Abstract
Mitochondrial succinate-coenzyme Q reductase (complex II) consists of four subunits, SDHA, SDHB, SDHC and SDHD. Heterozygous germline mutations in SDHB, SDHC, SDHD and SDHAF2 [encoding for succinate dehydrogenase (SDH) complex assembly factor 2] cause hereditary paragangliomas and pheochromocytomas. Surprisingly, no genetic link between SDHA and paraganglioma/pheochromocytoma syndrome has ever been established. We identified a heterozygous germline SDHA mutation, p.Arg589Trp, in a woman suffering from catecholamine-secreting abdominal paraganglioma. The functionality of the SDHA mutant was assessed by studying SDHA, SDHB, HIF-1α and CD34 protein expression using immunohistochemistry and by examining the effect of the mutation in a yeast model. Microarray analyses were performed to study gene expression involved in energy metabolism and hypoxic pathways. We also investigated 202 paragangliomas or pheochromocytomas for loss of heterozygosity (LOH) at the SDHA, SDHB, SDHC and SDHD loci by BAC array comparative genomic hybridization. In vivo and in vitro functional studies demonstrated that the SDHA mutation causes a loss of SDH enzymatic activity in tumor tissue and in the yeast model. Immunohistochemistry and transcriptome analyses established that the SDHA mutation causes pseudo-hypoxia, which leads to a subsequent increase in angiogenesis, as other SDHx gene mutations. LOH was detected at the SDHA locus in the patient's tumor but was present in only 4.5% of a large series of paragangliomas and pheochromocytomas. The SDHA gene should be added to the list of genes encoding tricarboxylic acid cycle proteins that act as tumor suppressor genes and can now be considered as a new paraganglioma/pheochromocytoma susceptibility gene.
INTRODUCTION
Paraganglioma and pheochromocytoma are rare tumors of chromaffin tissue that may secrete catecholamines. They arise in the adrenal medulla (pheochromocytoma proper) or in extra-adrenal regions in the thorax, abdomen or pelvis. They may also be derived from parasympathetic tissue of the head and neck. Paragangliomas and pheochromocytomas occur either sporadically or in the context of several inherited syndromes: multiple endocrine neoplasia type 2, von Hippel–Lindau disease, neurofibromatosis type 1 and familial paraganglioma-1 (PGL1), PGL2, PGL3 or PGL4 caused, respectively, by germline mutations in the RET, VHL, NF1 and SDHD, SDHAF2, SDHC or SDHB genes (1,2). SDHD, SDHB and SDHC encode three of the four subunits of succinate-coenzyme Q reductase (complex II, succinate dehydrogenase, SDH), a mitochondrial enzyme located at the crossroads between the tricarboxylic acid (TCA) cycle and the respiratory chain. SDH catalyzes the oxidation of succinate to fumarate and transfers electrons directly to the ubiquinone pool. SDHD, SDHB and SDHC (SDHx) mutations cause a cascade of molecular events leading to the abnormal stabilization of hypoxia-inducible factors (HIF) under normoxic conditions (2) or pseudo-hypoxia (via inactivation of SDH, accumulation of succinate, inhibition of prolyl-4-hydroxylases and subsequent impairment of HIF hydroxylation) (3,4), thereby promoting cell proliferation, angiogenesis and tumorigenesis (5).
Mutations in SDHA, which encodes the fourth subunit of SDH, have never been described in hereditary paraganglioma/pheochromocytoma. Biallelic SDHA mutations have been shown to cause an early onset encephalopathy known as Leigh syndrome (6–9). Reports of patients with complex II deficiency but lacking mutations in any of the four SDHx genes suggested the existence of additional nuclear genes involved in the synthesis, assembly or maintenance of SDH (8). This was confirmed by the recent description of homozygous mutations in the SDHAF1 (SDH assembly factor 1) gene, causing infantile leukoencephalopathy (10). A mutation in another gene, SDHAF2, involved in the assembly of SDH was also recently reported. Mutations in SDHAF2, encoding the SDH assembly factor 2, required for SDH activity and stability, were described in two families affected by head and neck paragangliomas (2,11). SDHA, the flavoprotein-containing subunit of SDH, contains a covalently attached flavin adenine dinucleotide (FAD) cofactor. The requirement of SDHAF2 for flavination is supported by a dramatic decrease in FAD in SDHA with a concomitant loss of SDH enzymatic activity in SDHAF2-related paragangliomas (2).
It has been a source of puzzlement that while paraganglioma/pheochromocytoma has been associated with mutations in SDHB, SDHC, SDHD and most recently in a gene involved in flavination of SDHA, none have been reported in SDHA. On the basis of the previous claim of the existence of two human SDHA isoforms encoded by two distinct genes (12), a possible explanation for the absence of SDHA-related paraganglioma/pheochromocytoma was that it would require tetra-allelic genetic events in two independent SDHA loci (3,12,13). This explanation, however, lost credibility in view of later evidence favoring the existence of a single highly polymorphic SDHA gene (14).
Here, based on the observation of a patient with an extra-adrenal paraganglioma resulting from a loss-of-function germline mutation in SDHA, we show that SDHA, like other SDHx genes, can act as a tumor suppressor gene and activate the pseudo-hypoxic pathway.
RESULTS
Identification of the SDHA mutation and loss of heterozygosity at the SDHA locus
Genetic testing was proposed to the patient affected by an extra-adrenal paraganglioma in accordance with the international recommendations (15).
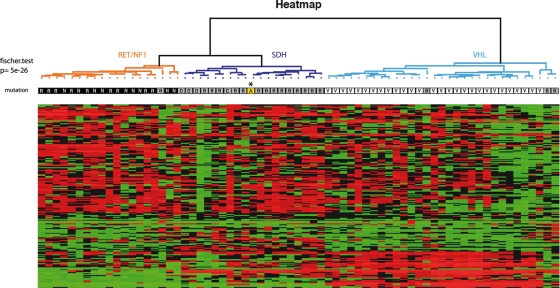
Mutation analyses in RET, VHL, SDHB, SDHC, SDHD and SDHAF2 genes were negative. Enzyme assays were performed on tumor tissue. Succinate cytochrome c reductase activity (complex II + III) measured in tumor homogenate indicated a total and selective loss of SDH activity (Supplementary Material, Fig. S1). At the same time, the tumor was added to a series of paraganglioma/pheochromocytoma transcriptome analyses as described previously (16). Unsupervised analysis of 103 genes' expression involved in energy metabolism (oxidative phosphorylation and glycolytic pathways) classified the patient's tumor in the subgroup of SDH-related paraganglioma/pheochromocytoma (Fig. 1). These data strongly suggested a defect in one of the SDHx genes. Sequence analysis was thus extended to SDHA.
Figure 1.
Microarray analysis of genes involved in oxidative phosphorylation and glycolysis pathways in SDHA-mutated paraganglioma and other inherited paraganglioma/pheochromocytoma. Unsupervised hierarchical clustering analysis of 69 samples according to the expression of 80 genes (150 probes) involved in oxidative phosphorylation and 23 glycolysis-associated genes (46 probes). Expression profiles are shown as a heat map indicating high (red) and low (green) expression according to a log2-transformed scale. The higher bipartition allows distinguishing VHL (white), SDH (grey), and RET and NF1 (black) tumors. The SDHA-tumor (in yellow, indicated by an asterisk) is classified with SDHx-tumors.
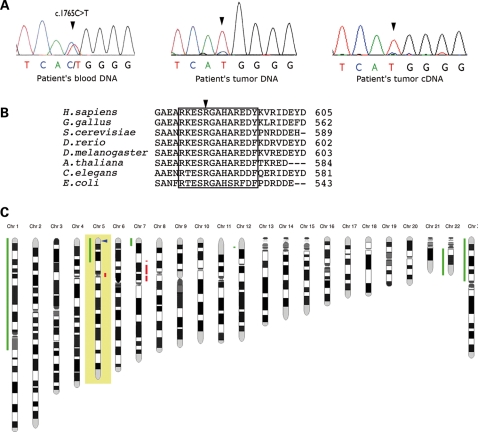
A missense mutation (c.1765C>T; p.Arg589Trp) was identified in germline DNA as well as in DNA and mRNA extracted from the tumor (Fig. 2A). Arginine 589 occurs in a highly conserved sequence of eukaryotic and prokaryotic SDHs, including Saccharomyces cerevisiae (Fig. 2B). The c.1765C>T variant was not found in 740 control chromosomes and in silico predictions indicated structural alterations in the protein as a result of the mutation. The predominance of the W589 mutant allele in tumor DNA (Fig. 2A, middle panel) and cDNA (Fig. 2A, right panel) suggested loss of heterozygosity (LOH) at the SDHA locus. By BAC array comparative genomic hybridization (CGH), we analyzed the LOH pattern in the patient's tumor included in a series of 202 paragangliomas and pheochromocytomas collected by the COMETE network. For the patient's tumor, we observed a 5p14–5p15 loss, confirming the LOH at the SDHA locus (Fig. 2C). The LOH pattern in the complete series showed that loss of 5p15 region (SDHA locus, 9/202, 4.5%) and 1q21 (SDHC locus, 9/202, 4.5%) are infrequent events compared with 1p36.1 (SDHB locus, 131/202, 64.9%) and 11q23 (SDHD locus, 55/202, 27.2%) losses (data not shown). The 9/202 tumors harboring a 5p15 loss included the patient's tumor, two NF1-, one VHL-, one RET-, one SDHB-, one SDHD-related paraganglioma/pheochromocytoma and two apparently sporadic tumors. No SDHA mutation was found in these two last samples.
Figure 2.
Identification of the SDHA mutation, associated with LOH at the SDHA locus. (A) Sequencing chromatograms of SDHA in the region of the mutation. Direct sequencing of the patient's DNA extracted from leukocytes (left panel) shows the SDHA heterozygous mutation c.1765C>T. Direct sequencing of the DNA extracted from the patient's tumor tissue (middle panel) and of the cDNA obtained by RT–PCR of RNA extracted from frozen tumor tissue (right panel) show the predominance of the mutant allele (T) in the tumor tissue suggesting an LOH at the SDHA locus. The faint persistence of a WT (C) peak is probably imputable to non-tumor cell contaminations (e.g. endothelial cells and stromal cells). (B) Alignment of SDHA homologues. The sequences surrounding arginine Arg589 (arrow) of human and other animal, plant and bacterial SDHA have been aligned by blastp. The short conserved sequence with Arg589 is enclosed by the box. (C) BAC array CGH analysis reveals 5p, 1p, 22q losses and partial 7p gain in SDHA tumor.
The human SDHA is a unique and highly polymorphic gene
Two distinct tissue-specific human SDHA cDNAs differing in a few nucleotides leading to two amino acid changes, ‘type I Fp’ (Y629 and V657) and ‘type II Fp’ (F629 and I657), were reported in 2003 (12). Subsequently, Baysal et al. (14) attributed the sequence differences to polymorphisms and concluded that only one SDHA gene exists in humans. To address the issue of two different SDHA genes, we analyzed SDHA cDNAs in different tissues. The tumor tissue of the patient contained only ‘type II Fp’ (F629 and I657). As we had previously observed the simultaneous presence of ‘type I Fp’ and ‘type II Fp’ in one pheochromocytoma sample (3), we extracted RNA from two additional pheochromocytomas and four fibroblast cultures from healthy control individuals. Y629 associated with V657 (corresponding to ‘type I Fp’) were found in three out of the four cultured fibroblasts and in the two pheochromocytoma controls. The fourth cultured fibroblasts contained both types (‘I Fp + II Fp’). Finally, we sequenced nucleotides at positions 1680, 1752, 1886, 1932 and 1969 (described as difference between type I Fp and type II Fp) (12) of SDHA in germline DNA of 216 healthy control subjects. For each of the five positions, two different nucleotides were observed. Haplotype analysis using the Thesias program showed six rare haplotypes and two common haplotypes: 1680G-1752A-1886A-1932G-1969G, corresponding to ‘type I Fp’ (f = 0.817) and 1680A-1752G-1886T-1932A-1969A to ‘type II Fp’ (f = 0.125) (Table 1). Strong linkage disequilibrium was identified among the five single-nucleotide polymorphisms with all the D′ values falling between 0.94 and 1.00 (P < 0.001). Together, these data constitute compelling evidence that SDHA, in agreement with the conclusions of Baysal et al. (14), is a unique polymorphic gene.
Table 1.
Genotype repartition and allelic frequencies of SDHA nucleotides 1680, 1752, 1886, 1932 and 1969 in a control population
| SDHA exon | Exon 13 |
Exon 14 | Exon 15 |
||
|---|---|---|---|---|---|
| Nucleotide position | c.1680 | c.1752 | c.1886 | c.1932 | c.1969 |
| Genotype repartition in the control population (n = 216) | GG: 66% | AA: 67% | AA: 68% | GG:68% | GG: 75% |
| GA: 31% | GA: 31% | AT: 30% | AG: 29% | GA: 24% | |
| AA: 3% | GG: 2% | TT: 2% | AA: 3% | AA: 1% | |
| Allelic frequencies | Ga: 81.9% (354/432) | Aa: 82.9% (358/432) | Aa: 83.3% (360/432) | Ga: 82.6% (357/432) | Ga: 86.8% (375/432) |
| Ab: 18.1% (78/432) | Gb: 17.1% (74/432) | Tb: 16.7% (72/432) | Ab: 17.4% (75/432) | Ab: 13.2% (57/432) | |
aNucleotide reported to be present in ‘type I Fp’.
bNucleotide reported to be present in ‘type II Fp’.
The SDHA mutation abolishes SDH activity in a yeast model
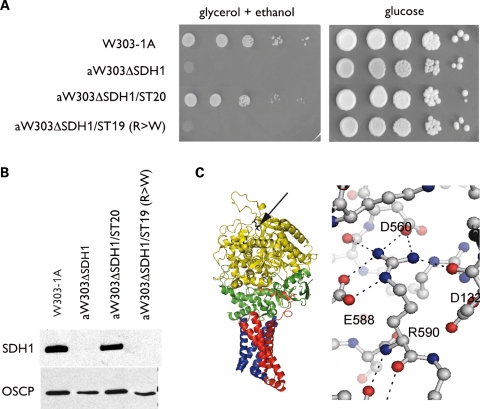
To assess the function of the mutant protein in vitro, we expressed the Arg589Trp in yeast. Arginine 582 of yeast Sdh1 protein corresponds to arginine 589 of human SDHA. This codon was changed to a tryptophan codon in the yeast SDH1 gene. The growth defect of the yeast SDH1 null mutant on a minimal ethanol/glycerol medium was not rescued by a chromosomally integrated copy of the Arg582Trp mutant gene (aW303ΔSDH1/ST19; Fig. 3A). In contrast, the null mutant transformed with the wild-type (WT) gene in the same vector (aW303ΔSDH1/ST20) grew as well as the WT on this medium. The inability of the mutant gene to restore SDH activity in the SDH1 null mutant (Table 2) confirmed Arg589Trp to be a loss-of-function mutation. As expected, integration of the WT gene in the SDH1 mutant restored enzyme activity to nearly WT levels.
Figure 3.
Functional consequences of the SDHA mutation expressed in yeast. (A) Growth phenotype on respiratory media (minimal glycerol/ethanol) and glycolytic media (minimal glucose). Ten-fold serial dilutions of the WT parent (W303-1A), the SDH1 null mutant (aW303ΔSDH1), the null mutant with an integrated copy of SDH1 (aW303ΔSDH1/ST20) and the null mutant with an integrated copy of the Arg582Trp mutant gene (aW303ΔSDH1/ST19) were plated on the minimal glycerol/ethanol and on the minimal glucose plates. Growth was recorded after 2 days on glucose and 4 days on glycerol/ethanol. (B) Western blot analysis of Sdh1 and OSCP, a subunit of mitochondrial ATP synthase. Mitochondria were prepared from cells grown in rich YPGal (2% galactose, 1% yeast extract and 2% peptone) by the method of Meisinger et al. (34). Mitochondrial proteins (40 µg) were separated on a 12% polyacrylamide gel. They were transferred to a nitrocellulose membrane and probed with polyclonal antibodies against Sdh1 and OSCP, used as a loading control. A rabbit polyclonal antibody was obtained against a fusion protein consisting of the N-terminal half of anthranilate synthase component 1 fused to residues 158–498 of SDH (35). No Sdh1 signal is detected in mitochondria of aW303ΔSDH1/ST19, the transformant harboring a chromosomally integrated copy of the mutant gene. (C) Structure of chicken complex II. The arginine 589 homologue is indicated by the arrow on the ribbon representation of chicken SDHA (in yellow) and is located distal to subunits SDHB (green), SDHC (red) and SDHD (blue). The environment of arginine 589 shows the conserved polar interactions in the structure of WT SDHA that are lost in the Arg582Trp mutant. R590, E588, D560 and D132 of chicken SDHA are homologous to R589, E587, D559 and D131 of the human protein, respectively.
Table 2.
Genotypes, sources and enzyme activities of S. cerevisiae strains
| Strain | Genotype | Source | % ρ−/o | SDH | SCCR |
|---|---|---|---|---|---|
| aW303 | MATa ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 | 31 | <2 | 178 ± 8 | 194 ± 40 |
| aW303ΔSDH1 | MATa ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 SDH1::HIS3 | This study | 47 | <1 | <1 |
| aW303ΔSDH1/ST20 | MATa ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 SDH1::HIS3 ura3-1:: SDH1 | This study | <2 | 141 ± 30 | 158 ± 7 |
| aW303ΔSDH1/ST19 | MATa ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 SDH1::HIS3 ura3-1:: SDH1 R → W | This study | 45 | <1 | <1 |
The SDH and succinate cytochrome c reductase (SCCR) activities are expressed, respectively, as nmol of DCPiP and cytochrome c reduced min−1 (mg of mitochondrial protein)−1. After 36 h of growth in liquid YPGal, the SDH1 mutant and the transformant with the Arg582Trp mutation accumulated ∼50% secondary ρ− and ρo mutants lacking part or all their mitochondrial genome. Under similar conditions of growth, there were <2% ρ− and ρo mutants in the WT and the transformant with the WT gene. The instability of mitochondrial DNA in the mutant is not surprising as it has also been reported in several TCA cycle mutants such as aconitase, isocitrate dehydrogenase and pyruvate dehydrogenase-deficient strains (36).
The steady-state abundance of SDH in the WT and mutant strains, assessed by western analysis of total mitochondrial proteins (Fig. 3B), indicated comparable amounts of Sdh1 in mitochondria of WT and the transformant with the WT gene. No Sdh1 was detected in the SDH1 null mutant and the mutant transformed with the Arg582Trp gene (Fig. 3B). Western analysis also failed to reveal the presence of Sdh1 in the cytosolic fraction of the four different strains. These results indicate that the structural change resulting from the Arg582Trp mutation renders the protein more susceptible to proteolysis.
The SDHA mutation leads to SDHA and SDHB protein losses in tumor tissue
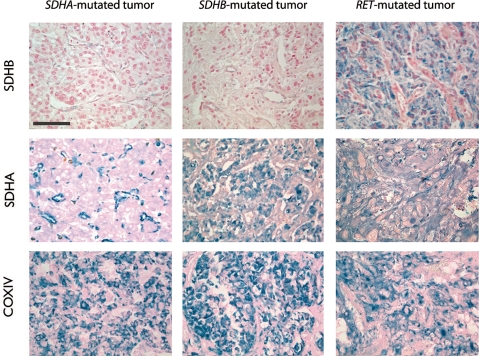
In order to confirm the instability of the mutant SDHA protein in human, we performed immunohistochemical analysis of the protein in the patient's tumor. Strong SDHA immunostaining was observed in the cytoplasm of tumor and endothelial cells in all paraganglioma/pheochromocytoma tissues used as controls (RET, NF1, SDHB and SDHD), but was only present in the endothelial cells and was not detected in tumor cells of the patient (Fig. 4). As recently reported for all SDHx-related tumors (17), we did not detect SDHB immunostaining in the SDHA-mutant tumor (Fig. 4). In contrast, COX-IV protein expression, used as an internal control, was positive in all samples.
Figure 4.
Expression of SDHA, SDHB and COX-IV in SDHA-mutated paraganglioma compared with other inherited paraganglioma/pheochromocytoma. SDHA-positive immunostaining is observed in the chromaffin cells of SDHB- and RET-related tumors. In the patient's tumor, it is detected solely in blood vessels and not in tumor cells. SDHB protein expression is lost in both SDHA- and SDHB-mutated tumors, whereas it is still present in RET-related pheochromocytomas. Subunit COX-IV of mitochondrial cytochrome c oxidase is expressed at comparable levels in all tumors. Calibration bar: 100 µm.
The SDHA mutation causes pseudo-hypoxia
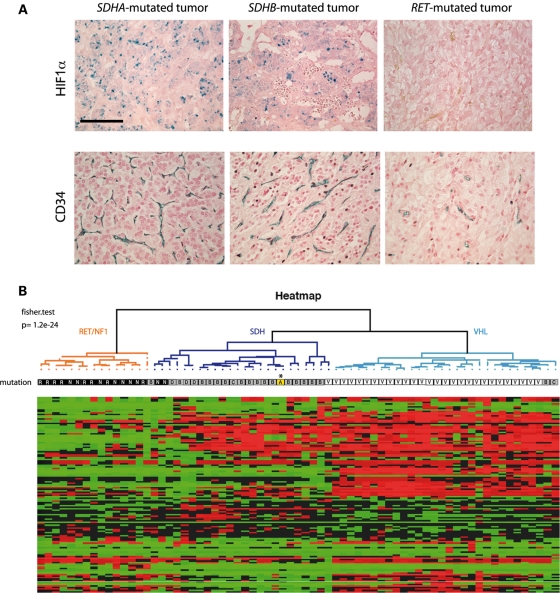
Pseudo-hypoxia is a known consequence of SDHB and SDHD mutations. We therefore compared HIF-1α expression by immunohistochemistry on the patient's tumor and on RET, SDHB and SDHD (data not shown) paraganglioma or pheochromocytoma tissues. HIF-1α was highly expressed in the SDHA- as well as in the SDHB- and SDHD-related tumors (Fig. 5A). In contrast, it was undetectable in the RET tumor. We also obtained expression data for 54 HIF target genes using transcriptome analysis. The expression profile of the corresponding probe sets was used to perform a clustering on 69 RET-, NF1-, SDH- and VHL-related tumors, including that of the patient. The unsupervised analysis using the hypoxic pathway enabled us to classify the tumors into two groups; the first one included all but three RET and NF1 tumors and the second one included all VHL- and SDHx-mutated paragangliomas and pheochromocytomas, including the SDHA tumor (Fig. 5B).
Figure 5.
Pseudohypoxia and angiogenesis in SDHA-mutated tumor compared with other inherited paraganglioma/pheochromocytoma. (A) HIF1-α nuclear immunostaining is detected in tumors with mutations in either SDHA- or SDHB- but not in RET-related pheochromocytomas. Calibration bar: 100 µm. CD34 immunohistochemistry was performed to evaluate angiogenesis in the SDHA-mutated paraganglioma and compared with other inherited paraganglioma/pheochromocytoma. Histogreen was used as a chromogen for detection (blue labeling). Quantification of vascular density reveals that the SDHA-mutated paraganglioma is highly vascularized, as are other SDHx-related tumors. (B) Unsupervised hierarchical clustering analysis of 69 samples according to the expression of 54 HIF-1 and/or HIF-2 targets (122 probes). Expression profiles are shown as a heat map indicating high (red) and low (green) expression according to a log2-transformed scale. The higher bipartition allows pseudo-hypoxic VHL- (white) and SDHx- (gray) to be distinguished from RET and NF1 (black) tumors. The SDHA paraganglioma (in yellow indicated by an asterisk) is classified with SDHx tumors.
Finally, we used CD34 immunohistochemistry to evaluate angiogenesis in the patient's tumor (Fig. 5A). We observed that vascular density in the SDHA tumor (34 ± 3 blood vessels/0.65 mm2) was comparable to that observed in eight SDHx-related tumors (mean = 31 ± 4 blood vessels/0.65 mm2) and higher than in five RET/NF1 tumors (mean = 9 ± 2 blood vessels/0.65 mm2), which we recently reported (16). All these data suggest that SDHA inactivation stimulates angiogenesis via the pseudo-hypoxic pathway as previously described for the other SDH subunits.
DISCUSSION
The observation of a patient with a paraganglioma resulting from a germline mutation in SDHA (p.Arg589Trp) associated with LOH in tumor provides the first evidence that mutations in SDHA can cause paraganglioma and that SDHA, like other SDHx genes, can act as a tumor suppressor gene in accordance to the Knudson's ‘two-hits’ model.
Immunohistochemistry revealed that SDHA was absent from tumor cells of the patient with the p.Arg589Trp mutation. This suggested that the p.Arg589Trp mutation leads to SDHA protein instability, a conclusion supported by the results obtained with the yeast model. Introduction of the homologous Arg582Trp in the Sdh1 subunit of the yeast complex abolished SDH activity with a concomitant reduction of mitochondrial Sdh1 to undetectable levels. This putative instability of the Trp589 SDHA is consistent with the amino acid substitution and its location in the protein. Arg589 is located at the apex of SDHA (18,19), about 20 Å removed from the flavin moiety and distal to the interface with SDHB (>30 Å) (Fig. 3C). This makes it unlikely that Arg589 plays a direct role in the activity or assembly of the complex. Arg589 contacts three highly conserved charged residues—Glu587, Asp602 and Asp131. A possible explanation is, thus, that Trp589 would destabilize the tertiary structure of SDHA by eliminating polar contacts and by introducing a bulky side chain into the constrained space formed by the side chains surrounding Arg589.
Using immunochemistry analyses in the patient's tumor, we showed that consequences of SDHA mutation are similar to those seen in other SDHx mutants but distinct from RET- or NF1-related tumors. All the SDHx-related tumors including the SDHA mutant reported here are deficient in SDHB expression, show HIF-1α protein stabilization and display high blood vessel density. By the same token, transcriptome analysis of gene expression in oxidative phosphorylation and glycolysis indicated a pattern in the SDHA-related tumor following that of other SDHx-related tumors. Consistent with the pheochromocytoma microarray study previously published by Dahia et al. (20), the microarray analysis we performed with HIF target genes showed two different clusters, one characteristic of the NF1- and RET-related pheochromocytomas and the second of SDHx- and VHL-related tumors. These data revealed that the SDHA patient's tumor was classifiable in the VHL- and SDHx-paraganglioma/pheochromocytoma group and that like other SDHx tumors, it also displays an activated hypoxic pathway.
The absence of any SDHA-associated paraganglioma or pheochromocytoma was previously thought to be the consequence of the possible existence of two different SDHA genes, ‘type I Fp’ and ‘type II Fp’, with tissue-specific expression (12). Although ‘type I Fp’ SDHA was located on chromosome 5 (5p15) (6), the location of the putative ‘type II Fp’ intronless gene was never identified. Our data are not in favor of the existence of an intronless gene encoding ‘type II Fp’ since we identified the nucleotides specific of ‘type II Fp’ even when intronic primers were used. They do not support either the tissue specificity of two isoforms since we observed the three possible conditions (‘only I Fp’, ‘only II Fp’ or ‘I + II Fp’) in pheochromocytoma of different individuals. Additionally, in agreement with another report (14), the two sequences displayed strong linkage disequilibrium in our control population. On the basis of these findings, we conclude that the ‘type I Fp’ and ‘type II Fp’ reported by Tomitsuka et al. (12) are most likely the result of frequent naturally occurring SDHA polymorphisms.
An alternative explanation was proposed by Guzy et al. to understand why SDHB, SDHC and SDHD mutations are associated with a tumor phenotype, whereas SDHA mutations are not (21). These authors suggested that mutations in SDHB, SDHC and SDHD would be expected to cause both a reactive oxygen species (ROS) (20) and succinate-mediated HIF-α stabilization leading to tumor progression. In contrast, SDHA mutations would act by increasing succinate alone. Using pharmacological inhibition or RNA interference in vitro, they did not detect any increase in normoxic ROS production, HIF-α stabilization or tumor progression in cells lacking SDHA as opposed to those lacking SDHB. Our data, however, show that the SDHA mutation does lead to HIF stabilization and the subsequent activation of the hypoxic pathway. This is consistent with Selak et al. (22) results, who demonstrated that siRNA inhibition of SDH expression stabilizes HIF-1α by a mechanism independent of ROS production and only mediated by succinate accumulation.
At present, the most credible explanation for the rarity of SDHA-related tumors is the relatively low frequency of 5p15 loss, the chromosomal region containing the SDHA locus, compared with the 1p36 (SDHB) and 11q23 (SDHD) loci that often undergo losses in tumor tissues. This was observed in our large series of 202 paraganglioma and pheochromocytoma samples analyzed by BAC array CGH and was previously reported by other groups (23–26). Interestingly, the rarity of SDHC-related tumors (27) may also be explained by the low frequency of LOH observed for the 1q21 chromosomal region containing SDHC locus.
This study adds SDHA to the list of genes coding for TCA cycle proteins involved in paraganglioma/pheochromocytoma tumorigenesis. SDHA mutations should be suspected in patients affected by paraganglioma or pheochromocytoma in the case of negative SDHA immunohistochemistry in the tumor tissue, when loss of SDH activity is found in tumor, despite negative SDHx genetic testing and/or when loss of 5p15 chromosome is found in tumor. Our study highlights the power of immunohistochemical and genomic techniques for the molecular characterization of paraganglioma/pheochromocytoma, approaches which can also provide valuable information for individually tailored genetic counseling.
MATERIALS AND METHODS
Case report
A 32-year-old woman developed pregnancy-induced hypertension at 30 weeks of gestation. No signs or symptoms of hyperadrenergic state were present during pregnancy. A few days after childbirth, hyperadrenergic symptoms appeared (dizziness, tachycardia and sweating) with hypertension (170/110 mmHg). High concentrations of urinary normetanephrine (12.12 µmol/24 h, normal range 0.5–2.40), urinary norepinephrine (4 287 nmol/24 h, normal range 70–500) and chromogranin A (944 mg/L, normal value <86.5) were measured. Urinary metanephrine, dopamine and plasmatic vanilmandelic acid levels were normal (Supplementary Material, Table S1). Magnetic resonance imaging disclosed a left adrenal mass (58 mm) suggestive of a pheochromocytoma or paraganglioma—iso-signal in T1-weighted images, high signal in fat-saturated T2-weighted images enhanced after gadolinium injection—(Supplementary Material, Fig. S2A). Computed tomography scan revealed a hypervascularized tumor (data not shown). Whole body meta-iodobenzylguanidine scintigraphy showed a single intense uptake in the left adrenal area (Supplementary Material, Fig. S2B) without any other localization. Following left adrenalectomy by laparoscopy, histology diagnosed a 47 mm extra-adrenal paraganglioma (Supplementary Material, Fig. S2C). Immediately after surgery, blood pressure and urinary normetanephrine returned to normal. There was no history of paraganglioma/pheochromocytoma known in her family or any syndromic lesion suggesting neurofibromatosis type 1, multiple endocrine neoplasia type 2 or von Hippel–Lindau disease.
Biological material
Patients signed a written informed consent for germline and somatic DNA analyses. Fresh tumor samples were collected during surgery and immediately frozen in liquid nitrogen. Germline DNAs were extracted from leukocytes according to standard protocols. Tumor DNAs and RNAs were extracted using AllPrep DNA/RNA Mini Kit (Qiagen). Total RNA was submitted to a DNAse I RNAse-free treatment (Roche) and retrotranscribed using M-MLV Reverse Transcriptase (Invitrogen) and random hexamers.
A Caucasian reference population was used to obtain 370 control DNAs. Control RNAs were extracted from fibroblast cultures from healthy control individuals and from pheochromocytoma tumor tissue of patients collected by the COMETE network (1). Ethical approval for the study was obtained from the institutional review board (CPP Paris-Cochin, January 2007).
Mutation analysis
Mutation analysis for RET, VHL, SDHB, SDHC, SDHD and SDHAF2 genes was performed by direct sequencing. VHL, SDHB, SDHC and SDHD were also analyzed for the presence of large deletions as described previously (1,27).
SDHA cDNA was sequenced in five overlapping amplicons. To amplify each fragment by PCR, at least one primer was systematically designed overlapping two exons. The 15 exons and the intron–exon boundaries of SDHA in the germline and tumor DNAs were sequenced with primers specific for the SDHA gene on chromosome 5p15 but not for the pseudo-genes known to be present on chromosomes 5 and 3q29 (Supplementary Material, Table S2). In silico predictions were performed, based on sequence similarity, amino acid composition, protein structure and function (SIFT predictor, http://sift.jcvi.org/ and PolyPhen predictor, http://coot.embl.de/PolyPhen/ websites).
Respiratory chain activities
Cytochrome c oxidase (complex IV), succinate cytochrome c reductase (complex II + III) and quinol cytochrome c reductase (complex III) activities were measured spectrophotometrically in pheochromocytoma or paraganglioma homogenates as described previously (28,29).
Microarray
Microarray analyses were performed as described previously (16). Tumor samples (20–30 mg) were powdered under liquid nitrogen. RNAs were extracted using RNeasy mini kit (Qiagen). Aliquots of the RNA were analyzed by electrophoresis on a Bioanalyser 2100 (Agilent Technologies) and quantified using Nano Drop ND-1000 (Labtech). Stringent criteria for RNA quality were applied to rule out degradation, especially a 28S/18S ratio above 1.5. Microarray analyses were performed on 3 µg of total RNA for each sample and 10 µg cRNA per hybridization (GeneChip Fluidics Station 400; Affymetrix, Santa Clara, CA, USA). Total RNA were amplified and labeled following the manufacturer's one-cycle target labeling protocol (http://www.affymetrix.com). The labeled cDNA were then hybridized to HG-U133 Plus 2.0 Affymetrix GeneChip arrays (Affymetrix). The chips were scanned with a GCOS 1.4.
BAC array CGH
Genomic DNA from paraganglioma or pheochromocytoma from 202 patients collected by the COMETE network was analyzed with IntegraChip (IntegraGen, Evry, France), a CGH microarray containing 4434 BACs. Raw log2-ratio feature values were filtered and remaining values were normalized using the lowess within-print tip group method (30). AWS smoothing technique was then applied to the normalized log2-ratio values [R package GLAD v1.8]. This yielded segments along the chromosome of homogeneous smoothed log2-ratios values. Thus, we assigned the smoothed CGH data into three different groups: gain, no change or loss.
Haplotype and linkage disequilibrium analysis
Linkage disequilibrium, carried out with THESIAS software (www.genecanvas.org), was deduced from the estimated haplotype frequencies, and its extent was expressed in terms of D′, this being the ratio of the non-standardized coefficient to its maximal and minimal values.
Studies on yeast SDH1 mutants
The SDH1 gene of S. cerevisiae is the homologue of human SDHA. An SDH1::HIS3 null allele was constructed by replacing the coding sequence between the two BglII sites internal to SDH1 with HIS3. The respiratory-deficient S. cerevisiae mutant aW303ΔSDH1 with the deleted SDH1 was obtained by the one-step gene replacement method (31).
Yeast SDH1 plus flanking 5′ and 3′ sequences was cloned in the integrative plasmid YIp352 (32). This construct was used to change the CGT codon 582 of SDH1 to TGG with the QuickChange II Site-Directed Mutagenesis Kit® (Stratagene). Complete sequences were obtained of WT SDH1 in plasmid pG52/ST20 and of the mutant SDH1 in plasmid pG52/ST19. The two sequences were identical to those reported in the database (http://www.yeastgenome.org/) except for the TGG codon of the gene in pG52/ST19. The WT and mutant SDH1 were integrated at the chromosomal URA3 locus of the null mutant aW303ΔSDH1.
Immunohistochemistry
Paraffin blocks were cut and 6 µm thick sections were mounted on Superfrost plus slides. Immunohistochemistry was performed as described using antibodies as follows (33): anti-SDHB (HPA002868, Sigma-Aldrich, 1/500), anti-SDHA (abcam, ab14715, 1/1000), anti-COX-IV (abcam, ab33985, 1/1000), anti-HIF1α (H1alpha67, abcam, 1/500) and anti-CD34 (Clone QBEND 10, Immunotech, 1/100). The protocol involved a biotinylated secondary antibody (Vector Laboratories), an avidin–biotin–peroxidase complex (Vectastain ABC Elite; Vector Laboratories) and Histogreen (Abcys) as a chromogen.
Quantification of vascular density
Vascular density was measured on sections after CD34 immunostaining as described previously (16). Total blood vessels were counted in eight randomly chosen fields of 0.65 mm2.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by the Programme Hospitalier de Recherche Clinique grant COMETE 3 (AOM 06 179 to J.B. and A.-P.G.-R.), the Agence Nationale de la Recherche (ANR 08 GENOPATH 029 MitOxy to P.R. and A.-P.G.-R.) and the National Institutes of Health (HL022174 to A.T.). This work is part of the national program Cartes d'Identité des Tumeurs® (CIT) funded and developed by the Ligue Nationale Contre le Cancer (website: http://cit.ligue-cancer.net).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr David Mueller, University of Chicago Medical School, for his generous gift of the antibody against OSCP. We thank the surgeon, Prof. Bertrand Dousset, the nuclear medicine physician, Dr Florence Tenenbaum and the staff of the Endocrinology and Radiology departments of Cochin hospital for the management of the patient, Fernande Rene-Corail for excellent technical assistance.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Amar L., Bertherat J., Baudin E., Ajzenberg C., Bressac-de Paillerets B., Chabre O., Chamontin B., Delemer B., Giraud S., Murat A., et al. Genetic testing in pheochromocytoma or functional paraganglioma. J. Clin. Oncol. 2005;23:8812–8818. doi: 10.1200/JCO.2005.03.1484. doi:10.1200/JCO.2005.03.1484. [DOI] [PubMed] [Google Scholar]
- 2.Hao H.X., Khalimonchuk O., Schraders M., Dephoure N., Bayley J.P., Kunst H., Devilee P., Cremers C.W., Schiffman J.D., Bentz B.G., et al. SDH5, a gene required for flavination of succinate dehydrogenase, is mutated in paraganglioma. Science. 2009;325:1139–1142. doi: 10.1126/science.1175689. doi:10.1126/science.1175689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Briere J.J., Favier J., Benit P., El Ghouzzi V., Lorenzato A., Rabier D., Di Renzo M.F., Gimenez-Roqueplo A.P., Rustin P. Mitochondrial succinate is instrumental for HIF1alpha nuclear translocation in SDHA-mutant fibroblasts under normoxic conditions. Hum. Mol. Genet. 2005;14:3263–3269. doi: 10.1093/hmg/ddi359. doi:10.1093/hmg/ddi359. [DOI] [PubMed] [Google Scholar]
- 4.Selak M.A., Armour S.M., MacKenzie E.D., Boulahbel H., Watson D.G., Mansfield K.D., Pan Y., Simon M.C., Thompson C.B., Gottlieb E. Succinate links TCA cycle dysfunction to oncogenesis by inhibiting HIF-alpha prolyl hydroxylase. Cancer Cell. 2005;7:77–85. doi: 10.1016/j.ccr.2004.11.022. doi:10.1016/j.ccr.2004.11.022. [DOI] [PubMed] [Google Scholar]
- 5.Gottlieb E., Tomlinson I.P. Mitochondrial tumour suppressors: a genetic and biochemical update. Nat. Rev. Cancer. 2005;5:857–866. doi: 10.1038/nrc1737. doi:10.1038/nrc1737. [DOI] [PubMed] [Google Scholar]
- 6.Bourgeron T., Rustin P., Chretien D., Birch-Machin M., Bourgeois M., Viegas-Pequignot E., Munnich A., Rotig A. Mutation of a nuclear succinate dehydrogenase gene results in mitochondrial respiratory chain deficiency. Nat. Genet. 1995;11:144–149. doi: 10.1038/ng1095-144. doi:10.1038/ng1095-144. [DOI] [PubMed] [Google Scholar]
- 7.Horvath R., Abicht A., Holinski-Feder E., Laner A., Gempel K., Prokisch H., Lochmuller H., Klopstock T., Jaksch M. Leigh syndrome caused by mutations in the flavoprotein (Fp) subunit of succinate dehydrogenase (SDHA) J. Neurol. Neurosurg. Psychiatry. 2006;77:74–76. doi: 10.1136/jnnp.2005.067041. doi:10.1136/jnnp.2005.067041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parfait B., Chretien D., Rotig A., Marsac C., Munnich A., Rustin P. Compound heterozygous mutations in the flavoprotein gene of the respiratory chain complex II in a patient with Leigh syndrome. Hum. Genet. 2000;106:236–243. doi: 10.1007/s004390051033. doi:10.1007/s004390051033. [DOI] [PubMed] [Google Scholar]
- 9.Van Coster R., Seneca S., Smet J., Van Hecke R., Gerlo E., Devreese B., Van Beeumen J., Leroy J.G., De Meirleir L., Lissens W. Homozygous Gly555Glu mutation in the nuclear-encoded 70 kDa flavoprotein gene causes instability of the respiratory chain complex II. Am. J. Med. Genet. A. 2003;120A:13–18. doi: 10.1002/ajmg.a.10202. doi:10.1002/ajmg.a.10202. [DOI] [PubMed] [Google Scholar]
- 10.Ghezzi D., Goffrini P., Uziel G., Horvath R., Klopstock T., Lochmuller H., D'Adamo P., Gasparini P., Strom T.M., Prokisch H., et al. SDHAF1, encoding a LYR complex-II specific assembly factor, is mutated in SDH-defective infantile leukoencephalopathy. Nat. Genet. 2009 doi: 10.1038/ng.378. doi:10.1038/ng.378. [DOI] [PubMed] [Google Scholar]
- 11.Bayley J.P., Kunst H.P., Cascon A., Sampietro M.L., Gaal J., Korpershoek E., Hinojar-Gutierrez A., Timmers H.J., Hoefsloot L.H., Hermsen M.A., et al. SDHAF2 mutations in familial and sporadic paraganglioma and phaeochromocytoma. Lancet Oncol. 2010 doi: 10.1016/S1470-2045(10)70007-3. doi:10.1016/S1470-2045(10)70007-3. [DOI] [PubMed] [Google Scholar]
- 12.Tomitsuka E., Goto Y., Taniwaki M., Kita K. Direct evidence for expression of type II flavoprotein subunit in human complex II (succinate-ubiquinone reductase) Biochem. Biophys. Res. Commun. 2003;311:774–779. doi: 10.1016/j.bbrc.2003.10.065. doi:10.1016/j.bbrc.2003.10.065. [DOI] [PubMed] [Google Scholar]
- 13.Tomitsuka E., Hirawake H., Goto Y., Taniwaki M., Harada S., Kita K. Direct evidence for two distinct forms of the flavoprotein subunit of human mitochondrial complex II (succinate-ubiquinone reductase) J. Biochem. 2003;134:191–195. doi: 10.1093/jb/mvg144. doi:10.1093/jb/mvg144. [DOI] [PubMed] [Google Scholar]
- 14.Baysal B.E., Lawrence E.C., Ferrell R.E. Sequence variation in human succinate dehydrogenase genes: evidence for long-term balancing selection on SDHA. BMC Biol. 2007;5:12. doi: 10.1186/1741-7007-5-12. doi:10.1186/1741-7007-5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pacak K., Eisenhofer G., Ahlman H., Bornstein S.R., Gimenez-Roqueplo A.P., Grossman A.B., Kimura N., Mannelli M., McNicol A.M., Tischler A.S. Pheochromocytoma: recommendations for clinical practice from the First International Symposium. October 2005. Nat. Clin. Pract. Endocrinol. Metab. 2007;3:92–102. doi: 10.1038/ncpendmet0396. doi:10.1038/ncpendmet0396. [DOI] [PubMed] [Google Scholar]
- 16.Favier J., Briere J.J., Burnichon N., Riviere J., Vescovo L., Benit P., Giscos-Douriez I., De Reynies A., Bertherat J., Badoual C., et al. The warburg effect is genetically determined in inherited pheochromocytomas. PLoS One. 2009;4:e7094. doi: 10.1371/journal.pone.0007094. doi:10.1371/journal.pone.0007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Nederveen F.H., Gaal J., Favier J., Korpershoek E., Oldenburg R.A., de Bruyn E.M., Sleddens H.F., Derkx P., Riviere J., Dannenberg H., et al. An immunohistochemical procedure to detect patients with paraganglioma and phaeochromocytoma with germline SDHB, SDHC, or SDHD gene mutations: a retrospective and prospective analysis. Lancet Oncol. 2009;10:764–771. doi: 10.1016/S1470-2045(09)70164-0. doi:10.1016/S1470-2045(09)70164-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun F., Huo X., Zhai Y., Wang A., Xu J., Su D., Bartlam M., Rao Z. Crystal structure of mitochondrial respiratory membrane protein complex II. Cell. 2005;121:1043–1057. doi: 10.1016/j.cell.2005.05.025. doi:10.1016/j.cell.2005.05.025. [DOI] [PubMed] [Google Scholar]
- 19.Yankovskaya V., Horsefield R., Tornroth S., Luna-Chavez C., Miyoshi H., Leger C., Byrne B., Cecchini G., Iwata S. Architecture of succinate dehydrogenase and reactive oxygen species generation. Science. 2003;299:700–704. doi: 10.1126/science.1079605. doi:10.1126/science.1079605. [DOI] [PubMed] [Google Scholar]
- 20.Dahia P.L., Ross K.N., Wright M.E., Hayashida C.Y., Santagata S., Barontini M., Kung A.L., Sanso G., Powers J.F., Tischler A.S., et al. A HIF1alpha regulatory loop links hypoxia and mitochondrial signals in pheochromocytomas. PLoS Genet. 2005;1:72–80. doi: 10.1371/journal.pgen.0010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guzy R.D., Sharma B., Bell E., Chandel N.S., Schumacker P.T. Loss of the SdhB, but Not the SdhA, subunit of complex II triggers reactive oxygen species-dependent hypoxia-inducible factor activation and tumorigenesis. Mol. Cell. Biol. 2008;28:718–731. doi: 10.1128/MCB.01338-07. doi:10.1128/MCB.01338-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Selak M.A., Duran R.V., Gottlieb E. Redox stress is not essential for the pseudo-hypoxic phenotype of succinate dehydrogenase deficient cells. Biochim. Biophys. Acta. 2006;1757:567–572. doi: 10.1016/j.bbabio.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 23.August C., August K., Schroeder S., Bahn H., Hinze R., Baba H.A., Kersting C., Buerger H. CGH and CD 44/MIB-1 immunohistochemistry are helpful to distinguish metastasized from nonmetastasized sporadic pheochromocytomas. Mod. Pathol. 2004;17:1119–1128. doi: 10.1038/modpathol.3800160. doi:10.1038/modpathol.3800160. [DOI] [PubMed] [Google Scholar]
- 24.Cascon A., Ruiz-Llorente S., Rodriguez-Perales S., Honrado E., Martinez-Ramirez A., Leton R., Montero-Conde C., Benitez J., Dopazo J., Cigudosa J.C., et al. A novel candidate region linked to development of both pheochromocytoma and head/neck paraganglioma. Genes Chromosomes Cancer. 2005;42:260–268. doi: 10.1002/gcc.20139. doi:10.1002/gcc.20139. [DOI] [PubMed] [Google Scholar]
- 25.Dannenberg H., Speel E.J., Zhao J., Saremaslani P., van Der Harst E., Roth J., Heitz P.U., Bonjer H.J., Dinjens W.N., Mooi W.J., et al. Losses of chromosomes 1p and 3q are early genetic events in the development of sporadic pheochromocytomas. Am. J. Pathol. 2000;157:353–359. doi: 10.1016/S0002-9440(10)64547-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Edstrom E., Mahlamaki E., Nord B., Kjellman M., Karhu R., Hoog A., Goncharov N., Teh B.T., Backdahl M., Larsson C. Comparative genomic hybridization reveals frequent losses of chromosomes 1p and 3q in pheochromocytomas and abdominal paragangliomas, suggesting a common genetic etiology. Am. J. Pathol. 2000;156:651–659. doi: 10.1016/S0002-9440(10)64769-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burnichon N., Rohmer V., Amar L., Herman P., Leboulleux S., Darrouzet V., Niccoli P., Gaillard D., Chabrier G., Chabolle F., et al. The succinate dehydrogenase genetic testing in a large prospective series of patients with paragangliomas. J. Clin. Endocrinol. Metab. 2009;94:2817–2827. doi: 10.1210/jc.2008-2504. doi:10.1210/jc.2008-2504. [DOI] [PubMed] [Google Scholar]
- 28.Benit P., Goncalves S., Philippe Dassa E., Briere J.J., Martin G., Rustin P. Three spectrophotometric assays for the measurement of the five respiratory chain complexes in minuscule biological samples. Clin. Chim. Acta. 2006;374:81–86. doi: 10.1016/j.cca.2006.05.034. doi:10.1016/j.cca.2006.05.034. [DOI] [PubMed] [Google Scholar]
- 29.Rustin P., Chretien D., Bourgeron T., Gerard B., Rotig A., Saudubray J.M., Munnich A. Biochemical and molecular investigations in respiratory chain deficiencies. Clin. Chim. Acta. 1994;228:35–51. doi: 10.1016/0009-8981(94)90055-8. doi:10.1016/0009-8981(94)90055-8. [DOI] [PubMed] [Google Scholar]
- 30.Yang Y.H., Dudoit S., Luu P., Lin D.M., Peng V., Ngai J., Speed T.P. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 2002;30:e15. doi: 10.1093/nar/30.4.e15. doi:10.1093/nar/30.4.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rothstein R.J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. doi:10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- 32.Hill J.E., Myers A.M., Koerner T.J., Tzagoloff A. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast. 1986;2:163–167. doi: 10.1002/yea.320020304. doi:10.1002/yea.320020304. [DOI] [PubMed] [Google Scholar]
- 33.Favier J., Kempf H., Corvol P., Gasc J.M. Cloning and expression pattern of EPAS1 in the chicken embryo. Colocalization with tyrosine hydroxylase. FEBS Lett. 1999;462:19–24. doi: 10.1016/s0014-5793(99)01476-3. doi:10.1016/S0014-5793(99)01476-3. [DOI] [PubMed] [Google Scholar]
- 34.Meisinger C., Sommer T., Pfanner N. Purification of Saccharomcyes cerevisiae mitochondria devoid of microsomal and cytosolic contaminations. Anal. Biochem. 2000;287:339–342. doi: 10.1006/abio.2000.4868. doi:10.1006/abio.2000.4868. [DOI] [PubMed] [Google Scholar]
- 35.Koerner T.J., Hill J.E., Myers A.M., Tzagoloff A. High-expression vectors with multiple cloning sites for construction of trpE fusion genes: pATH vectors. Methods Enzymol. 1991;194:477–490. doi: 10.1016/0076-6879(91)94036-c. doi:10.1016/0076-6879(91)94036-C. [DOI] [PubMed] [Google Scholar]
- 36.Chen X.J., Wang X., Kaufman B.A., Butow R.A. Aconitase couples metabolic regulation to mitochondrial DNA maintenance. Science. 2005;307:714–717. doi: 10.1126/science.1106391. doi:10.1126/science.1106391. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.