Abstract
Holoprosencephaly (HPE) is the most common forebrain and craniofacial malformation syndrome in humans. The genetics of HPE suggest that it often stems from a synergistic interaction of mutations in independent loci. In mice, several combinations of mutations in Nodal signaling pathway components can give rise to HPE, but it is not clear whether modest deficits of Nodal signaling along with lesions in other pathways might also cause such defects. We find that HPE results from simultaneous reduction of Nodal signaling and an organizer BMP (bone morphogenetic protein) antagonist, either Chordin or Noggin. These defects result from reduced production of tissues that promote forebrain and craniofacial development. Nodal promotes the expression of genes in the anterior primitive streak that are important for the development of these tissues, whereas BMP inhibits their expression. Pharmacological and transgenic manipulation of these signaling pathways suggests that BMP and Nodal antagonize each other prior to intracellular signal transduction. Biochemical experiments in vitro indicate that secreted Bmp2 and Nodal can form extracellular complexes, potentially interfering with receptor activation. Our results reveal that the patterning of forebrain and medial craniofacial elements requires a fine balance between BMP and Nodal signaling during primitive streak development, and provide a potential mechanistic basis for a new multigenic model of HPE.
INTRODUCTION
Holoprosencephaly (HPE), a partial or complete failure of forebrain bifurcation, is the most common anomaly of forebrain development in humans. HPE is often associated with other anomalies, including variable medial craniofacial deficiencies and occasional laterality defects (1). Heterozygosity for mutations in any of several loci has been associated with HPE, often in genes functioning in the SHH and NODAL intercellular signaling pathways (2). However, the extreme phenotypic variability in HPE patients with particular gene mutations cannot be explained by single-gene haploinsufficiency. Such considerations have led to a ‘multiple hit’ hypothesis for HPE pathogenesis, in which HPE might frequently result from two or more independent genetic lesions impacting common or interacting developmental pathways during forebrain formation (3).
An understanding of the cellular and molecular causes of HPE has been garnered largely from functional studies of early forebrain patterning (4). In the mouse, forebrain initiation occurs in the distal epiblast during gastrulation and requires reinforcing signals from the gastrula organizer, located at the anterior end of the primitive streak (APS) (5). The APS gives rise to the anterior-most axial mesendoderm (AME), including the prechordal plate (PCP) and anterior definitive endoderm (ADE) (6). The PCP and ADE migrate to underlie the developing anterior neural plate, reinforcing and refining an initial anterior identity (7). Defects in these midline tissues can result in forebrain mispatterning, leading to HPE, as well as craniofacial or laterality defects.
The transforming growth factor β (TGFβ) ligand Nodal plays a pivotal role in specifying the APS and its derivatives. Nodal is expressed in the proximal posterior of the embryo as the primitive streak forms, and regulates its fates: the ADE and PCP are most sensitive to decreases in Nodal activity (8). Nodal acts through a cell-surface receptor complex that phosphorylates its intracellular effectors Smad2 and Smad3, which in turn activate the transcription of target genes, including Nodal itself (9,10). Decreased signaling via compound mutations in this pathway, such as Nodal+/−;Smad2+/−, Smad2+/−;Smad3−/− or ActRIIA−/−;Nodal+/−, results in anterior midline defects (11–14) similar to those of a Nodal hypomorph (15). Similarly, HPE also occurs in embryos carrying mutations in Nodal and Gdf1 (16), a co-ligand for Nodal (17), or Nodal and Foxa2 (18), a downstream target of Nodal signaling in the APS (19). These data provide evidence for the validity of the multiple-hit model for HPE pathogenesis (3) in the mouse when two mutations occur in the Nodal pathway; but they do not resolve whether defects in other signaling pathways might also interact with Nodal pathway lesions to cause HPE.
Antagonists of bone morphogenetic proteins (BMPs), namely Chordin (Chrd) and Noggin (Nog), are expressed in the AME and promote early forebrain patterning (20–22). Decreasing the gene dosage of both Chrd and Nog (Chrd−/−;Nog+/−) results in a range of HPE phenotypes remarkably similar to those seen in human, caused at least in part by defective PCP function (23).
Thus, Nodal and the BMP antagonists Chrd and Nog exhibit intriguing similarities in that all are expressed in or around the developing organizer; all are required for normal development of the forebrain, mediated at least in part by a role in promoting AME; and loss-of-function mutations in all three have been associated with HPE in murine digenic models. Accordingly, we hypothesized that Nodal and BMP antagonism act synergistically during mammalian forebrain formation. Here, we use genetics to probe possible interactions of Nodal signaling and BMP antagonism in this context. These studies are complemented by tissue culture, gene misexpression and co-immunoprecipitation (IP) experiments to gain mechanistic insight into the interactions of the BMP and Nodal pathways.
RESULTS
To begin testing our hypothesis of synergistic interaction between Nodal signaling and BMP antagonism during anterior midline development, we generated loss-of-function double-mutants. Because the spatiotemporal expression of Nodal and Chrd partially overlaps during anterior primitive streak development (24), we first generated compound mutations of these genes.
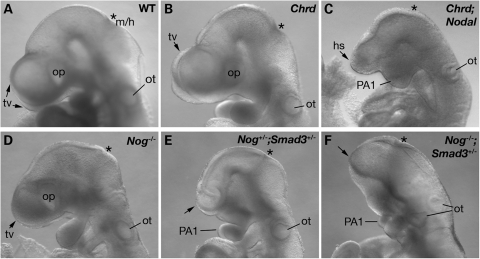
Nodal−/− embryos show severe defects prior to gastrulation and die at gastrulation, whereas Nodal+/− mice appear normal (25). In the outbred background used here, Chrd−/− embryos are viable, with no anterior truncations or midline deletions (20). In contrast, 23% (19/83) of E9.5–E10.5 Chrd−/−;Nodal+/− embryos showed morphological defects in anterior midline tissues. Among these embryos, 74% (14/19) showed HPE in association with anterior truncation and fused first pharyngeal arches (Fig. 1C). Five also had a cardiac laterality defect (data not shown). These findings indicate a genetic interaction between Chrd and Nodal during anterior patterning. They also demonstrate that their simultaneous reduction can result in forebrain, craniofacial and laterality defects similar to human HPE and associated malformations.
Figure 1.
HPE in compound mutant embryos. Lateral view of E9.5 embryos. (A and B) WT and Chrd−/− embryos have normal bilateral telencephalic vesicles (tv) and optic vesicles (op). (C) Chrd−/−;Nodal+/− embryo with a single holospheric vesicle (hs) and fused, hypoplastic pharyngeal arch 1 (PA1). (D) Nog−/− embryo with normal morphology of anterior structures. (E) Nog+/−;Smad3+/− embryo with a lack of the telecephalon (arrow) and abnormal BA1 morphology. (F) Nog−/−;Smad3+/− embryo showing severe forebrain truncation. Asterisks mark midbrain/hindbrain boundary (m/h). ot, otic vesicle.
The Chrd;Nodal genotype implies reduced Nodal signaling and BMP antagonism. If this combination can cause HPE, it might be possible to replicate such defects by analogous double-mutants that reduce Nodal signaling and organizer BMP antagonism. For example, a similar impact on Nodal signaling and BMP antagonism might result from simultaneously reducing gene dosage of the Nodal signaling effector Smad3 and the BMP antagonist Nog. Based on this logic, we prepared a series of additional, analogous compound mutants, Nog−/+(−);Nodal+/−, Chrd−/−;Smad3−/+(−) and Nog−/+(−);Smad3−/+(−) (Table 1). Smad3 expression occurs throughout gastrulation, strong in the axial midline at E7.5 (11). Although Smad3 mutants have no obvious defects in embryogenesis (26), Smad3 interacts synergistically with Smad2 during mesodermal and endodermal patterning (11,12). Nog is expressed in the node and AME at E7.5 (21), and Nog−/− embryos show posterior abnormalities but no rostral truncations in an outbred genetic background (27). Indeed, we saw no HPE phenotypes in Nog+/− (n = 68) or Nog−/− embryos (n = 39) (Table 1). Nog+/−;Smad3−/− embryos with HPE were found occasionally (2/16, Fig. 1E), whereas a much higher penetrance of HPE occurred in Nog−/−;Smad3+/− and Nog−/−;Smad3−/− embryos (29 and 60%, Fig. 1F). These data indicate that the penetrance of the HPE phenotype is associated with a decrease in Nog and Smad3 alleles in a dose-dependent manner. In contrast, we saw no significant penetrance of rostral defects in Chrd−/−;Smad3+/−, Chrd−/−;Smad3−/− or Nog−/−;Nodal+/− mutants (Table 1); this can be explained at least in part by the spatiotemporal expression and functional differences of these pairs of genes, as considered below and in Discussion. Nonetheless, the genetic interactions revealed by Chrd and Nodal double-mutants or by Nog and Smad3 double-mutants provide independent evidence that the balance of BMP signaling and Nodal signaling plays a key role in anterior development. In the analysis of these mutants, we focus on Chrd−/−;Nodal+/− and Nog−/−;Smad3+/− embryos specifically, referred to from here on as Chrd;Nodal and Nog;Smad3 embryos.
Table 1.
Distribution of anterior defects among genotypic classes (E9.5)
| Genotype | Chrd−/− | Nog+/− | Nog−/− | Nodal+/− | Smad3−/− |
| Sample no. | 36 | 68 | 39 | 18 | 44 |
| Anterior defects | 0 | 0 | 0 | 0 | 0 |
| Genotype | Chrd−/−;Nodal+/− | Nog−/−;Nodal+/− | Chrd−/−;Smad3+/− | Chrd−/−;Smad3−/− | |
| Sample no. | 83 | 22 | 37 | 25 | |
| Anterior defects | 19 (23%) | 0 | 0 | 0 | |
| Genotype | Nog+/−;Smad3+/− | Nog+/−;Smad3−/− | Nog−/−;Smad3+/− | Nog−/−;Smad3−/− | |
| Sample no. | 61 | 16 | 58 | 30 | |
| Anterior defects | 3 (5%) | 2 (13%) | 17 (29%) | 18 (60%) |
Synergistic function of Nodal and BMP antagonism patterns the APS and anterior AME
To determine the mechanisms underlying anterior malformations in Chrd;Nodal and Nog;Smad3 embryos, we assessed the expression of several key genes required for the normal anterior development. These studies were done at stages when the initial cellular defects might be occurring, well before any overt phenotype distinguishes affected from unaffected mutants. Because the phenotypes are partially penetrant, we expected meaningful molecular changes to occur at a penetrance comparable with the phenotypic penetrance observed at later stages.
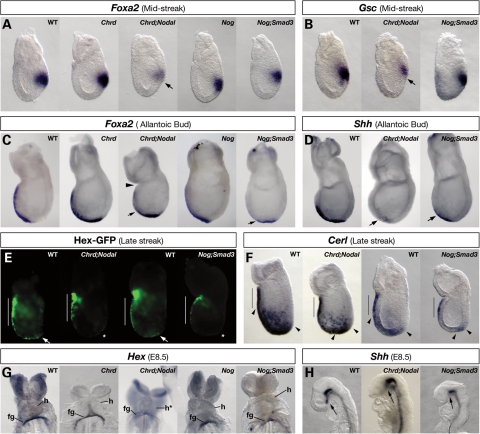
The anterior visceral endoderm (AVE) promotes initial forebrain identity (22,28) and is involved in forebrain patterning (29); therefore, we examined the expression of AVE genes at E6.5, including Cerl and Hex. All Chrd;Nodal and Nog;Smad3 embryos examined showed normal Hex and Cerl expression in the VE cells located adjacent to the anterior epiblast (Fig. 2E and F; Hex-GFP, n = 4 and 3; Cerl, n = 4 and 3). Such results suggest that the AVE forms normally in these double-mutant embryos.
Figure 2.
APS and AME marker expression in compound mutant embryos. WMISH (A–D, F–H) or optically sectioned GFP fluorescence (E) of APS or AME markers. (A) At MS, Foxa2 is expressed normally in the APS of WT, Chrd−/−, Nog−/− and Nog−/−;Smad3+/− embryos, but is significantly decreased in Chrd−/−;Nodal+/− embryos (arrow). (B) At MS, Gsc expression is normal in WT and Nog−/−;Smad3+/− embryos but decreased in Chrd−/−;Nodal+/− embryos. (C) At the bud stage, Foxa2 is expressed in the extending AME in WT, Chrd−/− and Nog−/− embryos, but is lost in the anterior AME of Chrd−/−;Nodal+/− and Nog−/−;Smad3+/− embryos (arrows). Chrd−/−;Nodal+/− embryos are sometimes constricted at the extra-embryonic–embryonic junction (arrowhead). (D) At the bud stage, Shh expression in anterior AME is lost in Chrd−/−;Nodal+/− and Nog−/−;Smad3+/− embryos (arrows). (E) In LS WT embryos, Hex-GFP is expressed in the AVE (white bar) and in the ADE (arrows). In Chrd−/−;Nodal+/− and Nog−/−;Smad3+/− embryos, AVE expression of Hex-GFP is normal, but ADE expression is decreased (asterisks). (F) At LS, Cerl is expressed in the AVE (black bars) as well as in ADE cells (between arrowheads). Relative to WT, ADE expression in Chrd−/−;Nodal+/− and Nog−/−;Smad3+/− embryos is markedly decreased and located only distally. (G) Hex is expressed in the foregut endoderm pocket (fg) of WT, Chrd−/− and Nog−/− embryos, but this is decreased or lost in Chrd−/−;Nodal+/− and Nog−/−;Smad3+/− embryos. (H) In WT embryos, the PCP expresses Shh (arrow). Shh expression is lost from the PCP area of Chrd−/−;Nodal+/− and Nog−/−;Smad3+/− embryos, but is retained in the rest of the midline. h, heart.
The APS, whose derivatives promote forebrain identity, is first identifiable by the expression of Foxa2 and Gsc at the mid-streak (MS) stage. Both genes function in the APS for normal anterior–posterior axis patterning (30,31). We found that some Chrd;Nodal embryos showed decreased Foxa2 (5/22) and Gsc expression (4/13) relative to Chrd−/− littermates (0/9; Fig. 2A and B). We saw no significant change in early Foxa2 or Gsc expression in Nog−/− (0/4) or Nog;Smad3 embryos (0/8). These data suggest that APS development is defective in Chrd;Nodal, but not in Nog;Smad3 embryos. The difference in APS marker expression between these double-mutants is consistent with Chrd and Nodal expression in and around the APS during its early development, whereas Nog is not expressed until the APS reaches the distal end of the embryo and begins to form the node. Therefore, even though both classes of mutants possess similar rostral deficiencies by mid-gestation, the initial defects leading to their terminal phenotypes are likely different.
Next, we examined the development of key APS derivatives, the AME and the PCP, at E7.5–E8.25. Foxa2, whose expression is essential in the AME for normal head development (32), is diminished in the anterior AME of some Chrd;Nodal (5/20) and Nog;Smad3 embryos (5/10; Fig. 2C). Shh expression is required in the AME for ventral forebrain development (33), and is significantly decreased in the most anterior domain of embryos in both mutant classes (5/12 and 2/3; Fig. 2D). At E8.25, the anterior-most derivative of the AME, the PCP, secretes inductive cues such as Shh to the overlying forebrain region (33). PCP expression of Shh is greatly reduced in both Chrd;Nodal (2/5) and Nog;Smad3 (4/10) mutant embryos (Fig. 2H). Together, these results suggest that the anterior AME and the PCP are abnormally patterned and functionally deficient in both mutant classes.
Another relevant derivative of the APS is the ADE, which also plays an important role in promoting anterior neural development (34). It emerges from either side of the midline to displace the visceral endoderm anteriorly. The ADE is marked by the expression of Hex and Cerl. Expression levels of each are decreased in the distal epiblast of both classes of double-mutants (n= 4 and 3; Fig. 2E and F), revealing that ADE specification is significantly impaired. Hex expression in the ADE is required for forebrain development (35), and its expression in the ADE-derived foregut pocket is diminished in embryos of both classes (2/5 and 2/4; Fig. 2G); this is further evidence that the ADE is deficient.
These results demonstrate expression changes of key anterior patterning genes at frequencies similar to the phenotypic penetrance of HPE in these digenic mouse models, but with a key difference between the two genotypes. Chrd;Nodal embryos, but not Nog;Smad3, lack normal expression of APS genes, suggesting that the APS is compromised in its ability to form derivatives. Both Chrd;Nodal and Nog;Smad3 mutants have defective anterior AME specification that likely compromises the subsequent patterning of the PCP and ADE.
Alteration of BMP and Nodal signaling levels in double-mutants
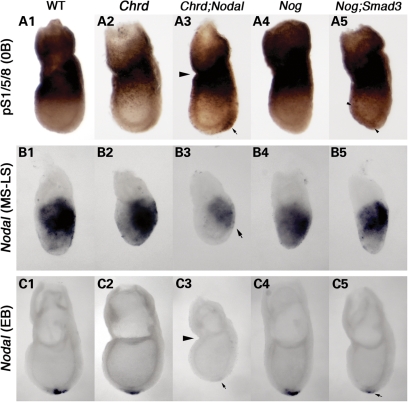
Genetic interactions between BMP antagonists and Nodal, or its intracellular effector Smad3, suggest a synergistic function to promote Nodal signaling while simultaneously inhibiting BMP signaling. Accordingly, we determined whether the patterns of BMP and Nodal signaling were altered in the compound mutants during the development of the APS and its derivatives. We examined the distribution of active BMP signaling using immunohistochemistry with an antibody specific to the activated BMP signaling effector, phosphorylated (p)-Smad1/5/8 (22). At MS–0B stages, we found ectopic and elevated levels of p-Smad1/5/8 in the APS of some Chrd;Nodal embryos (7/18; Fig. 3A3, arrow), whereas Chrd−/− embryos showed a pattern similar to wild-type embryos (n = 16; Fig. 3A2). Nog−/− embryos showed no significant difference in p-Smad1/5/8 staining during gastrulation (Fig. 3A4), though they show increased levels at the neurulation stage (Y.-P.Y and J.K., unpublished data). Double null embryos for Chrd and Nog (Chrd−/−;Nog−/−) show ectopic p-Smad1/5/8 in the APS, very similar to that seen in Chrd;Nodal (Supplementary Material, Fig. S2). In contrast, some Nog;Smad3 embryos displayed ectopic p-Smad1/5/8 throughout the distal epiblast (3/13; Fig. 3A5, arrowheads), indicating ectopic but less focused activation of BMP signaling. Therefore, like Chrd;Nog embryos, which lack two organizer BMP antagonists, both Chrd;Nodal and Nog;Smad3 double-mutants have expanded activation of BMP signaling during initial stages of axial midline development.
Figure 3.
Alteration of BMP and Nodal signaling levels in compound mutant embryos. (A) Phospho-Smad1/5/8 (pS1/5/8) localization at the no-bud (0B) stage. pS1/5/8 is seen in extra-embryonic tissues and at the embryonic/extra-embryonic junction, but is absent in the distal region of wild-type (WT) (A1), Chrd−/− (A2) and Nog−/− embryos (A4). Ectopic pS1/5/8 is detected in the APS region of the distal epiblast of Chrd−/−;Nodal embryos (arrow in A3) and faintly throughout the epiblast in some Nog−/−;Smad3+/− embryos (arrowheads in A5). (B) Nodal expression at the MS–LS stage. Expression is similar in the primitive streak of WT and all mutant classes except Chrd−/−;Nodal+/− embryo (B3), where it is decreased (arrow). (C) Nodal expression at the EB stage. Nodal is expressed around the node, at the distal tip of the WT embryo (C1). Nodal expression is normal in Chrd−/− and Nog−/− embryos (C2 and C4), but it is lost or decreased in Chrd−/−;Nodal+/− (C3) and Nog−/−;Smad3+/− (C5) embryos. Arrowheads in A3 and C3 denote the constriction found in many Chrd−/−;Nodal+/− embryos.
There is a paucity of known, direct transcriptional targets relevant to the APS for Nodal signaling; however, Nodal transcription itself is positively regulated by Nodal signaling through a positive feedback mechanism (9). We therefore used Nodal expression per se as an indicator of Nodal signaling. Nodal expression in the primitive streak looked the same in wild-type, Nodal+/− and Chrd−/− embryos (Fig. 3B1 and 2), as indicated by whole-mount in situ hybridization (WMISH). In contrast, most Chrd;Nodal embryos showed significantly diminished Nodal expression, both in the APS (11/11, Fig.3B3) and in the node (6/9, Fig. 3C2 and 3). In Nog;Smad3 embryos, no change was detected in APS Nodal expression (0/3), consistent with our finding of the normal expression of other APS genes in these mutants. However, we found decreased levels of Nodal expression around the node of some Nog;Smad3 embryos (2/9; Fig. 3C5). These data are further evidence that Nog functions at a later developmental stage than Chrd with regard to Nodal signaling. Overall, these findings suggest that BMP antagonists and Nodal synergistically create an environment that promotes Nodal signaling while inhibiting BMP signaling.
Nodal antagonizes the inhibitory effect of BMP on APS gene expression
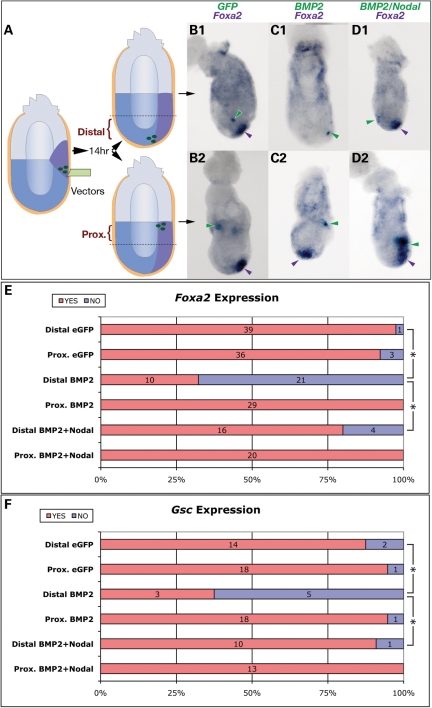
APS specification can be severely compromised by decreased Nodal expression (8). To determine whether it can be similarly diminished by ectopic BMP activity, we utilized an embryo culture system that allows for the introduction of ectopic gene expression vectors into the primitive streak area via microinjection (36,37). Wild-type, E6.5 embryos were injected with liposomes containing an experimental transgene into the nascent primitive streak region (Fig. 4A). An eGFP transgene (encoding enhanced green fluorescent protein) was co-injected as a marker to localize transfected cells. After culture, eGFP-positive embryos were sorted with respect to eGFP localization in either the proximal or distal embryo (Fig. 4A). Embryos without eGFP expression, or with eGFP expression only in the extra-embryonic portion of the conceptus, were excluded. We then used two-colored WMISH to assay the expression of the transgene and the endogenous APS marker Foxa2 (see Materials and Methods). Most embryos injected with eGFP alone showed normal Foxa2 expression (Fig. 4B1 and 2), suggesting that injection and culturing per se did not affect endogenous Foxa2 expression.
Figure 4.
Nodal rescues Bmp2-mediated suppression of APS gene expression. (A) Diagram of the experimental strategy. eGFP alone, or eGFP with experimental transgene expression vectors, was introduced into the APS region of WT E6.5 embryos. After 14 h culture, transgenic embryos were sorted by location of GFP expression (distal or proximal). Two-colored WMISH was used to detect ectopic gene expression (light blue) and endogenous APS gene expression (purple). eGFP expression (green arrowheads) in neither the distal (B1) nor the proximal epiblast (B2) alters Foxa2 expression. Foxa2 expression is inhibited by ectopic Bmp2 in the distal epiblast (C1) but not in the proximal epiblast (C2). Co-expression of Bmp2 and Nodal either distally (D1) or proximally (D2) does not affect Foxa2 expression. (E and F) Summary of the effects of transgene expression on Foxa2 and Gsc expression. The numbers of embryos with normal (red) or reduced expression (blue) are indicated. Asterisks denote a significant difference (P < 0.001) by Student's t-test between the indicated categories.
In embryos transfected with eGFP and Bmp2, Foxa2 expression was lost or significantly decreased when the transfected cells were present in the distal embryo (Fig. 4C1), but unchanged if transfected cells were proximal (Fig. 4C2). Similar results were found for Gsc expression (summarized in Fig. 4F). These results indicate that ectopic Bmp2 expression inhibits APS gene expression when present in or around the anterior but not the posterior primitive streak.
Our genetic data imply an antagonistic relationship between Nodal and BMP signaling in APS development. If so, one would expect that increased Nodal might relieve the inhibitory effect of BMP on APS gene expression. To test this, we co-injected Bmp2 and Nodal vectors, along with eGFP vector, into the primitive streak. Ectopic Bmp2 and Nodal expression in the distal epiblast partially rescued Foxa2 expression (Fig. 4D1), when compared with the effects of Bmp2 injection alone (16/20 versus 10/31; Fig. 4E). Similar results were observed for Gsc expression (Fig. 4F). These results suggest that Nodal suppresses the diminished expression of APS genes caused by ectopic Bmp2.
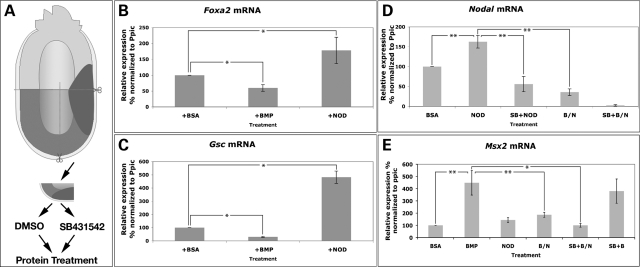
To further probe the mechanisms underlying the antagonistic relationship of Nodal and BMP activity on APS patterning, we isolated APS explants from MS stage embryos, and cultured them in medium supplemented with recombinant human BMP2 or Nodal protein (Fig. 5A). After culture, gene expression was assayed by quantitative PCR (qPCR). As expected, each protein treatment increased the expression of a corresponding, known target gene; BMP2 increased Msx2 expression, whereas Nodal protein led to a modest but consistent increase in Nodal transcription levels (Fig. 5D and E). In the same samples, transcript levels of the APS genes Foxa2 and Gsc were significantly decreased by BMP2 treatment, but increased by Nodal treatment (Fig. 5B and C). Thus, the expression of the APS genes Foxa2 and Gsc is promoted by Nodal but inhibited by BMP activity.
Figure 5.
Mutual antagonism of BMP and Nodal does not require Nodal signal transduction. (A) Diagram of the APS explant assay. APS fragments were isolated from E6.5 embryos and incubated in culture medium with DMSO or the Nodal inhibitor SB-431542 (SB), followed by protein treatment and then analyzed with qPCR. Protein treatments included BSA, BMP2 (BMP), Nodal (NOD) and BMP2/Nodal co-treatment (B/N). (B–E) Relative mRNA levels of Foxa2, Gsc, Nodal and Msx2 in explants as measured by qPCR after the indicated treatments. Results were normalized to the mouse ‘house-keeping’ gene peptidylprolyl isomerase C (Ppic). The asterisks denote a significant difference between the indicated categories as determined by Student's t-test: *P < 0.01; **P < 0.005.
We then considered the consequences of simultaneous treatment of APS explants with BMP2 and Nodal proteins (BMP–Nodal). Nodal mRNA was decreased in the explants treated with BMP/Nodal, but significantly increased when Nodal protein was used alone at the same concentration. Conversely, Msx2 expression was greatly reduced after combined BMP/Nodal treatments, relative to the treatment with BMP2 alone (Fig. 5E). Thus, BMP2 antagonizes Nodal signaling induced by Nodal protein, whereas Nodal inhibits the activation of BMP signaling by BMP2.
Inhibition of Nodal signal transduction does not diminish Nodal protein's attenuation of BMP activity
We used the Nodal inhibitor SB-431542 to probe the nature of the interaction between BMP and Nodal signaling in our explant system. In cell culture, this compound inhibits the activation of TGFβ signaling by blocking activity of activin type I receptors Alk4, 5 and 7, without affecting BMP signaling activation (38). In vivo, SB-431542 can block both endogenous and exogenous signaling activation via Smad2 phosphorylation in frog and fish embryos, further supporting its specificity (39). Similarly, in chick axial mesoderm explants, SB-431542 can block the transcription of Nodal signaling target genes (40). In cultured mouse embryos, SB-431542 prevents the expression of Nodal targets in the left lateral plate mesoderm (LPM) (36). We tested the efficiency and specificity of SB-431542 in our APS explant system, by treating explants with the compound, or the carrier DMSO as a control, followed by incubation with Nodal or BMP2 proteins and measurement of target gene expression levels by qPCR. We used a concentration of SB-431542 (100 μg/ml) that blocked the expression of Nodal target genes in the left LPM of cultured embryos (36). In control explants, Nodal protein induced an increase in Nodal expression of about 1.5-fold; however, in explants pretreated with SB-431542, Nodal was unable to induce ectopic expression of Nodal mRNA; moreover, these explants displayed a decrease in natural Nodal levels (Fig. 5D). In contrast, SB-431542 treatment did not affect the induction of Msx2 by BMP2 (Fig. 5E). These results confirm that, in this assay system, SB-431542 is able to significantly inhibit Nodal signaling, but not BMP signaling.
We then tested whether BMP and Nodal still exhibit an antagonistic interaction when the Nodal receptor is inhibited by SB-431542. If Nodal protein were unable to inhibit the induction of Msx2 by BMP2 protein upon SB-431542 treatment, this would suggest that Nodal signal transduction via its receptor is required to inhibit BMP signaling. In contrast, if Nodal protein were still able to inhibit Msx2 expression at levels comparable with when the inhibitor is absent, this would indicate that Nodal can inhibit BMP signaling even if Nodal signaling transduction is severely inhibited. To test these possibilities, APS explants were treated with SB-431542 and then cultured with BMP and Nodal proteins together (SB/BMP/Nodal treatment). Msx2 expression was significantly reduced in explants after SB/BMP/Nodal treatment relative to those treated only with BMP (Fig. 5E), despite the severe attenuation of Nodal's ability to induce target gene transcription upon SB-431542 treatment (Fig. 5D). Moreover, the level of Msx2 expression upon SB/BMP/Nodal treatment was very similar to that upon BMP/Nodal treatment. Thus, SB-431542 does not affect Nodal protein's ability to antagonize BMP's activation of a transcriptional target, though it does largely block Nodal target gene expression. These data suggest that Nodal protein is able to antagonize BMP signaling even when Nodal signal transduction is severely attenuated due to dysfunctional receptors.
Formation of an extracellular BMP–nodal heteromeric complex
In considering potential mechanism(s) by which BMP and Nodal might antagonize each other's signaling pathways during early head development, we considered their expression at primitive streak stages. Nodal and Bmp2 are both expressed in the forming streak and posterior visceral endoderm (Supplementary Material, Fig. S1), in which tissue Nodal function is required for early head formation (18,41). Our Nog;Smad3 results suggest that an additional relevant interaction of BMP and Nodal occurs in or around the forming node at the anterior end of the fully elongated primitive streak, where Nodal, Bmp2, Bmp7 and Nog are expressed in close proximity (22,25,42). Thus, BMP and Nodal signaling are potentially active in the same or in closely juxtaposed tissues in the relevant spatiotemporal contexts for the genetic and embryological effects we observe.
Because the mutually inhibitory BMP–Nodal interaction occurs even when Nodal signal transduction is severely attenuated at the receptor level, it is conceivable that the relevant antagonism of Nodal and BMP pathways occurs via an extracellular interaction of the ligands. One possibility is that Nodal and Bmp2 proteins form heteromeric complexes that fail to signal, given that Xenopus BMP7 precursor protein heterodimerizes with mouse Nodal precursor protein when co-expressed (43). We therefore tested whether mouse Nodal and Bmp2 form protein complexes when they are co-expressed in the same cells, and/or after they are secreted into the extracellular space.
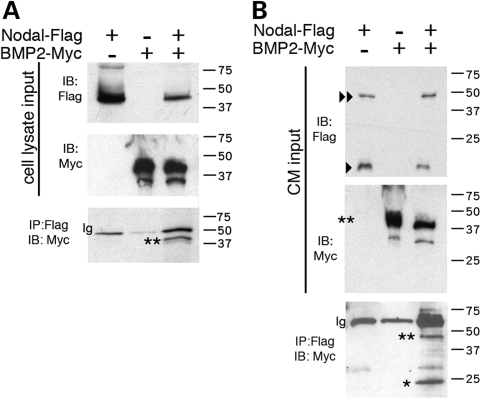
To determine whether Nodal and Bmp2 interact directly, we performed reciprocal co-IP assays. Flag-tagged Nodal protein and Myc-tagged Bmp2 were co-expressed in COS cells, with cell lysates subjected to IP using anti-Flag or anti-Myc antibodies. Unprocessed Nodal-Flag and Bmp2-Myc precursor proteins were successfully detected in cell lysates (expected sizes for both: ∼45 kDa; Fig. 6A). Bmp2-Myc precursor protein co-immunoprecipitated with Nodal-Flag precursor protein (Fig. 6A). In control experiments using the same conditions, we saw no co-IP in lysates from cells co-expressing Bmp2-Myc and an irrelevant Flag-tagged protein, or those co-expressing Nodal-Flag and an irrelevant Myc-tagged protein, indicating that neither the epitope tags nor the Bmp2 and Nodal peptides exhibit evidence of non-specific binding in these conditions (Supplementary Material, Fig. S3). These data collectively indicate that Bmp2 and Nodal proteins can bind each other when expressed in the same cells, and this interaction is likely the result of specific interactions between the Bmp2 and Nodal proteins.
Figure 6.
Direct interaction of Nodal and Bmp2 protein. IP of Nodal and Bmp2 proteins from COS cells transfected with Nodal-Flag and/or Bmp2-Myc. Transfection with either construct is indicated by a plus (+), and absence by a minus (−). IP, immunoprecipitation; IB, immunoblot. (A) Cell lysates blotted and incubated with anti-Flag or anti-Myc demonstrate expression of both constructs. IPs were performed with anti-Flag (for Nodal) and blotted for anti-Myc (for Bmp2). Only precursor forms were detected (**). Expected sizes for tagged Nodal and Bmp2 are both around ∼45 kDa. (B) IP of Bmp2-Myc by Nodal-Flag from the CM of COS cells, in which separate cell populations were transfected with Nodal-Flag or Bmp2-Myc and then cultured. In the CM input, Bmp2-Myc precursor form (**) was detected, as well as Nodal-Flag precursor (<<) and mature forms (<; ∼18 kDa). Both precursor and mature forms of Bmp2-Myc were immunoprecipitated by Nodal-Flag [∼45 kDa (**) and ∼20 kDa (*), respectively].
We then tested whether Nodal and Bmp2 proteins can form a heteromeric complex extracellularly, i.e. after the proteins are secreted from the cells that express them. Nodal-Flag or Bmp2-Myc proteins were expressed individually in different cultures of COS cells. Both the precursor and mature forms of Nodal-Flag (∼45 and ∼18 kDa, respectively) were detected in the conditioned medium (CM) (Fig. 6B). Only precursor proteins of Bmp2-Myc were detected in the CM (Fig. 6B), consistent with previous findings that Bmp2 processing may be inefficient in COS cells (44). We then performed IPs on either mixed or individual CM from cultured cells expressing either Nodal-Flag or Bmp2-Myc. The data indicate that Bmp2 and Nodal proteins can form a heteromeric complex when present in media from separate cell cultures (Fig. 6B). Thus, a Bmp2–Nodal complex can form when these proteins are expressed in either the same or separate cells. Given that BMPs and Nodal are expressed in close proximity during early stages of head development, our results support the possibility of a direct, antagonistic BMP–Nodal protein interaction in the cellular interactions underlying the establishment of head primordia.
DISCUSSION
The results of this study demonstrate an antagonistic relationship between Nodal and BMP signaling at the earliest stages of head development in mammalian embryos, acting to regulate the development of the APS and its derivatives. Our molecular analysis suggests a previously unidentified role for BMP antagonism in head development: to protect Nodal from an inhibitory effect of BMP in regulating key genes directing the development of the APS, ADE and AME. This in turn provides a mechanistic explanation for the HPE, craniofacial and laterality defects observed in Chrd;Nodal and Nog;Smad3 mutants.
Antagonistic interactions between Nodal and BMP signaling in mouse head development
Our work reveals the in vivo significance of BMP–Nodal interactions during mouse gastrulation. With regard to embryological function, it demonstrates that (i): a genetic interaction between BMP antagonists and Nodal pathway components occurs and is required during anterior formation; (ii) mutual antagonism between BMP and Nodal signaling patterns the APS and anterior AME; and (iii) BMP antagonists Chrd and Nog have spatiotemporally unique functions during their interaction with Nodal signaling.
It has been shown previously that Nodal is required in the VE to establish the AVE and initiate anterior specification (45); however, attenuated Nodal-Smad2/3 signals, such as in embryos carrying a Nodal hypomorph mutation (8) or Smad2;Smad3 compound mutations (11), do not affect AVE patterning. Furthermore, embryos lacking Chrd and Nog do not display defects in AVE specification (21). Similarly, in Chrd;Nodal and Nog;Smad3 embryos, the AVE forms normally, and the anterior cell fate is imposed in the epiblast. Thus, the anterior defects in these mutants are not due to failed AVE specification. Rather, these phenotypes result from deficient generation of ADE and AME, and reflect a need for balanced BMP and Nodal signaling during APS patterning.
Although Chrd;Nodal and Nog;Smad3 embryos often showed HPE and associated rostral defects, we found no such malformations in Nog−/−;Nodal+/− and Chrd−/−;Smad3−/+(−). This leads us to consider that Chrd and Nog may have unique functions interacting with Nodal signaling during early gastrulation. Although previous studies demonstrate redundant function of Chrd and Nog in mouse anterior formation after gastrulation (20,21), the spatiotemporal difference in their expression suggests distinct functions during gastrulation. Early Chrd expression is seen at E6.5 in the APS (24), within or very close to the tissue expressing Nodal (25). In Chrd;Nodal mutants, ectopic p-Smad1/5/8 and decreased Nodal expression in the APS region correlate with the mispatterning of the APS. These data reveal a novel synergism of Nodal and BMP antagonism, via Chrd, in APS development.
In contrast, Nog expression is first found in the node from bud stages (21), after the cessation of Nodal expression in the primitive streak. Nodal expression around the node does not appear to be necessary for head development, and it is not expressed in the AME (46). Not surprisingly, Nog;Nodal mutants have no defects in anterior patterning. Similarly, we also found no evidence of an interaction between Nog and Smad3 in APS development. In contrast, we did find that Nog and Smad3 interact to promote development specifically of the AME. Given that Smad3 is downstream of all TGFβ ligands (but not BMP ligands), this suggests that TGFβ signaling has a positive role in concert with BMP antagonism in promoting AME development in the context of the node or AME itself. By extension, we suggest that such a role is fulfilled by some TGFβ ligand other than Nodal per se. GDF1, a TGFβ homolog and Nodal co-ligand (17), is expressed around the node and axial midline and functions together with Nodal to promote head development (16,47). Although neither Nodal nor Gdf1 individually appear to be necessary in the node or axial derivatives for head development (17,46), it is conceivable that a Nodal-Gdf1 heteromer (17) is active in this context.
As with Nog, the expression and function of Chrd overlap with that of Smad3 in the AME. Unexpectedly, we found no evidence of a genetic interaction between Chrd and Smad3. This probably reflects the greater sensitivity of AME development to decreased dosage of Nog as opposed to Chrd, as demonstrated previously (20).
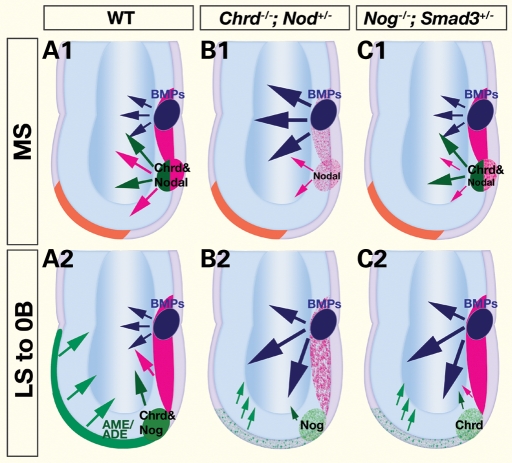
Altogether, our data lead us to propose that the crosstalk between BMP and Nodal signaling occurs via the cooperative activity of (i): Chrd and Nodal in the APS at the MS stage (Fig. 7A1), and (ii) Nog and Smad3 in the anterior AME at the LS to 0B stage (Fig. 7A2). This reveals two distinct phases of midline axial patterning, which both require Nodal signaling and suppression of BMP signaling. In Chrd;Nodal embryos, APS specification is deficient due to an excess of BMP signaling and reduced Nodal signaling (Fig. 7B1). This aberrant APS gives rise to a deficient AME that lacks its most anterior region, and thus the critical cues that emanate from this tissue (Fig. 7B2). In Nog;Smad3 embryos, APS patterning appears normal, possibly because Chrd, Nodal and Smad2 are still expressed (Fig. 7C1). However, the AME, emerging from the compromised APS, lacks both Nog and Smad3 functions and can no longer sufficiently antagonize BMPs (Fig. 7C2). In embryos of both mutant classes, failure in maintaining balanced BMP–Nodal levels shifts the boundaries between mutually antagonistic signals in the APS and/or the anterior AME, which in turn leads to the gross anterior patterning defects.
Figure 7.
Model for the interaction of Nodal and anti-BMP activities during the formation of the anterior primitive streak and its derivatives. (A) BMP signaling activation (blue) is restricted, and Nodal signaling enhanced, by the synergistic interaction of Chrd and Nodal in the APS (green/red) at the MS (A1) stage, and of Nog and Smad3 in the anterior AME (green) at the 0B stage (A2). These interactions promote the specification of the APS and/or its derivatives, the AME and ADE, and the BMP antagonism they convey to promote forebrain initiation in the ectoderm (green arrows). (B) In Chrd−/−;Nodal+/− embryos, the APS is defective due to excess BMP activation and reduced Nodal signaling (B1), which results in AME/ADE deficiency (B2). (C) In Nog−/−;Smad3+/− embryos, early APS appears normal (C1), but AME emerging from the compromised distal APS (early node) is defective due to ectopic BMP activation and decreased Nodal pathway signaling (C2).
Molecular interactions of BMP and Nodal pathways in patterning the anterior primitive streak
Our data show that Nodal and BMP can each diminish the other's signaling activity. Use of the Nodal receptor inhibitor SB-431542 in tissue culture studies revealed that even when Nodal signal transduction is severely inhibited, Nodal protein can still efficiently attenuate BMP signaling. Because SB-431542 acts at the level of the TGFβ type I receptors (38,39), this indicates that Nodal protein can interfere with BMP signaling in a manner that does not fully depend on the activation of its own receptors. Our explant studies indicated that steady-state levels of Nodal or BMP protein were sufficient to activate their own signaling pathways robustly, but also to strongly inhibit the ability of the other ligand to activate its own signaling pathway. We note, however, that the endogenous amount of Nodal or BMP molecules required to activate downstream signal transduction may differ from those effective in a BMP–Nodal mutual antagonism.
Previous studies in Xenopus have suggested an antagonistic interaction between BMP and Nodal. The Nodal family member Xnr3 promotes neural induction by antagonizing BMP4 even though it lacks mesodermal-inducing capability via Smad2/3 activity (48,49). Moreover, Nodal can antagonize the endogenous activation of Smad1 by BMP7 signals, and BMP7 can antagonize the activation of Smad2 by Nodal, at least when Nodal and Bmp7 are co-expressed (43). A possible molecular explanation for the mutually inhibitory interaction of BMP and Nodal in these frog assays is the formation of signaling-inactive BMP–Nodal (or BMP–Xnr3) heterodimers (43,49).
We considered whether a physical interaction of Nodal and Bmp2 could potentially occur to account for our findings. The genes are indeed expressed in a spatiotemporal pattern consistent with a potential molecular interaction. For example, we found that Nodal and Bmp2 are both expressed in the extra-embryonic endoderm around the forming APS, where Chrd is expressed (Supplementary Material, Fig. S1). We then used biochemical experiments to assess whether mouse Nodal and BMP ligands can physically interact. Bmp2 and Nodal were able to form complexes containing their immature as well as their mature, processed forms. These complexes formed both when Bmp2 and Nodal were expressed in the same cells, and when they were expressed in different cells and secreted into the extracellular environment (CM).
Other mechanistic contributions to the mutual antagonistic interaction of BMP and Nodal signaling are not ruled out by our results. For instance, the titration of or the competition for common signaling components could contribute to the observed outcome of mutual antagonism, such as Smad4 and/or Actr2a/b as suggested by previous studies in left–right patterning and early distal visceral endoderm formation, respectively (50,51). Nonetheless, we believe that the model most consistent with our work and prior experiments in Xenopus is that Bmp2 and Nodal proteins form a heteromeric complex that is defective in activating either BMP or Nodal receptors. This would occur in the cells around the developing APS. A similar mechanism may be acting in the node region, where Bmp2 and/or Bmp7 could interact with Nodal and/or GDF1, or perhaps some other TGFβ homolog. The presence of the BMP antagonist Chrd in the early APS would protect APS gene expression from the repressive role of BMP, and protect Nodal signaling from inhibition by BMP protein. In the node region, our genetic results suggest that Nog is the more relevant BMP antagonist. All of these are secreted factors, and the molecular interactions are likely to occur extracellularly.
A new multigenic model for HPE
The genetic analysis of HPE in humans has led to a hypothesis that many cases of HPE result from two or more independent genetic lesions impacting common or interacting developmental pathways (3). The present work not only establishes the significance of BMP–Nodal interaction during mammalian anterior development, but also demonstrates that lesions in Nodal signaling can interact with lesions in BMP antagonists to cause the malformation; thus, HPE can arise from the co-occurrence of mutations in Chrd and Nodal, as well as Nog and Smad3.
However, in both Chrd;Nodal and Nog;Smad3 double-mutants, we observed the variability of phenotypic severity: from no morphological anterior deficits to severe anterior truncation. The phenotypic variability may reflect a combination of an inexact quantitative requirement of the level of BMP versus Nodal signaling for head development, compounded by partial genetic redundancy in both BMP antagonism (20) and Nodal signaling (11) during early stages of AME development. An alternative explanation is also possible; because these studies were conducted in an outbred genetic background, cryptic mutations in other genes may play a role in modifying the severity of the rostral phenotypes observed in these otherwise double-mutant embryos. Indeed, this is likely to be the case in humans: the consequences of any particular pair of unlinked mutations are likely to be influenced in many cases by unknown independent loci playing lesser roles. Thus, we use the term ‘multigenic’ rather than ‘digenic’ to describe these HPE mouse models; although only mice mutant for specific pairs of genes (Chrd;Nodal or Nog;Smad3) show head phenotypes, we cannot exclude the potential involvement of additional, unknown mutation(s).
Nevertheless, given that mutations in several Nodal signaling pathway components are associated with human HPE (52), it is possible that some of these cases also involve mutations in BMP antagonists. Whether mutations in CHRD or NOG, alone or in combination with mutations in Nodal signaling components, are associated with cases of human HPE remains to be addressed.
MATERIALS AND METHODS
Mouse genetics and embryo collection
Using random outbred stock, we crossed Chrd (Chrdtm1Emdr) homozygotes (53) or Nog (Nogtm1Amc) heterozygotes (54) to Nodaltm1Rob (25) or Smad3tm1Xfw (26) heterozygotes to generate intermediates that were crossed to produce the experimental genotypic classes. Tg(Hex-eGFP)ARbe (55) was crossed in to visualize Hex expression. Embryos were genotyped by PCR (21,22,25,26,54) and staged as described (56). All animal handling was done under the supervision and detailed protocol approval of the Duke University Institutional Animal Care and Use Committee.
WMISH and immunohistochemistry
Double- and single-colored WMISH followed an established protocol (57). Whole-mount immunohistochemistry for anti-p-Smad1/5/8 (Cell Signaling) was performed as described (22).
Introduction of expression vectors into the APS
Expression vectors were introduced into the APS of E6.5 embryos as described (36,37). Three sets of liposome solutions with a total of 1 μg expression vector(s) were prepared for injection: eGFP only (1 μg of eGFP vector), eGFP/Bmp2 (0.65 μg of eGFP and 0.35 μg of Bmp2 vector) and eGFP/Bmp2/Nodal (0.3 μg of eGFP, 0.35 μg of Bmp2 and Nodal vector each). Injected embryos were cultured for 14 h at 37°C in medium containing DMEM (Gibco) supplemented with 50% rat serum (Biomeda) with rotation. Transfection was initially confirmed by GFP visualization, followed by WMISH using probes against GFP (control) or BMP2 (experimental embryos).
Explant studies and qPCR
APS explants from MS stage embryos were isolated using glass needles and cultured in 96-well plates at 37°C. Recombinant BMP2 and/or Nodal proteins (R&D Systems) were used at 400 ng/ml and 2 μg/ml, respectively. SB-431542 (Sigma) was used at 100 μg/ml in 0.1% DMSO (Sigma). SB-431542 or DMSO pretreatment was for 30 min, followed by 1.5 h protein treatment. BSA (Sigma, 1 μg/ml) and DMSO were used in control explants. Total RNA was prepared from three explants for cDNA synthesis and qPCR procedures as described (22). Two to three independent sets of cDNA samples were prepared, and qPCR was performed three times on each sample to determine each gene expression level. Error bars presents standard error of the mean, and results were subjected to Student's t-test analysis.
Protein preparation, IP and western blots
Epitope-tagged constructs were generated with the pCS2+ vector using PCR-based cloning from E7.0 total cDNA and confirmed by sequencing. In pCS-Nodal-Flag (Nodal-Flag), three repeats of Flag epitope were fused after Ser251 of Nodal. In pCS-Bmp2-Myc (Bmp2-Myc), six repeats of Myc epitope were fused after His284 of Bmp2. The epitope is located within the mature protein domain (seven or four amino acids from the processing site of Nodal or Bmp2, respectively), allowing the detection of both precursor and mature forms.
For preparing proteins from cell lysates, Nodal-Flag and/or Bmp2-Myc plasmids were transfected into COS cells using FuGene (Roche). Cells were harvested and lysed in a buffer containing 50 mm Tris–Cl (pH7.4), 150 mm NaCl, 1 mm EGTA, 1% NP-40, 0.25% deoxycholate and protease inhibitors (Complete Mini, Roche). After centrifugation, supernatants were immunoprecipitated. αFlag (Stratagene) and αMyc (clone 9E10, Biomol) antibodies were used in IP and western blot analysis.
For preparing proteins from conditioned media (CM), Nodal-Flag or Bmp2-Myc was transfected separately into independent cultures of cells. Each culture was trypsinized, washed extensively with PBS and then combined and plated. After 48 h, CM was harvested and centrifuged to remove any cells. Similar results were obtained by collecting CM from separately transfected cultures of cells, centrifuging to remove any cells and then mixing CM samples when still warm. Antibodies were added directly to the CM for IP. For loading crude CM as input, 1 ml of CM was concentrated 2-fold by Centricon-3 filtration.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by a grant from the NIH to J.K. (R01DE013674).
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Drs X.-F. Wang, E. Robertson and T. Rodriguez for mice, and G. Oliver and J. Pearce for plasmids. We thank Drs A. Levine, N. Mine and S. Wu for technical advice and Dr D. McClay for his support. Drs B. Hogan, E. Robertson and our laboratory colleagues provided helpful discussion and comments.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Cohen M.M., Jr Holoprosencephaly: clinical, anatomic, and molecular dimensions. Birth Defects Res. Part A: Clin. Mol. Teratol. 2006;76:658–673. doi: 10.1002/bdra.20295. doi:10.1002/bdra.20295. [DOI] [PubMed] [Google Scholar]
- 2.Roessler E., Muenke M. Midline and laterality defects: left and right meet in the middle. Bioessays. 2001;23:888–900. doi: 10.1002/bies.1130. doi:10.1002/bies.1130. [DOI] [PubMed] [Google Scholar]
- 3.Ming J.E., Muenke M. Multiple hits during early embryonic development: digenic diseases and holoprosencephaly. Am. J. Hum. Genet. 2002;71:1017–1032. doi: 10.1086/344412. doi:10.1086/344412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Geng X., Oliver G. Pathogenesis of holoprosencephaly. J. Clin. Invest. 2009;119:1403–1413. doi: 10.1172/JCI38937. doi:10.1172/JCI38937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kinder S.J., Tsang T.E., Wakamiya M., Sasaki H., Behringer R.R., Nagy A., Tam P.P. The organizer of the mouse gastrula is composed of a dynamic population of progenitor cells for the axial mesoderm. Development. 2001;128:3623–3634. doi: 10.1242/dev.128.18.3623. [DOI] [PubMed] [Google Scholar]
- 6.Lawson K.A. Fate mapping the mouse embryo. Int. J. Dev. Biol. 1999;43:773–775. [PubMed] [Google Scholar]
- 7.Stern C.D. Initial patterning of the central nervous system: how many organizers? Nat. Rev. Neurosci. 2001;2:92–98. doi: 10.1038/35053563. doi:10.1038/35053563. [DOI] [PubMed] [Google Scholar]
- 8.Vincent S.D., Dunn N.R., Hayashi S., Norris D.P., Robertson E.J. Cell fate decisions within the mouse organizer are governed by graded Nodal signals. Genes Dev. 2003;17:1646–1662. doi: 10.1101/gad.1100503. doi:10.1101/gad.1100503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shen M.M. Nodal signaling: developmental roles and regulation. Development. 2007;134:1023–1034. doi: 10.1242/dev.000166. doi:10.1242/dev.000166. [DOI] [PubMed] [Google Scholar]
- 10.Whitman M. Nodal signaling in early vertebrate embryos: themes and variations. Dev. Cell. 2001;1:605–617. doi: 10.1016/s1534-5807(01)00076-4. doi:10.1016/S1534-5807(01)00076-4. [DOI] [PubMed] [Google Scholar]
- 11.Dunn N.R., Vincent S.D., Oxburgh L., Robertson E.J., Bikoff E.K. Combinatorial activities of Smad2 and Smad3 regulate mesoderm formation and patterning in the mouse embryo. Development. 2004;131:1717–1728. doi: 10.1242/dev.01072. doi:10.1242/dev.01072. [DOI] [PubMed] [Google Scholar]
- 12.Liu Y., Festing M., Thompson J.C., Hester M., Rankin S., El-Hodiri H.M., Zorn A.M., Weinstein M. Smad2 and Smad3 coordinately regulate craniofacial and endodermal development. Dev. Biol. 2004;270:411–426. doi: 10.1016/j.ydbio.2004.03.017. doi:10.1016/j.ydbio.2004.03.017. [DOI] [PubMed] [Google Scholar]
- 13.Nomura M., Li E. Smad2 role in mesoderm formation, left-right patterning and craniofacial development. Nature. 1998;393:786–790. doi: 10.1038/31693. [DOI] [PubMed] [Google Scholar]
- 14.Song J., Oh S.P., Schrewe H., Nomura M., Lei H., Okano M., Gridley T., Li E. The type II activin receptors are essential for egg cylinder growth, gastrulation, and rostral head development in mice. Dev. Biol. 1999;213:157–169. doi: 10.1006/dbio.1999.9370. doi:10.1006/dbio.1999.9370. [DOI] [PubMed] [Google Scholar]
- 15.Lowe L.A., Yamada S., Kuehn M.R. Genetic dissection of nodal function in patterning the mouse embryo. Development. 2001;128:1831–1843. doi: 10.1242/dev.128.10.1831. [DOI] [PubMed] [Google Scholar]
- 16.Andersson O., Reissmann E., Jornvall H., Ibanez C.F. Synergistic interaction between Gdf1 and Nodal during anterior axis development. Dev. Biol. 2006;293:370–381. doi: 10.1016/j.ydbio.2006.02.002. doi:10.1016/j.ydbio.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 17.Tanaka C., Sakuma R., Nakamura T., Hamada H., Saijoh Y. Long-range action of Nodal requires interaction with GDF1. Genes Dev. 2007;21:3272–3282. doi: 10.1101/gad.1623907. doi:10.1101/gad.1623907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Varlet I., Collignon J., Robertson E.J. Nodal expression in the primitive endoderm is required for specification of the anterior axis during mouse gastrulation. Development. 1997;124:1033–1044. doi: 10.1242/dev.124.5.1033. [DOI] [PubMed] [Google Scholar]
- 19.Hoodless P.A., Pye M., Chazaud C., Labbe E., Attisano L., Rossant J., Wrana J.L. FoxH1 (Fast) functions to specify the anterior primitive streak in the mouse. Genes Dev. 2001;15:1257–1271. doi: 10.1101/gad.881501. doi:10.1101/gad.881501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anderson R.M., Lawrence A.R., Stottmann R.W., Bachiller D., Klingensmith J. Chordin and noggin promote organizing centers of forebrain development in the mouse. Development. 2002;129:4975–4987. doi: 10.1242/dev.129.21.4975. [DOI] [PubMed] [Google Scholar]
- 21.Bachiller D., Klingensmith J., Kemp C., Belo J.A., Anderson R.M., May S.R., McMahon J.A., McMahon A.P., Harland R.M., Rossant J., et al. The organizer factors Chordin and Noggin are required for mouse forebrain development. Nature. 2000;403:658–661. doi: 10.1038/35001072. [DOI] [PubMed] [Google Scholar]
- 22.Yang Y.P., Klingensmith J. Roles of organizer factors and BMP antagonism in mammalian forebrain establishment. Dev. Biol. 2006;296:458–475. doi: 10.1016/j.ydbio.2006.06.014. doi:10.1016/j.ydbio.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 23.Klingensmith J., Matsui M., Yang Y.P., Anderson R.M. Roles of bone morphogenetic protein signaling and its antagonism in holoprosencephaly. Am. J. Med. Genet. C Semin. Med. Genet. 2010;154C:43–51. doi: 10.1002/ajmg.c.30256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klingensmith J., Ang S.L., Bachiller D., Rossant J. Neural induction and patterning in the mouse in the absence of the node and its derivatives. Dev. Biol. 1999;216:535–549. doi: 10.1006/dbio.1999.9525. doi:10.1006/dbio.1999.9525. [DOI] [PubMed] [Google Scholar]
- 25.Collignon J., Varlet I., Robertson E.J. Relationship between asymmetric nodal expression and the direction of embryonic turning. Nature. 1996;381:155–158. doi: 10.1038/381155a0. doi:10.1038/381155a0. [DOI] [PubMed] [Google Scholar]
- 26.Datto M.B., Frederick J.P., Pan L., Borton A.J., Zhuang Y., Wang X.F. Targeted disruption of Smad3 reveals an essential role in transforming growth factor beta-mediated signal transduction. Mol. Cell. Biol. 1999;19:2495–2504. doi: 10.1128/mcb.19.4.2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stottmann R.W., Berrong M., Matta K., Choi M., Klingensmith J. The BMP antagonist Noggin promotes cranial and spinal neurulation by distinct mechanisms. Dev. Biol. 2006;295:647–663. doi: 10.1016/j.ydbio.2006.03.051. doi:10.1016/j.ydbio.2006.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kimura C., Yoshinaga K., Tian E., Suzuki M., Aizawa S., Matsuo I. Visceral endoderm mediates forebrain development by suppressing posteriorizing signals. Dev. Biol. 2000;225:304–321. doi: 10.1006/dbio.2000.9835. doi:10.1006/dbio.2000.9835. [DOI] [PubMed] [Google Scholar]
- 29.Beddington R.S., Robertson E.J. Axis development and early asymmetry in mammals. Cell. 1999;96:195–209. doi: 10.1016/s0092-8674(00)80560-7. doi:10.1016/S0092-8674(00)80560-7. [DOI] [PubMed] [Google Scholar]
- 30.Ang S.-L., Rossant J. HNF-3[beta] is essential for node and notochord formation in mouse development. Cell. 1994;78:561. doi: 10.1016/0092-8674(94)90522-3. doi:10.1016/0092-8674(94)90522-3. [DOI] [PubMed] [Google Scholar]
- 31.Belo J.A., Leyns L., Yamada G., De Robertis E.M. The prechordal midline of the chondrocranium is defective in Goosecoid-1 mouse mutants. Mech. Dev. 1998;72:15–25. doi: 10.1016/s0925-4773(97)00204-9. doi:10.1016/S0925-4773(97)00204-9. [DOI] [PubMed] [Google Scholar]
- 32.Hallonet M., Kaestner K.H., Martin-Parras L., Sasaki H., Betz U.A., Ang S.L. Maintenance of the specification of the anterior definitive endoderm and forebrain depends on the axial mesendoderm: a study using HNF3beta/Foxa2 conditional mutants. Dev. Biol. 2002;243:20–33. doi: 10.1006/dbio.2001.0536. doi:10.1006/dbio.2001.0536. [DOI] [PubMed] [Google Scholar]
- 33.Chiang C., Litingtung Y., Lee E., Young K.E., Corden J.L., Westphal H., Beachy P.A. Cyclopia and defective axial patterning in mice lacking Sonic hedgehog gene function. Nature. 1996;383:407–413. doi: 10.1038/383407a0. doi:10.1038/383407a0. [DOI] [PubMed] [Google Scholar]
- 34.Lewis S.L., Tam P.P. Definitive endoderm of the mouse embryo: formation, cell fates, and morphogenetic function. Dev. Dyn. 2006;235:2315–2329. doi: 10.1002/dvdy.20846. doi:10.1002/dvdy.20846. [DOI] [PubMed] [Google Scholar]
- 35.Martinez Barbera J.P., Clements M., Thomas P., Rodriguez T., Meloy D., Kioussis D., Beddington R.S. The homeobox gene Hex is required in definitive endodermal tissues for normal forebrain, liver and thyroid formation. Development. 2000;127:2433–2445. doi: 10.1242/dev.127.11.2433. [DOI] [PubMed] [Google Scholar]
- 36.Mine N., Anderson R.M., Klingensmith J. BMP antagonism is required in both the node and lateral plate mesoderm for mammalian left-right axis establishment. Development. 2008;135:2425–2434. doi: 10.1242/dev.018986. doi:10.1242/dev.018986. [DOI] [PubMed] [Google Scholar]
- 37.Yamamoto M., Saijoh Y., Perea-Gomez A., Shawlot W., Behringer R.R., Ang S.L., Hamada H., Meno C. Nodal antagonists regulate formation of the anteroposterior axis of the mouse embryo. Nature. 2004;428:387–392. doi: 10.1038/nature02418. doi:10.1038/nature02418. [DOI] [PubMed] [Google Scholar]
- 38.Inman G.J., Nicolas F.J., Callahan J.F., Harling J.D., Gaster L.M., Reith A.D., Laping N.J., Hill C.S. SB-431542 is a potent and specific inhibitor of transforming growth factor-beta superfamily type I activin receptor-like kinase (ALK) receptors ALK4, ALK5 and ALK7. Mol. Pharmacol. 2002;62:65–74. doi: 10.1124/mol.62.1.65. doi:10.1124/mol.62.1.65. [DOI] [PubMed] [Google Scholar]
- 39.Ho D.M., Chan J., Bayliss P., Whitman M. Inhibitor-resistant type I receptors reveal specific requirements for TGF-beta signaling in vivo. Dev. Biol. 2006;295:730–742. doi: 10.1016/j.ydbio.2006.03.050. doi:10.1016/j.ydbio.2006.03.050. [DOI] [PubMed] [Google Scholar]
- 40.Chu J., Ding J., Jeays-Ward K., Price S.M., Placzek M., Shen M.M. Non-cell-autonomous role for Cripto in axial midline formation during vertebrate embryogenesis. Development. 2005;132:5539–5551. doi: 10.1242/dev.02157. doi:10.1242/dev.02157. [DOI] [PubMed] [Google Scholar]
- 41.Coucouvanis E., Martin G.R. BMP signaling plays a role in visceral endoderm differentiation and cavitation in the early mouse embryo. Development. 1999;126:535–546. doi: 10.1242/dev.126.3.535. [DOI] [PubMed] [Google Scholar]
- 42.Solloway M.J., Robertson E.J. Early embryonic lethality in Bmp5;Bmp7 double mutant mice suggests functional redundancy within the 60A subgroup. Development. 1999;126:1753–1768. doi: 10.1242/dev.126.8.1753. [DOI] [PubMed] [Google Scholar]
- 43.Yeo C., Whitman M. Nodal signals to Smads through Cripto-dependent and Cripto-independent mechanisms. Mol. Cell. 2001;7:949–957. doi: 10.1016/s1097-2765(01)00249-0. doi:10.1016/S1097-2765(01)00249-0. [DOI] [PubMed] [Google Scholar]
- 44.Constam D.B., Robertson E.J. Regulation of bone morphogenetic protein activity by pro domains and proprotein convertases. J. Cell Biol. 1999;144:139–149. doi: 10.1083/jcb.144.1.139. doi:10.1083/jcb.144.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brennan J., Lu C.C., Norris D.P., Rodriguez T.A., Beddington R.S., Robertson E.J. Nodal signalling in the epiblast patterns the early mouse embryo. Nature. 2001;411:965–969. doi: 10.1038/35082103. doi:10.1038/35082103. [DOI] [PubMed] [Google Scholar]
- 46.Brennan J., Norris D.P., Robertson E.J. Nodal activity in the node governs left–right asymmetry. Genes Dev. 2002;16:2339–2344. doi: 10.1101/gad.1016202. doi:10.1101/gad.1016202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wall N.A., Craig E.J., Labosky P.A., Kessler D.S. Mesendoderm induction and reversal of left–right pattern by mouse Gdf1, a Vg1-related gene. Dev. Biol. 2000;227:495–509. doi: 10.1006/dbio.2000.9926. doi:10.1006/dbio.2000.9926. [DOI] [PubMed] [Google Scholar]
- 48.Hansen C.S., Marion C.D., Steele K., George S., Smith W.C. Direct neural induction and selective inhibition of mesoderm and epidermis inducers by Xnr3. Development. 1997;124:483–492. doi: 10.1242/dev.124.2.483. [DOI] [PubMed] [Google Scholar]
- 49.Haramoto Y., Tanegashima K., Onuma Y., Takahashi S., Sekizaki H., Asashima M. Xenopus tropicalis nodal-related gene 3 regulates BMP signaling: an essential role for the pro-region. Dev. Biol. 2004;265:155–168. doi: 10.1016/j.ydbio.2003.09.015. doi:10.1016/j.ydbio.2003.09.015. [DOI] [PubMed] [Google Scholar]
- 50.Furtado M.B., Solloway M.J., Jones V.J., Costa M.W., Biben C., Wolstein O., Preis J.I., Sparrow D.B., Saga Y., Dunwoodie S.L., et al. BMP/SMAD1 signaling sets a threshold for the left/right pathway in lateral plate mesoderm and limits availability of SMAD4. Genes Dev. 2008;22:3037–3049. doi: 10.1101/gad.1682108. doi:10.1101/gad.1682108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yamamoto M., Beppu H., Takaoka K., Meno C., Li E., Miyazono K., Hamada H. Antagonism between Smad1 and Smad2 signaling determines the site of distal visceral endoderm formation in the mouse embryo. J. Cell Biol. 2009;184:323–334. doi: 10.1083/jcb.200808044. doi:10.1083/jcb.200808044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roessler E., Ouspenskaia M.V., Karkera J.D., Velez J.I., Kantipong A., Lacbawan F., Bowers P., Belmont J.W., Towbin J.A., Goldmuntz E., et al. Reduced NODAL signaling strength via mutation of several pathway members including FOXH1 is linked to human heart defects and holoprosencephaly. Am. J. Hum. Genet. 2008;83:18–29. doi: 10.1016/j.ajhg.2008.05.012. doi:10.1016/j.ajhg.2008.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stottmann R.W., Anderson R.M., Klingensmith J. The BMP antagonists Chordin and Noggin have essential but redundant roles in mouse mandibular outgrowth. Dev. Biol. 2001;240:457–473. doi: 10.1006/dbio.2001.0479. doi:10.1006/dbio.2001.0479. [DOI] [PubMed] [Google Scholar]
- 54.McMahon J.A., Takada S., Zimmerman L.B., Fan C.M., Harland R.M., McMahon A.P. Noggin-mediated antagonism of BMP signaling is required for growth and patterning of the neural tube and somite. Genes Dev. 1998;12:1438–1452. doi: 10.1101/gad.12.10.1438. doi:10.1101/gad.12.10.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rodriguez T.A., Casey E.S., Harland R.M., Smith J.C., Beddington R.S. Distinct enhancer elements control Hex expression during gastrulation and early organogenesis. Dev. Biol. 2001;234:304–316. doi: 10.1006/dbio.2001.0265. doi:10.1006/dbio.2001.0265. [DOI] [PubMed] [Google Scholar]
- 56.Downs K.M., Davies T. Staging of gastrulating mouse embryos by morphological landmarks in the dissecting microscope. Development. 1993;118:1255–1266. doi: 10.1242/dev.118.4.1255. [DOI] [PubMed] [Google Scholar]
- 57.Davis S., Miura S., Hill C., Mishina Y., Klingensmith J. BMP receptor IA is required in the mammalian embryo for endodermal morphogenesis and ectodermal patterning. Dev. Biol. 2004;270:47–63. doi: 10.1016/j.ydbio.2004.01.048. doi:10.1016/j.ydbio.2004.01.048. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.