differences among these species. In the future, the integration of array and sequencing-based results and further improvement in these technologies could provide a more complete picture of splicing evolution in the primate brain transcriptomes.
Among the 33 events validated by RT–PCR, we observed both exons with substantial changes in splicing patterns between species (e.g. HIP14L, MAGOH, DDX42), and exons with consistent but subtle inter-species differences in exon inclusion levels (see Supplementary Material, Table S2). For subtle differences in splicing between species, there could be several possible evolutionary implications. Some of these events may result from minor, nonfunctional changes in splicing activities that have been tolerated during evolution. These may include evolutionary intermediates that have the potential to evolve into novel functions, as hypothesized by Lee and colleagues (22–24). On the other hand, we note that minor changes in exon inclusion levels (as low as a few percent) sometimes could have significant functional impacts in the brain, as highlighted by studies of several neurological diseases caused by splicing defects (100,110).
MATERIALS AND METHODS
Total RNA preparation and HJAY array profiling of human, chimpanzee and rhesus cerebellum tissues
Postmortem cerebellum samples of eight adult chimpanzees (four males and four females) and eight adult rhesus macaques (two males and six females) were generously provided by the Southwest National Primate Research Center (San Antonio, TX, USA). These animals died of natural causes or non-brain-related diseases. Total RNAs were extracted using TRIzol (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. Two pooled human cerebellum total RNA samples were purchased from Clontech (Mountain View, CA, USA). These two samples (designated as Clontech-1 and Clontech-2 in this manuscript) were pooled from 24 and 10 donors, respectively. Two single-donor human cerebellum total RNA samples were purchased from Ambion (Austin, TX, USA) and BioChain (Hayward, CA, USA). For a detailed description of RNA samples, see Supplementary Material, Table S1.
We used the HJAY array (GEO platform ID: GPL8444) to profile cerebellum tissues from humans, chimpanzees and rhesus macaques. The HJAY array is a next-generation exon array from Affymetrix, with a significantly improved probe design for alternative splicing events over the Affymetrix Exon 1.0 array (38–41). This array averages eight probes per probe set for 315 137 exons and 260 488 exon–exon junctions in the human genome. It covers all alternative splicing events with mRNA/EST evidence in the UCSC/Ensembl databases, including 13 151 exon-skipping events, 6518 alternative splice site events and 1146 events of mutually exclusive exon usage. The HJAY arrays were purchased from Affymetrix as a Technology Access product. Gene, probe set and probe annotations were provided by Affymetrix.
In total, we performed HJAY array hybridization of 18 samples, with 6 samples per species. Specifically, we analyzed three technical replicates for each of the two pooled human cerebellum RNAs (Clontech-1 and Clontech-2). We also examined cerebellum RNAs from six chimpanzees and six rhesus macaques. All RNA samples were processed using the GeneChip Whole Transcript Sense Target Labeling Assay (Affymetrix, Santa Clara, CA, USA) according to the manufacturer's instructions.
Identification of differential splicing events between human, chimpanzee and rhesus cerebellums from the HJAY profiles
We have developed a series of statistical methods for the detection of differential splicing events from the Affymetrix HJAY array data (38,41–45). In this work, these methods were applied to HJAY array data of human, chimpanzee and rhesus cerebellums, after modifications to enable comparisons of splicing patterns between species. Briefly, we identified HJAY array probes perfectly matching exons or exon–exon junctions of orthologous human, chimpanzee and rhesus mRNA transcripts. We then used microarray signals of these probes to identify differential splicing events between humans and chimpanzees or between humans and rhesus macaques. A detailed description of our HJAY array analysis procedure is provided in the Supplementary Material, Methods.
RT–PCR validation of differentially spliced exons between human, chimpanzee and rhesus cerebellums
Single-pass cDNA was synthesized using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA) according to manufacturer's instructions. Two micrograms of total RNA were used for each 20 μl cDNA synthesis reaction. For each tested exon, a pair of forward and reverse PCR primers targeting flanking constitutive exons were designed using PRIMER3 (111). Primer sequences are described in Supplementary Material, Table S6. For each RT–PCR reaction, 15 ng of total RNA equivalent of cDNA were used for the amplification in a 10 μl PCR reaction using the Phire® Hot Start DNA Polymerase (NEB, Ipswich, MA, USA). PCR reactions were run between 25 and 35 cycles (depending on target transcript abundance; optimized for each exon) in a Bio-Rad thermocycler with an annealing temperature of 62–66°C (optimized for each exon). The reaction products were resolved on 5% TBE polyacrylamide gels and visualized by ethidium bromide staining. Each gel picture shown in Supplementary Material, Table S2 was a representation of three to six RT–PCR replications.
For exons with subtle changes in inclusion levels, a fluorescently labeled RT–PCR protocol was used to accurately quantify exon inclusion levels. This protocol was modified from the method described in Schuelke (112). Briefly, a 22 nt universal tag sequence (designated as ‘GFPN’), 5′-CGTCGCCGTCCAGCTCGACCAG-3′ derived from GFP N-terminal region, was added to the 5′ end of the forward or reverse primer during oligo synthesis, while the other primer remained un-tagged. A fluorescently labeled universal primer 5′-FAM-CGTCGCCGTCCAGCTCGACCAG-3′ was added as a third primer in the RT–PCR reaction. The reaction products were resolved on 5% TBE-urea polyacrylamide gels and visualized using a Typhoon 9200 phosphorimager (GE Healthcare, Piscataway, NJ, USA). Bands were quantified using the Quantity One software (Bio-Rad, Hercules, CA, USA). The exon inclusion level of each exon was calculated as the intensity of the exon inclusion band divided by the total intensity of the exon inclusion and skipping bands.
GO term enrichment analysis of genes with identified splicing differences between human, chimpanzee and rhesus cerebellums
We used the online tool DAVID (113,114) to identify significantly enriched GO terms in genes with splicing differences between human, chimpanzee and rhesus cerebellums as identified by our HJAY array analysis. Of all the cassette exons queried by the HJAY array, 1842 exons passed our filters for gene expression level in the cerebellum, probe intensity and number of perfect-match probes for orthologous chimpanzee/rhesus transcripts (see Supplementary Material, Methods). These 1842 cassette exons were included in our pairwise comparisons of splicing patterns between humans and nonhuman primates. From these 1842 exons, we collected 1056 genes with GO terms and used these genes as our background set in the GO term analysis. Using MADS+, we identified a total of 567 cassette exons with substantial splicing changes between humans and chimpanzees or humans and rhesus macaques. From these 567 exons, we collected 389 genes with GO terms. These 389 genes were analyzed by DAVID for the enrichment of GO terms against our background set of 1056 genes as the control.
Analysis of exon–intron nucleotide differences between human, chimpanzee and rhesus genomes
For all the cassette exons included in our HJAY array analysis, we calculated the rates of nucleotide differences between the human, chimpanzee and rhesus genomes within the exons, 100 nt upstream intronic regions and 100 nt downstream intronic regions. Orthologous exonic and intronic regions between these species were identified using the UCSC pairwise genome alignments of the human genome (hg18) to the genomes of chimpanzee (panTro2) and rhesus macaque (rheMac2) (46,47). For each exon, the rates of nucleotide differences (including substitutions and indels) were calculated separately for exonic and flanking intronic regions using the global alignment program NEEDLE from the EMBOSS package (115).
Calculation of exonic synonymous substitution rate (Ks rate)
For all the cassette exons included in our HJAY array analysis, we calculated their exonic synonymous substitution rate (Ks rate) between the human, chimpanzee and rhesus genomes. We computed the Ks rate between orthologous exon pairs following the approach that we used previously (116). Briefly, orthologous exon sequences from human and chimpanzee (or human and rhesus macaque) were retrieved from the UCSC genome alignments and translated in all three possible reading frames. Translations containing stop codons were removed, and the resulting protein sequences were aligned in all possible combinations of reading frames. We computed sequence identities in all resulting alignments using the global sequence alignment program NEEDLE (115). After removing alignments with <50% protein sequence identity, we selected the reading-frame pair with the highest sequence identity and re-aligned these two protein sequences using CLUSTALW (117,118) under default parameters. The resulting CLUSTALW alignment was used to align corresponding nucleotide sequences (codons), and gaps in the alignment were trimmed. We calculated Ks from the codon-based nucleotide sequence alignment using the Yang–Nielsen maximum-likelihood method (119) implemented in the yn00 program of the PAML package (120). For each group of exons (i.e. exons with or without identified splicing changes between species), we summed up the total numbers of synonymous substitutions/sites over all sequences to calculate its overall Ks rate.
Vector construction for luciferase reporter assay
The psiCHECK2 (Promega, Madison, WI, USA) plasmid was linearized through NheI restriction digestion. The exon 2 inclusion and skipping isoforms of DDX42 5′-UTR were cloned into the NheI-linearized psiCHECK2 vector through homologous recombination using the In-Fusion PCR Cloning System (Clontech) according to the manufacturer's protocol. The resulting final constructs contained the DDX42 5′-UTR region directly upstream of the Renilla luciferase start codon without the linker sequence. Structures of the tested vectors are illustrated in Figure 3A.
Transient transfection and dual-luciferase reporter assay
HeLa cells were grown at 37°C with 5% CO2 in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 2 mm glutamine and the antibiotics penicillin and streptomycin. Transfection was done in 24-well plates using Lipofectamine™ 2000 (Invitrogen) according to manufacturer's instructions. Twenty-four hours after transfection, cells were treated with passive lysis buffer (Promega). The firefly and Renilla luciferase activities were assayed using the Dual-luciferase Reporter Assay Kit (Promega) according to the manufacturer's instructions with a GloMax® 96 Microplate Luminometer (Promega).
Quantitative real-time PCR assay
Total RNA was prepared from transfected cells using the TRIzol (Invitrogen) according to manufacturer's instructions. Single-pass cDNA was synthesized using High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems) according to manufacturer's instructions. Quantitative real-time polymerase chain reaction (qRT-PCR) was performed using Power SYBR Green PCR Master Mix (Applied Biosystems) for the luciferase mRNA level. Primers used for the qRT-PCR analysis are Renilla luciferase: forward: 5′-ACAAGTACCTCACCGCTTGG-3′, reverse: 5′-CGATGGCCTTGATCTTGTCT-3′; firefly luciferase: forward: 5′-GGACATCACCTATGCCGAGT-3′, reverse: 5′-GTTCTCAGAGCACACCACGA-3′. Using a mathematical method described by Pfaffl (121), we calculated the average expression fold change of Renilla luciferase and used mRNA concentrations of firefly luciferase for normalization.
Minigene construction and site-directed mutagenesis
Exon 3 of MAGOH gene homologs and its partial flanking introns were amplified from the human and chimpanzee genomic DNAs using PfuUltra Fusion II HS DNA polymerase (Stratagene, La Jolla, CA, USA). PCR products were subcloned into the NheI site of the pI-11-H3 minigene vector (91) (kindly provided by Dr Russ P. Carstens, University of Pennsylvania, Philadelphia, PA, USA) using the In-Fusion Advantage PCR Cloning Kit (Clontech). Site-directed mutagenesis was done using PfuUltra Fusion II HS DNA polymerase. All sequences and mutations were verified by DNA sequencing.
In vitro minigene splicing reporter assay
HeLa cells were grown in DMEM (Invitrogen) with 10% FBS (Invitrogen). Cells were plated in 24-well plates and transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. RNA was purified 16 h after transfection and reverse-transcribed into single-pass cDNA. Fluorescently labeled RT–PCR was done as described in the previous section. The pI-11-H3 minigene-specific primer sequences were pI11-F: 5′-GCTGTCTGCGAGGTACCCTA-3′; pI11-R: 5′-CGTCGCCGTCCAGCTCGACCAGCGTTCGGAGGATGCATAGAG-3′.
SUPPLEMENTARY MATERIAL
FUNDING
We thank David Eichmann, Ben Rogers and the University of Iowa Institute for Clinical and Translational Science (NIH grant UL1 RR024979) for computer support. This study is supported by NIH grants R01HG004634 and R01GM088342, a junior faculty grant from the Edward Mallinckrodt Jr Foundation, a Basil O'Connor Starter Scholar Research Award from the March of Dimes Foundation and a research startup fund from the University of Iowa. This study used biological materials obtained from the Southwest National Primate Research Center, which is supported by NIH-NCRR grant P51 RR013986.
Supplementary Material
ACKNOWLEDGEMENTS
We wish to thank Jerilyn Pecotte, Mary Jo Aivaliotis for assistance, Russ Carstens and Peter Stoilov for discussions and comments on this work, Douglas Black and Peter Stoilov for sharing of the FAM-labeling RT–PCR protocol and James Cai for SNP-based scans of positive selection.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Varki A. A chimpanzee genome project is a biomedical imperative. Genome Res. 2000;10:1065–1070. doi: 10.1101/gr.10.8.1065. [DOI] [PubMed] [Google Scholar]
- 2.King M.C., Wilson A.C. Evolution at two levels in humans and chimpanzees. Science. 1975;188:107–116. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- 3.Khaitovich P., Enard W., Lachmann M., Paabo S. Evolution of primate gene expression. Nat. Rev. Genet. 2006;7:693–702. doi: 10.1038/nrg1940. [DOI] [PubMed] [Google Scholar]
- 4.Blekhman R., Oshlack A., Chabot A.E., Smyth G.K., Gilad Y. Gene regulation in primates evolves under tissue-specific selection pressures. PLoS Genet. 2008;4:e1000271. doi: 10.1371/journal.pgen.1000271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gilad Y., Oshlack A., Smyth G.K., Speed T.P., White K.P. Expression profiling in primates reveals a rapid evolution of human transcription factors. Nature. 2006;440:242–245. doi: 10.1038/nature04559. [DOI] [PubMed] [Google Scholar]
- 6.Enard W., Khaitovich P., Klose J., Zollner S., Heissig F., Giavalisco P., Nieselt-Struwe K., Muchmore E., Varki A., Ravid R., et al. Intra- and interspecific variation in primate gene expression patterns. Science. 2002;296:340–343. doi: 10.1126/science.1068996. [DOI] [PubMed] [Google Scholar]
- 7.Khaitovich P., Hellmann I., Enard W., Nowick K., Leinweber M., Franz H., Weiss G., Lachmann M., Paabo S. Parallel patterns of evolution in the genomes and transcriptomes of humans and chimpanzees. Science. 2005;309:1850–1854. doi: 10.1126/science.1108296. [DOI] [PubMed] [Google Scholar]
- 8.Khaitovich P., Muetzel B., She X., Lachmann M., Hellmann I., Dietzsch J., Steigele S., Do H.H., Weiss G., Enard W., et al. Regional patterns of gene expression in human and chimpanzee brains. Genome Res. 2004;14:1462–1473. doi: 10.1101/gr.2538704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clark T.A., Schweitzer A.C., Chen T.X., Staples M.K., Lu G., Wang H., Williams A., Blume J.E. Discovery of tissue-specific exons using comprehensive human exon microarrays. Genome Biol. 2007;8:R64. doi: 10.1186/gb-2007-8-4-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu X., Yang D., Ding J.H., Wang W., Chu P.H., Dalton N.D., Wang H.Y., Bermingham J.R., Jr, Ye Z., Liu F., et al. ASF/SF2-regulated CaMKIIdelta alternative splicing temporally reprograms excitation–contraction coupling in cardiac muscle. Cell. 2005;120:59–72. doi: 10.1016/j.cell.2004.11.036. [DOI] [PubMed] [Google Scholar]
- 11.Boutz P.L., Stoilov P., Li Q., Lin C.H., Chawla G., Ostrow K., Shiue L., Ares M., Jr, Black D.L. A post-transcriptional regulatory switch in polypyrimidine tract-binding proteins reprograms alternative splicing in developing neurons. Genes Dev. 2007;21:1636–1652. doi: 10.1101/gad.1558107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ip J.Y., Tong A., Pan Q., Topp J.D., Blencowe B.J., Lynch K.W. Global analysis of alternative splicing during T-cell activation. RNA. 2007;13:563–572. doi: 10.1261/rna.457207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee J.A., Xing Y., Nguyen D., Xie J., Lee C.J., Black D.L. Depolarization and CaM kinase IV modulate NMDA receptor splicing through two essential RNA elements. PLoS Biol. 2007;5:e40. doi: 10.1371/journal.pbio.0050040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cartegni L., Chew S.L., Krainer A.R. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat. Rev. Genet. 2002;3:285–298. doi: 10.1038/nrg775. [DOI] [PubMed] [Google Scholar]
- 15.Wang G.S., Cooper T.A. Splicing in disease: disruption of the splicing code and the decoding machinery. Nat. Rev. Genet. 2007;8:749–761. doi: 10.1038/nrg2164. [DOI] [PubMed] [Google Scholar]
- 16.Modrek B., Lee C. A genomic view of alternative splicing. Nat. Genet. 2002;30:13–19. doi: 10.1038/ng0102-13. [DOI] [PubMed] [Google Scholar]
- 17.Johnson J.M., Castle J., Garrett-Engele P., Kan Z., Loerch P.M., Armour C.D., Santos R., Schadt E.E., Stoughton R., Shoemaker D.D. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- 18.Wang E.T., Sandberg R., Luo S., Khrebtukova I., Zhang L., Mayr C., Kingsmore S.F., Schroth G.P., Burge C.B. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pan Q., Shai O., Lee L.J., Frey B.J., Blencowe B.J. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 20.Black D.L. Protein diversity from alternative splicing: a challenge for bioinformatics and post-genome biology. Cell. 2000;103:367–370. doi: 10.1016/s0092-8674(00)00128-8. [DOI] [PubMed] [Google Scholar]
- 21.Smith C.W.J., Valcarcel J. Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem. Sci. 2000;25:381–388. doi: 10.1016/s0968-0004(00)01604-2. [DOI] [PubMed] [Google Scholar]
- 22.Xing Y., Lee C. Alternative splicing and RNA selection pressure – evolutionary consequences for eukaryotic genomes. Nat. Rev. Genet. 2006;7:499–509. doi: 10.1038/nrg1896. [DOI] [PubMed] [Google Scholar]
- 23.Sorek R. The birth of new exons: mechanisms and evolutionary consequences. RNA. 2007;13:1603–1608. doi: 10.1261/rna.682507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Modrek B., Lee C. Alternative splicing in the human, mouse and rat genomes is associated with an increased rate of exon creation/loss. Nat. Genet. 2003;34:177–180. doi: 10.1038/ng1159. [DOI] [PubMed] [Google Scholar]
- 25.Kan Z., States D., Gish W. Selecting for functional alternative splices in ESTs. Genome Res. 2002;12:1837–1845. doi: 10.1101/gr.764102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thanaraj T.A., Clark F., Muilu J. Conservation of human alternative splice events in mouse. Nucleic Acids Res. 2003;31:2544–2552. doi: 10.1093/nar/gkg355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nurtdinov R.N., Artamonova I.I., Mironov A.A., Gelfand M.S. Low conservation of alternative splicing patterns in the human and mouse genomes. Hum. Mol. Genet. 2003;12:1313–1320. doi: 10.1093/hmg/ddg137. [DOI] [PubMed] [Google Scholar]
- 28.Pan Q., Bakowski M.A., Morris Q., Zhang W., Frey B.J., Hughes T.R., Blencowe B.J. Alternative splicing of conserved exons is frequently species-specific in human and mouse. Trends Genet. 2005;21:73–77. doi: 10.1016/j.tig.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 29.Lev-Maor G., Goren A., Sela N., Kim E., Keren H., Doron-Faigenboim A., Leibman-Barak S., Pupko T., Ast G. The ‘alternative’ choice of constitutive exons throughout evolution. PLoS Genet. 2007;3:e203. doi: 10.1371/journal.pgen.0030203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ast G. How did alternative splicing evolve? Nat. Rev. Genet. 2004;5:773–782. doi: 10.1038/nrg1451. [DOI] [PubMed] [Google Scholar]
- 31.Lin L., Shen S., Tye A., Cai J.J., Jiang P., Davidson B.L., Xing Y. Diverse splicing patterns of exonized Alu elements in human tissues. PLoS Genet. 2008;4:e1000225. doi: 10.1371/journal.pgen.1000225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sorek R. When new exons are born. Heredity. 2009;103:279–280. doi: 10.1038/hdy.2009.62. [DOI] [PubMed] [Google Scholar]
- 33.Lev-Maor G., Sorek R., Shomron N., Ast G. The birth of an alternatively spliced exon: 3′ splice-site selection in Alu exons. Science. 2003;300:1288–1291. doi: 10.1126/science.1082588. [DOI] [PubMed] [Google Scholar]
- 34.Gerber A., O'Connell M.A., Keller W. Two forms of human double-stranded RNA-specific editase 1 (hRED1) generated by the insertion of an Alu cassette. RNA. 1997;3:453–463. [PMC free article] [PubMed] [Google Scholar]
- 35.Calarco J.A., Xing Y., Caceres M., Calarco J.P., Xiao X., Pan Q., Lee C., Preuss T.M., Blencowe B.J. Global analysis of alternative splicing differences between humans and chimpanzees. Genes Dev. 2007;21:2963–2975. doi: 10.1101/gad.1606907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blekhman R., Marioni J.C., Zumbo P., Stephens M., Gilad Y. Sex-specific and lineage-specific alternative splicing in primates. Genome Res. 2010;20:180–189. doi: 10.1101/gr.099226.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hedges S.B. The origin and evolution of model organisms. Nat. Rev. Genet. 2002;3:838–849. doi: 10.1038/nrg929. [DOI] [PubMed] [Google Scholar]
- 38.Shen S., Warzecha C.C., Carstens R.P., Xing Y. MADS+: discovery of differential splicing events from Affymetrix exon junction array data. Bioinformatics. 2010;26:268–269. doi: 10.1093/bioinformatics/btp643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Licatalosi D.D., Mele A., Fak J.J., Ule J., Kayikci M., Chi S.W., Clark T.A., Schweitzer A.C., Blume J.E., Wang X., et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamamoto M.L., Clark T.A., Gee S.L., Kang J.A., Schweitzer A.C., Wickrema A., Conboy J.G. Alternative pre-mRNA splicing switches modulate gene expression in late erythropoiesis. Blood. 2009;113:3363–3370. doi: 10.1182/blood-2008-05-160325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin L., Liu S., Brockway H., Seok J., Jiang P., Wong W.H., Xing Y. Using high-density exon arrays to profile gene expression in closely related species. Nucleic Acids Res. 2009;37:e90. doi: 10.1093/nar/gkp420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kapur K., Jiang H., Xing Y., Wong W.H. Cross-hybridization modeling on Affymetrix exon arrays. Bioinformatics. 2008;24:2887–2893. doi: 10.1093/bioinformatics/btn571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kapur K., Xing Y., Ouyang Z., Wong W.H. Exon arrays provide accurate assessments of gene expression. Genome Biol. 2007;8:R82. doi: 10.1186/gb-2007-8-5-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xing Y., Kapur K., Wong W.H. Probe selection and expression index computation of Affymetrix exon arrays. PLoS ONE. 2006;1:e88. doi: 10.1371/journal.pone.0000088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xing Y., Stoilov P., Kapur K., Han A., Jiang H., Shen S., Black D.L., Wong W.H. MADS: a new and improved method for analysis of differential alternative splicing by exon-tiling microarrays. RNA. 2008;14:1470–1479. doi: 10.1261/rna.1070208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kuhn R.M., Karolchik D., Zweig A.S., Wang T., Smith K.E., Rosenbloom K.R., Rhead B., Raney B.J., Pohl A., Pheasant M., et al. The UCSC Genome Browser database: update 2009. Nucleic Acids Res. 2009;37:D755–D761. doi: 10.1093/nar/gkn875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miller W., Rosenbloom K., Hardison R.C., Hou M., Taylor J., Raney B., Burhans R., King D.C., Baertsch R., Blankenberg D., et al. 28-way vertebrate alignment and conservation track in the UCSC Genome Browser. Genome Res. 2007;17:1797–1808. doi: 10.1101/gr.6761107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dorus S., Vallender E.J., Evans P.D., Anderson J.R., Gilbert S.L., Mahowald M., Wyckoff G.J., Malcom C.M., Lahn B.T. Accelerated evolution of nervous system genes in the origin of Homo sapiens. Cell. 2004;119:1027–1040. doi: 10.1016/j.cell.2004.11.040. [DOI] [PubMed] [Google Scholar]
- 49.Bakewell M.A., Shi P., Zhang J. More genes underwent positive selection in chimpanzee evolution than in human evolution. Proc. Natl Acad. Sci. USA. 2007;104:7489–7494. doi: 10.1073/pnas.0701705104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shi P., Bakewell M.A., Zhang J. Did brain-specific genes evolve faster in humans than in chimpanzees? Trends Genet. 2006;22:608–613. doi: 10.1016/j.tig.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 51.Huang da W., Sherman B.T., Tan Q., Collins J.R., Alvord W.G., Roayaei J., Stephens R., Baseler M.W., Lane H.C., Lempicki R.A. The DAVID Gene Functional Classification Tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007;8:R183. doi: 10.1186/gb-2007-8-9-r183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bustamante C.D., Fledel-Alon A., Williamson S., Nielsen R., Hubisz M.T., Glanowski S., Tanenbaum D.M., White T.J., Sninsky J.J., Hernandez R.D., et al. Natural selection on protein-coding genes in the human genome. Nature. 2005;437:1153–1157. doi: 10.1038/nature04240. [DOI] [PubMed] [Google Scholar]
- 53.Sabeti P.C., Schaffner S.F., Fry B., Lohmueller J., Varilly P., Shamovsky O., Palma A., Mikkelsen T.S., Altshuler D., Lander E.S. Positive natural selection in the human lineage. Science. 2006;312:1614–1620. doi: 10.1126/science.1124309. [DOI] [PubMed] [Google Scholar]
- 54.Kelley J.L., Swanson W.J. Positive selection in the human genome: from genome scans to biological significance. Annu. Rev. Genomics Hum. Genet. 2008;9:143–160. doi: 10.1146/annurev.genom.9.081307.164411. [DOI] [PubMed] [Google Scholar]
- 55.Huttley G.A., Smith M.W., Carrington M., O'Brien S.J. A scan for linkage disequilibrium across the human genome. Genetics. 1999;152:1711–1722. doi: 10.1093/genetics/152.4.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Akey J.M., Zhang G., Zhang K., Jin L., Shriver M.D. Interrogating a high-density SNP map for signatures of natural selection. Genome Res. 2002;12:1805–1814. doi: 10.1101/gr.631202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Carlson C.S., Thomas D.J., Eberle M.A., Swanson J.E., Livingston R.J., Rieder M.J., Nickerson D.A. Genomic regions exhibiting positive selection identified from dense genotype data. Genome Res. 2005;15:1553–1565. doi: 10.1101/gr.4326505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.International-HapMap-Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nielsen R., Williamson S., Kim Y., Hubisz M.J., Clark A.G., Bustamante C. Genomic scans for selective sweeps using SNP data. Genome Res. 2005;15:1566–1575. doi: 10.1101/gr.4252305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Voight B.F., Kudaravalli S., Wen X., Pritchard J.K. A map of recent positive selection in the human genome. PLoS Biol. 2006;4:e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang E.T., Kodama G., Baldi P., Moyzis R.K. Global landscape of recent inferred Darwinian selection for Homo sapiens. Proc. Natl Acad. Sci. USA. 2006;103:135–140. doi: 10.1073/pnas.0509691102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Frazer K.A., Ballinger D.G., Cox D.R., Hinds D.A., Stuve L.L., Gibbs R.A., Belmont J.W., Boudreau A., Hardenbol P., Leal S.M., et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kimura R., Fujimoto A., Tokunaga K., Ohashi J. A practical genome scan for population-specific strong selective sweeps that have reached fixation. PLoS ONE. 2007;2:e286. doi: 10.1371/journal.pone.0000286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sabeti P.C., Varilly P., Fry B., Lohmueller J., Hostetter E., Cotsapas C., Xie X., Byrne E.H., McCarroll S.A., Gaudet R., et al. Genome-wide detection and characterization of positive selection in human populations. Nature. 2007;449:913–918. doi: 10.1038/nature06250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tang K., Thornton K.R., Stoneking M. A new approach for using genome scans to detect recent positive selection in the human genome. PLoS Biol. 2007;5:e171. doi: 10.1371/journal.pbio.0050171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Williamson S.H., Hubisz M.J., Clark A.G., Payseur B.A., Bustamante C.D., Nielsen R. Localizing recent adaptive evolution in the human genome. PLoS Genet. 2007;3:e90. doi: 10.1371/journal.pgen.0030090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Oleksyk T.K., Zhao K., De La Vega F.M., Gilbert D.A., O'Brien S.J., Smith M.W. Identifying selected regions from heterozygosity and divergence using a light-coverage genomic dataset from two human populations. PLoS ONE. 2008;3:e1712. doi: 10.1371/journal.pone.0001712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Huang K., Sanders S., Singaraja R., Orban P., Cijsouw T., Arstikaitis P., Yanai A., Hayden M.R., El-Husseini A. Neuronal palmitoyl acyl transferases exhibit distinct substrate specificity. FASEB J. 2009;23:2605–2615. doi: 10.1096/fj.08-127399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hong G.S., Heun R., Jessen F., Popp J., Hentschel F., Kelemen P., Schulz A., Maier W., Kolsch H. Gene variations in GSTM3 are a risk factor for Alzheimer's disease. Neurobiol. Aging. 2009;30:691–696. doi: 10.1016/j.neurobiolaging.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 70.Buxbaum J.D., Georgieva L., Young J.J., Plescia C., Kajiwara Y., Jiang Y., Moskvina V., Norton N., Peirce T., Williams H., et al. Molecular dissection of NRG1-ERBB4 signaling implicates PTPRZ1 as a potential schizophrenia susceptibility gene. Mol. Psychiatry. 2008;13:162–172. doi: 10.1038/sj.mp.4001991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li J., Mahajan A., Tsai M.D. Ankyrin repeat: a unique motif mediating protein–protein interactions. Biochemistry. 2006;45:15168–15178. doi: 10.1021/bi062188q. [DOI] [PubMed] [Google Scholar]
- 72.Gehring N.H., Lamprinaki S., Kulozik A.E., Hentze M.W. Disassembly of exon junction complexes by PYM. Cell. 2009;137:536–548. doi: 10.1016/j.cell.2009.02.042. [DOI] [PubMed] [Google Scholar]
- 73.Scheper G.C., van der Knaap M.S., Proud C.G. Translation matters: protein synthesis defects in inherited disease. Nat. Rev. Genet. 2007;8:711–723. doi: 10.1038/nrg2142. [DOI] [PubMed] [Google Scholar]
- 74.Wang G., Guo X., Floros J. Differences in the translation efficiency and mRNA stability mediated by 5′-UTR splice variants of human SP-A1 and SP-A2 genes. Am. J. Physiol. Lung Cell. Mol. Physiol. 2005;289:L497–L508. doi: 10.1152/ajplung.00100.2005. [DOI] [PubMed] [Google Scholar]
- 75.Shalev A., Blair P.J., Hoffmann S.C., Hirshberg B., Peculis B.A., Harlan D.M. A proinsulin gene splice variant with increased translation efficiency is expressed in human pancreatic islets. Endocrinology. 2002;143:2541–2547. doi: 10.1210/endo.143.7.8920. [DOI] [PubMed] [Google Scholar]
- 76.Lin L., Jiang P., Shen S., Sato S., Davidson B.L., Xing Y. Large-scale analysis of exonized mammalian-wide interspersed repeats in primate genomes. Hum. Mol. Genet. 2009;18:2204–2214. doi: 10.1093/hmg/ddp152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang X.H., Chasin L.A. Comparison of multiple vertebrate genomes reveals the birth and evolution of human exons. Proc. Natl Acad. Sci. USA. 2006;103:13427–13432. doi: 10.1073/pnas.0603042103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Watatani Y., Ichikawa K., Nakanishi N., Fujimoto M., Takeda H., Kimura N., Hirose H., Takahashi S., Takahashi Y. Stress-induced translation of ATF5 mRNA is regulated by the 5′-untranslated region. J. Biol. Chem. 2008;283:2543–2553. doi: 10.1074/jbc.M707781200. [DOI] [PubMed] [Google Scholar]
- 79.Arora A., Dutkiewicz M., Scaria V., Hariharan M., Maiti S., Kurreck J. Inhibition of translation in living eukaryotic cells by an RNA G-quadruplex motif. RNA. 2008;14:1290–1296. doi: 10.1261/rna.1001708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang Z., Burge C.B. Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. RNA. 2008;14:802–813. doi: 10.1261/rna.876308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hurst L.D., Pal C. Evidence for purifying selection acting on silent sites in BRCA1. Trends Genet. 2001;17:62–65. doi: 10.1016/s0168-9525(00)02173-9. [DOI] [PubMed] [Google Scholar]
- 82.Chamary J.V., Parmley J.L., Hurst L.D. Hearing silence: non-neutral evolution at synonymous sites in mammals. Nat. Rev. Genet. 2006;7:98–108. doi: 10.1038/nrg1770. [DOI] [PubMed] [Google Scholar]
- 83.Resch A.M., Carmel L., Marino-Ramirez L., Ogurtsov A.Y., Shabalina S.A., Rogozin I.B., Koonin E.V. Widespread positive selection in synonymous sites of mammalian genes. Mol. Biol. Evol. 2007;24:1821–1831. doi: 10.1093/molbev/msm100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ke S., Zhang X.H., Chasin L.A. Positive selection acting on splicing motifs reflects compensatory evolution. Genome Res. 2008;18:533–543. doi: 10.1101/gr.070268.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lu H., Lin L., Sato S., Xing Y., Lee C.J. Predicting functional alternative splicing by measuring RNA selection pressure from multigenome alignments. PLoS Comput. Biol. 2009;5:e1000608. doi: 10.1371/journal.pcbi.1000608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yeo G., Burge C.B. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J. Comput. Biol. 2004;11:377–394. doi: 10.1089/1066527041410418. [DOI] [PubMed] [Google Scholar]
- 87.Fairbrother W.G., Yeh R.F., Sharp P.A., Burge C.B. Predictive identification of exonic splicing enhancers in human genes. Science. 2002;297:1007–1013. doi: 10.1126/science.1073774. [DOI] [PubMed] [Google Scholar]
- 88.Wang Z., Rolish M.E., Yeo G., Tung V., Mawson M., Burge C.B. Systematic identification and analysis of exonic splicing silencers. Cell. 2004;119:831–845. doi: 10.1016/j.cell.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 89.Blencowe B.J. Alternative splicing: new insights from global analyses. Cell. 2006;126:37–47. doi: 10.1016/j.cell.2006.06.023. [DOI] [PubMed] [Google Scholar]
- 90.Li Q., Lee J.A., Black D.L. Neuronal regulation of alternative pre-mRNA splicing. Nat. Rev. Neurosci. 2007;8:819–831. doi: 10.1038/nrn2237. [DOI] [PubMed] [Google Scholar]
- 91.Warzecha C.C., Sato T.K., Nabet B., Hogenesch J.B., Carstens R.P. ESRP1 and ESRP2 are epithelial cell-type-specific regulators of FGFR2 splicing. Mol. Cell. 2009;33:591–601. doi: 10.1016/j.molcel.2009.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ramskold D., Wang E.T., Burge C.B., Sandberg R. An abundance of ubiquitously expressed genes revealed by tissue transcriptome sequence data. PLoS Comput. Biol. 2009;5:e1000598. doi: 10.1371/journal.pcbi.1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pan Q., Shai O., Misquitta C., Zhang W., Saltzman A.L., Mohammad N., Babak T., Siu H., Hughes T.R., Morris Q.D., et al. Revealing global regulatory features of mammalian alternative splicing using a quantitative microarray platform. Mol. Cell. 2004;16:929–941. doi: 10.1016/j.molcel.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 94.Mortazavi A., Williams B.A., McCue K., Schaeffer L., Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 95.Guo J.H., Huang Q., Studholme D.J., Wu C.Q., Zhao Z. Transcriptomic analyses support the similarity of gene expression between brain and testis in human as well as mouse. Cytogenet. Genome Res. 2005;111:107–109. doi: 10.1159/000086378. [DOI] [PubMed] [Google Scholar]
- 96.Guo J., Zhu P., Wu C., Yu L., Zhao S., Gu X. In silico analysis indicates a similar gene expression pattern between human brain and testis. Cytogenet. Genome Res. 2003;103:58–62. doi: 10.1159/000076290. [DOI] [PubMed] [Google Scholar]
- 97.Lorson C.L., Hahnen E., Androphy E.J., Wirth B. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc. Natl Acad. Sci. USA. 1999;96:6307–6311. doi: 10.1073/pnas.96.11.6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hutton M., Lendon C.L., Rizzu P., Baker M., Froelich S., Houlden H., Pickering-Brown S., Chakraverty S., Isaacs A., Grover A., et al. Association of missense and 5′-splice-site mutations in tau with the inherited dementia FTDP-17. Nature. 1998;393:702–705. doi: 10.1038/31508. [DOI] [PubMed] [Google Scholar]
- 99.Ingram E.M., Spillantini M.G. Tau gene mutations: dissecting the pathogenesis of FTDP-17. Trends Mol. Med. 2002;8:555–562. doi: 10.1016/s1471-4914(02)02440-1. [DOI] [PubMed] [Google Scholar]
- 100.Garcia-Blanco M.A., Baraniak A.P., Lasda E.L. Alternative splicing in disease and therapy. Nat. Biotechnol. 2004;22:535–546. doi: 10.1038/nbt964. [DOI] [PubMed] [Google Scholar]
- 101.Kwan T., Benovoy D., Dias C., Gurd S., Provencher C., Beaulieu P., Hudson T.J., Sladek R., Majewski J. Genome-wide analysis of transcript isoform variation in humans. Nat. Genet. 2008;40:225–231. doi: 10.1038/ng.2007.57. [DOI] [PubMed] [Google Scholar]
- 102.Kwan T., Benovoy D., Dias C., Gurd S., Serre D., Zuzan H., Clark T.A., Schweitzer A., Staples M.K., Wang H., et al. Heritability of alternative splicing in the human genome. Genome Res. 2007;17:1210–1218. doi: 10.1101/gr.6281007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhang W., Duan S., Bleibel W.K., Wisel S.A., Huang R.S., Wu X., He L., Clark T.A., Chen T.X., Schweitzer A.C., et al. Identification of common genetic variants that account for transcript isoform variation between human populations. Hum. Genet. 2009;125:81–93. doi: 10.1007/s00439-008-0601-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Su W.L., Modrek B., GuhaThakurta D., Edwards S., Shah J.K., Kulkarni A.V., Russell A., Schadt E.E., Johnson J.M., Castle J.C. Exon and junction microarrays detect widespread mouse strain- and sex-bias expression differences. BMC Genomics. 2008;9:273. doi: 10.1186/1471-2164-9-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gilad Y., Borevitz J. Using DNA microarrays to study natural variation. Curr. Opin. Genet. Dev. 2006;16:553–558. doi: 10.1016/j.gde.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 106.Gilad Y., Rifkin S.A., Bertone P., Gerstein M., White K.P. Multi-species microarrays reveal the effect of sequence divergence on gene expression profiles. Genome Res. 2005;15:674–680. doi: 10.1101/gr.3335705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wang Z., Gerstein M., Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Blencowe B.J., Ahmad S., Lee L.J. Current-generation high-throughput sequencing: deepening insights into mammalian transcriptomes. Genes Dev. 2009;23:1379–1386. doi: 10.1101/gad.1788009. [DOI] [PubMed] [Google Scholar]
- 109.Warzecha C.C., Shen S., Xing Y., Carstens R.P. The epithelial splicing factors ESRP1 and ESRP2 positively and negatively regulate diverse types of alternative splicing events. RNA Biol. 2009;6:546–562. doi: 10.4161/rna.6.5.9606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Buchner D.A., Trudeau M., Meisler M.H. SCNM1, a putative RNA splicing factor that modifies disease severity in mice. Science. 2003;301:967–969. doi: 10.1126/science.1086187. [DOI] [PubMed] [Google Scholar]
- 111.Rozen S., Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 112.Schuelke M. An economic method for the fluorescent labeling of PCR fragments. Nat. Biotechnol. 2000;18:233–234. doi: 10.1038/72708. [DOI] [PubMed] [Google Scholar]
- 113.Huang da W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 114.Dennis G., Jr, Sherman B.T., Hosack D.A., Yang J., Gao W., Lane H.C., Lempicki R.A. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 115.Rice P., Longden I., Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 116.Xing Y., Lee C. Evidence of functional selection pressure for alternative splicing events that accelerate evolution of protein subsequences. Proc. Natl Acad. Sci. USA. 2005;102:13526–13531. doi: 10.1073/pnas.0501213102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R., et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 118.Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yang Z., Nielsen R. Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol. Biol. Evol. 2000;17:32–43. doi: 10.1093/oxfordjournals.molbev.a026236. [DOI] [PubMed] [Google Scholar]
- 120.Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- 121.Pfaffl M.W. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.