Abstract
The insulin-like growth factor (IGF) pathway has been implicated in prostate development and carcinogenesis. We conducted a comprehensive analysis, utilizing a resequencing and tagging single-nucleotide polymorphism (SNP) approach, between common genetic variation in the IGF1, IGF binding protein (BP) 1, and IGFBP3 genes with IGF-I and IGFBP-3 blood levels, and prostate cancer (PCa) risk, among Caucasians in the NCI Breast and Prostate Cancer Cohort Consortium. We genotyped 14 IGF1 SNPs and 16 IGFBP1/IGFBP3 SNPs to capture common [minor allele frequency (MAF) ≥ 5%] variation among Caucasians. For each SNP, we assessed the geometric mean difference in IGF blood levels (N = 5684) across genotypes and the association with PCa risk (6012 PCa cases/6641 controls). We present two-sided statistical tests and correct for multiple comparisons. A non-synonymous IGFBP3 SNP in exon 1, rs2854746 (Gly32Ala), was associated with IGFBP-3 blood levels (Padj = 8.8 × 10−43) after adjusting for the previously established IGFBP3 promoter polymorphism A-202C (rs2854744); IGFBP-3 blood levels were 6.3% higher for each minor allele. For IGF1 SNP rs4764695, the risk estimates among heterozygotes was 1.01 (99% CI: 0.90–1.14) and 1.20 (99% CI: 1.06–1.37) for variant homozygotes with overall PCa risk. The corrected allelic P-value was 8.7 × 10−3. IGF-I levels were significantly associated with PCa risk (Ptrend = 0.02) with a 21% increase of PCa risk when compared with the highest quartile to the lowest quartile. We have identified SNPs significantly associated with IGFBP-3 blood levels, but none of these alter PCa risk; however, a novel IGF1 SNP, not associated with IGF-I blood levels, shows preliminary evidence for association with PCa risk among Caucasians.
INTRODUCTION
The role of the insulin-like growth factor (IGF) pathway has been studied extensively in both normal and transformed cells. Both in vivo (1–3) and in vitro (4–6) studies demonstrate that IGF-I binding to the IGF type 1 receptor modulates cellular proliferation, differentiation and apoptosis—important characteristics in tumorigenesis (7–11). Circulating levels of IGF-I derive mostly from the liver; more than 90% is complexed with IGF-binding protein 3 (IGFBP-3) and an acid-labile subunit thus reducing bioavailability (12,13). However, many types of tissues, including certain neoplasms (10), are capable of producing IGF-I locally. Although the main effect of IGFBP-3 is thought to be inhibition of cell growth and proliferation due to sequestration of the IGF-I ligand, recent research suggests that IGFBP-3 has antiproliferative and proapoptotic effects independent of IGF-I (14,15).
Elevated blood levels of IGF-I have been associated with several cancers, most commonly with prostate cancer (PCa), although later studies have found weaker associations than initially reported (16–23). A recent meta-analysis of 12 prospective studies reported a 38% increased risk of developing PCa when comparing the highest to lowest quartile of IGF-I levels (24). Although circulating IGFBP-3 levels were inversely associated with PCa risk in earlier studies, recent findings have been mostly null (23–27).
Nutrition remains a key determinant of circulating IGF-I levels (28,29), but heritability studies have estimated that the proportion of variance explained by inherited genetic variation ranges from 38 to >80% for IGF-I and IGFBP-3 blood levels, respectively (30–34). The specific genetic variants that contribute to heritable risk are not well defined. Results between an upstream IGF1 repeat sequence (CA)n and IGF-I blood levels have varied (35–38), and most studies, including a recently published meta-analysis (39), have reported a null association (40–44). In a case–control study, Johansson et al. (45) recently reported a marginally significant association (P = 0.02) between an IGF1 haplotype, previously reported associated with PCa risk, and IGF-I blood levels among controls in the Cancer Prostate in Sweden (CAPS) study (46); however, the authors were unable to replicate this haplotype–IGF-I association in a prospective study. In contrast, the significant decreased IGFBP-3 blood levels among carriers of the C allele of the IGFBP3 A-202C promoter polymorphism (rs2854744) has been observed across multiple studies of both men and women (39,41,43,47–54). Additionally, an in vitro transient transfection assay demonstrated that the C allele had 50% lower activity than the A allele (55).
Most previous genetic studies of the IGF1 locus with PCa risk have focused on an upstream IGF1 (CA)n repeat with equivocal results (56–60). Recently, two investigations comprehensively examined IGF1 genetic variation with PCa risk selecting single-nucleotide polymorphisms (SNPs) by public databases (i.e. HapMap), exonic resequencing or both. In the Multiethnic Cohort (MEC) study, Cheng et al. (61) identified an association with an upstream IGF1 SNP (rs7965399) and PCa, whereas in the CAPS study, Johansson et al. (45) used public databases to report a significant increased risk of PCa for single-copy carriers of an IGF1 haplotype spanning intron 2 through the 3′ UTR. The relationship between common genetic variation in the IGFBPs and PCa risk has only been thoroughly examined by Cheng et al. (62) in the MEC; no association was found between IGFBP1 and IGFBP3 polymorphisms with PCa risk.
We conducted a comprehensive haplotype tag-SNP analysis of the common genetic variation in IGF1, IGFBP1 and IGFBP3 in relation to IGF-I and IGFBP-3 blood levels and PCa risk among Caucasians in the NCI Breast and Prostate Cancer Cohort Consortium (BPC3), a pooled nested case–control study from seven cohorts (63). The large sample size of the BPC3 having 6012 prospective PCa cases and 6641 controls allows us to detect modest genetic effects and to assess effect modification. In addition, we are able to examine risk in clinically important subgroups of PCa defined by stage and Gleason score at diagnosis.
RESULTS
Characteristics of the studies within the BPC3 are presented in Table 1. Briefly, the cases and controls were comparable across cohorts with respect to demographic and other potentially PCa-related factors, with the exception of height where the ATBC Study (Finnish population) and EPIC cohorts (eight European countries) are shorter in stature. Family history was not available for the EPIC and PHS cohorts. PCa clinical information such as stage (63%) and Gleason score (56%) was available for more than half of the cases.
Table 1.
Characteristics of Caucasians in the Breast and Prostate Cancer Cohort Consortium (BPC3) by cohort and overall
| CPS-II |
ATBC |
EPIC |
HPFS |
MEC |
PHS |
PLCO |
Overall |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases | Controls | Cases | Controls | Cases | Controls | Cases | Cases | Cases | Controls | Cases | Controls | Cases | Controls | Cases | Controls | |
| Number | 1162 | 1163 | 987 | 965 | 733 | 1116 | 659 | 654 | 457 | 452 | 843 | 987 | 1171 | 1304 | 6012 | 6641 |
| Age at diagnosis (mean, year) | 70 | 70 | 70 | 69 | 64 | 65 | 69 | 69 | 68 | 66 | 70 | 70 | 67 | 67 | 68 | 68 |
| BMI (mean, kg/m2) | 26 | 26 | 26 | 26 | 27 | 27 | 25 | 26 | 26 | 27 | 25 | 25 | 27 | 27 | 26 | 26 |
| Height (mean, cm) | 178 | 179 | 174 | 174 | 173 | 173 | 178 | 178 | 177 | 177 | 179 | 178 | 178 | 178 | 177 | 177 |
| Family history availablea (n) | 1162 | 1163 | 865 | 852 | 0 | 0 | 659 | 654 | 432 | 431 | 0 | 0 | 1171 | 1304 | 4289 | 4404 |
| Family history (% yes) | 21 | 11 | 6 | 3 | n/a | n/a | 20 | 15 | 13 | 8 | n/a | n/a | 11 | 7 | 15 | 9 |
| For cases | ||||||||||||||||
| Years of diagnoses (range) | 1992–2002 | 1986–2003 | 1991–2003 | 1994–2000 | 1995–2002 | 1982–2000 | 1994–2001 | 1982–2003 | ||||||||
| Stage info available (n) | 1129 | 596 | 405 | 570 | 437 | 668 | 0 | 3805 | ||||||||
| Stage (%≥C) | 11 | 31 | 17 | 15 | 14 | 31 | n/a | 19 | ||||||||
| Gleason score available | 999 | 583 | 97 | 583 | 445 | 666 | 0 | 3373 | ||||||||
| Gleason score (%≥8) | 11 | 26 | 16 | 9 | 26 | 13 | n/a | 16 | ||||||||
aFamily history: at least one first-degree relative or two second-degree relatives diagnosed with prostate cancer.
The IGF1 locus was characterized by four haplotype blocks (Fig. 1, bottom panel). The IGFBP1 and IGFBP3 loci are near each other, separated by 19-kb, and were characterized by three haplotype blocks defined by 12 htSNPs (Fig. 2, bottom panel). We included four additional IGFBP3 SNPs. The average genotyping success rate across cohorts was 94.7% (ranging from 84.4 to 99.8%). No deviation from Hardy–Weinberg equilibrium was observed for any SNP (at the P < 0.01 level) among controls across more than one study. The controls minor allele frequencies were similar across the cohorts, albeit the ATBC (Finnish) cohort differed slightly for a few markers (IGF1: Supplementary Material, Table S1; IGFBP1 and IGFBP3: Supplementary Material, Table S2).
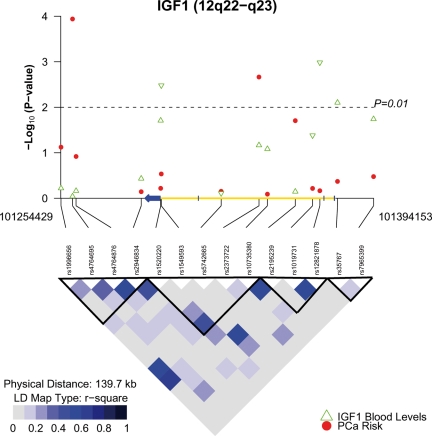
Figure 1.
IGF1 LD and SNP associations with IGF-I blood levels and prostate cancer risk among Caucasians in the BPC3. The upper panel is a plot of the −log10 P-values for IGF1 htSNP associations with IGF-I blood levels (green triangles; N = 5684) and prostate cancer risk (red circles; 6012 PCa cases/6641 controls). The direction of the green triangles indicates the direction of effect between the minor allele and IGF-I blood levels. The IGF1 locus is illustrated on the x-axis with the exons colored blue and the introns yellow. The arrow indicates the 5′ → 3′ direction. The P-values are plotted according to genetic position. The bottom panel contains an LD plot for the 14 IGF1 htSNPs among PLCO Caucasian controls. The r2 values are used for LD. The black triangles indicate the haplotype blocks. Corrected P-values for marginally associated SNPs with prostate cancer risk: rs2373722 = 0.14; rs4764695 = 8.7 × 10−3.
Figure 2.
IGFBP1 and IGFBP3 LD and SNP associations with IGFBP-3 blood levels and prostate cancer risk among Caucasians in the BPC3. The upper panel is a plot of the −log10 P-values for IGFBP1 and IGFBP3 SNP associations with IGFBP-3 blood levels (green triangles; N = 5667) and prostate cancer risk (red circles; 6012 PCa cases/6641 controls). The direction of the green triangles indicates the direction of effect between the minor allele and IGFBP-3 blood levels. The IGFBP1 and IGFBP3 loci are illustrated in the x-axis with the exons colored blue and the introns yellow. The arrow indicates the 5′ → 3′ direction. The P-values are plotted according to genetic position. The bottom panel contains an LD plot for the 16 IGFBP1 and IGFBP3 SNPs among PLCO Caucasian controls. The r2 values are used for LD. The black triangles indicate the haplotype blocks. Corrected P-values for blood level associations: rs2270628 = 8.6 × 10−3; rs3110697 = 1.1 × 10−16; rs2854746 = 8.8 × 10−43; rs2854744 = 8.1 × 10−34; rs2132570 = 1.8 × 10−4; rs2960436 = 4.4 × 10−34.
IGF1 genetic variation and IGF-I blood levels
Individual cohort and pooled values of log-transformed IGF-I blood levels were similar as were the case and control levels within each cohort (Supplementary Material, Fig. S1). In a pooled analysis, 3 of 14 htSNPs were nominally associated with IGF-I blood levels (Fig. 1, green triangles): rs35767 (Block 1; Puncorr = 7.9 × 10−3), rs12821878 (Block 2; Puncorr = 1.0 × 10−3) and rs1549593 (Block 3; Puncorr = 3.3 × 10−3). As shown in Figure 1, the pair-wise linkage disequilibrium (LD) for these three markers is negligible except between rs12821878 and rs1549593 (r2 = 0.27 among PLCO controls). The geometric means and 95% CIs by SNP and haplotypes are presented in Supplementary Material, Tables S3 and S4. After correcting for multiple comparisons, none of the IGF1 marker's associations remained statistically significant (Pcorr > 0.07) with IGF-I blood levels.
IGFBP1/IGFBP3 genetic variation and IGFBP-3 blood levels
The distributions of IGFBP-3 blood levels were not as uniform as IGF-I blood levels across the cohorts (Supplementary Material, Fig. S2). Within each cohort, case and control blood levels of IGFBP-3 were very similar. The geometric means and 95% CIs by SNP and haplotypes are presented in Supplementary Material, Tables S5 and S6.
Among the 12 htSNPs and 4 candidate SNPs in IGFBP1 and IGFBP3, six were significantly associated with blood levels before and after adjusting for multiple comparisons (Fig. 2, green triangles). For the most strongly associated htSNP, rs2854746 (Pcorr = 8.8 × 10−43), the mean IGFBP-3 blood level was 3046 ng/ml for wild-type homozygotes, 3263 ng/ml for heterozygotes and 3442 ng/ml for variant homozygotes. The extensively studied IGFBP3 promoter polymorphism rs2854744 (A-202C) was also strongly associated with IGFBP-3 blood levels in the univariate analysis (Pcorr = 8.1 × 10−34). However, after simultaneously including all six significant IGFBP3 SNPs in a multi-SNP linear regression, only rs2854746 remained statistically significant overall (Puncorr = 1.4 × 10−10) and among controls (Puncorr = 4.9 × 10−8), while rs2854744 (A-202C) was no longer associated with IGFBP-3 blood levels (P = 0.91). Stratified analysis of the IGFBP3 htSNPs rs2854746 and rs2854744 (A-202C) showed a consistent result such that across the three genotypes for rs2854746, the mean IGFBP-3 blood levels remained unchanged by rs2854744 (A-202C) genotypes (Table 2). However, the differences in mean IGFBP-3 blood levels were statistically significant across rs2854746 in the heterozygote (CA) and variant homozygote (AA) strata defined by rs2854744 (A-202C) genotypes (PCA = 2.7 × 10−6; PAA = 4.1 × 10−6).
Table 2.
Stratified associations between IGFBP3 SNPs and IGFBP-3 blood levels
| rs2854746 |
P* | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| GG |
GC |
CC |
||||||||
| N | Mean (95% CI)a | N | Mean (95% CI)a | N | Mean (95% CI)a | N | Mean (95% CI)a | |||
| Total | 1918 | 3046 (2975–3119) | 2597 | 3263 (3188–3339) | 875 | 3442 (3353–3533) | 5.48 × 10−44 | |||
| rs2854744 | CC | 1612 | 3031 (2960–3104) | 1573 | 3039 (2935–3146) | 4 | 3304 (2643–4132) | 0 | n/a | 0.32 |
| CA | 2732 | 3231 (3157–3306) | 301 | 3049 (2923–3181) | 2343 | 3251 (3136–3370) | 11 | 3456 (3005–3974) | 2.74 × 10−6 | |
| AA | 1158 | 3371 (3288–3456) | 26 | 3081 (2809–3379) | 222 | 3170 (3029–3317) | 858 | 3479 (3351–3613) | 4.10 × 10−6 | |
| P* | 5.08 × 10−35 | 0.93 | 0.28 | 0.92 | ||||||
aMean IGFBP-3 blood levels (nanograms per microliter) and 95% confidence intervals.
*SNP stratified P-value from linear regression adjusting for age at blood draw, assay batch, prostate cancer, cohort and country.
IGF1 genetic variation and prostate cancer risk
The associations between the 14 IGF1 htSNPs and overall PCa risk among Caucasians are presented in the upper panel of Figure 1 (red circles). The tests for heterogeneity (Supplementary Material, Table S7) across cohorts were not statistically significant, thus we present the pooled results for the main effect analyses. We found nominally significant associations between IGF1 SNPs rs2373722 (Puncorr = 2.0 × 10−3) and rs4764695 (Puncorr = 1.2 × 10−4) with overall PCa risk (Supplementary Material, Table S8). After controlling for multiple comparisons, the corrected P-value for rs2373722 was no longer significant (Pcorr = 0.14), whereas rs4764695 remained statistically significant (Pcorr = 8.7 × 10−3). For rs4764695, the overall risk estimate was 1.01 (99% CI: 0.90–1.14) for heterozygotes and 1.20 (99% CI: 1.06–1.37) for variant homozygotes, with consistent point estimates for heterozygotes and variant homozygotes across the cohorts (Fig. 3). The results remained unchanged when previously published data from the MEC were excluded from the analysis (Puncorr = 2.8 × 10−4; Supplementary Material, Table S8). IGF1 marker rs4764695 was not significantly associated with IGF-I blood levels. Testing for effect modification by several variables of interest [family history of PCa, age at diagnosis, body mass index (BMI) and height] revealed no statistically significant heterogeneity in any of the subgroup analyses for the 14 htSNPs (data not shown).
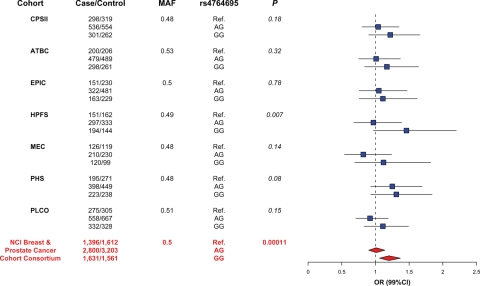
Figure 3.
Forest plot of risk estimates for rs4764695 with prostate cancer among Caucasians in the seven cohorts and combined. The left side list of the figure lists the cohort member, the case–control genotype distribution and the MAF. The blue squares and connected lines represent the cohort-specific genotype risk estimates and the 99% CI. The genotype summary risk estimates and 99% CIs are represented by the red diamonds. Corrected P-value = 8.7 × 10−3.
We examined the risk association of each SNP stratified by stage at diagnosis (high stage: C or D; low stage: A or B) and Gleason score at diagnosis (high grade: ≥8; low grade: <8) when compared with controls (data not shown). Among the 14 IGF1 SNPs, rs4764695 was the only marker remaining statistically significant (Puncorr < 0.01) across all of the strata except for stage (Table 3). The risk estimate was slightly greater for high-grade cancer (Gleason score ≥8, OR = 1.43) than low-grade cancer (Gleason score 2–7, OR = 1.19) for variant homozygotes, but the test of heterogeneity was only marginally statistically significant (P = 0.065).
Table 3.
Risk estimates for IGF1 SNP rs4764695 and prostate cancer aggressiveness among Caucasians in the NCI Breast and Prostate Cancer Cohort Consortium. Corrected P-value for overall PCa risk = 8.7 × 10−3
| Genotype | Case (%) | Control (%) | ORa (99% CIb) | Pc |
|---|---|---|---|---|
| AA | 1396 (24.0) | 1612 (25.3) | Ref. | 0.00011 |
| AG | 2800 (48.1) | 3203 (50.2) | 1.01 (0.9–1.14) | |
| GG | 1631 (28.0) | 1561 (24.5) | 1.20 (1.06–1.37) | |
| High grade (≥8) | ||||
| AA | 123 (23.6) | 1612 (25.3) | Ref. | 0.001 |
| AG | 234 (44.9) | 3203 (50.2) | 0.97 (0.71–1.31) | |
| GG | 164 (31.5) | 1561 (24.5) | 1.43 (1.02–1.99) | |
| Low grade (<8) | ||||
| AA | 697 (25.0) | 1612 (25.3) | Ref. | 0.014 |
| AG | 1331 (47.8) | 3203 (50.2) | 1.02 (0.87–1.19) | |
| GG | 756 (27.2) | 1561 (24.5) | 1.19 (1.00–1.42) | |
| High stage (C or D) | ||||
| AA | 172 (24.1) | 1612 (25.3) | Ref. | 0.107 |
| AG | 338 (47.4) | 3203 (50.2) | 1.01 (0.78–1.31) | |
| GG | 203 (28.5) | 1561 (24.5) | 1.22 (0.91–1.64) | |
| Low stage (A or B) | ||||
| AA | 736 (24.4) | 1612 (25.3) | Ref. | 0.002 |
| AG | 1449 (48.0) | 3203 (50.2) | 1.05 (0.90–1.22) | |
| GG | 834 (27.6) | 1561 (24.5) | 1.24 (1.04–1.47) | |
aConditional logistic regression models adjusted for age in 5-year intervals, cohort, and country.
b99% confidence intervals.
cP-value for association from 2 df LRT.
The cohort-specific haplotype frequencies for IGF1 among Caucasians in the MEC panel and other six cohorts are shown in Supplementary Material, Table S9, and the pooled and cohort-specific associations of these haplotypes and overall PCa risk are presented in Supplementary Material, Table S10. The tests for heterogeneity by cohort across the haplotype blocks were not statistically significant. We observed statistically significant global P-values for IGF1 block 3 (Puncorr = 0.002) and block 4 (Puncorr = 0.004). Since SNP rs2373722 resides within block 3 and rs4764695 resides within block 4, we removed both SNPs from their respective blocks and found that neither block remained statistically significant with PCa risk (data not shown), suggesting that the haplotype analysis did not provide any additional insight beyond the SNP analysis. None of the haplotype analyses were significant for effect modification by family history, age at diagnosis, BMI, height or PCa diagnostic variables (data not shown).
IGFBP1 and IGFBP3 genetic variation and prostate cancer risk
Figure 2 shows the results for overall PCa risk for the 12 htSNPs and 4 additional IGFBP1 and IGFBP3 SNPs (red circles). None of the 16 SNPs were nominally associated with PCa risk at the Puncorr < 0.01 level. The IGFBP3 markers significantly associated with blood levels were not associated with PCa risk. The cohort-specific results are presented in Supplementary Material, Table S11, and the IGFBP1 and IGFBP3 SNP analyses with and without the previously reported MEC samples are available in Supplementary Material, Table S12. None of the tests for heterogeneity (by cohort) or effect modification (family history, age at diagnosis, BMI and height in tertiles and quartiles) were statistically significant in any of the subgroup analyses (data not shown).
The haplotype frequencies within the MEC panel and by cohort are presented in Supplementary Material, Table S13. Similar to the IGFBP1 and IGFBP3 SNP analyses, neither the global tests for haplotype blocks nor the test for any individual haplotype were statistically significant (Supplementary Material, Table S14) in the pooled or cohort-specific analyses.
DISCUSSION
Substantial epidemiologic and experimental evidence exist implicating the IGF pathway in prostate carcinogenesis. Although many studies have demonstrated an increased risk of PCa, especially advanced disease among men with high IGF-I blood levels (16–18,27), comprehensive genetic analyses of the IGF pathway with PCa risk are limited. In this large consortium (n = 6012 PCa cases) from seven prospective studies, we conducted a thorough analysis of common genetic variation of three primary loci in the IGF pathway (IGF1, IGFBP1 and IGFBP3) and observed among Caucasians an association between an IGF1 SNP (rs4764695, MAF = 0.49, ∼34-kb downstream of IGF1 exon 4) and PCa risk following multiple testing corrections. Men carrying the homozygous variant had a 20% higher risk of developing PCa (OR = 1.20; 99% CI: 1.06–1.37; Padj = 8.69 × 10−3) compared to those with the wild-type controlling for age in 5-year intervals, cohort and country of residence for EPIC.
The IGF1 htSNPs presented here are a subset of the IGF1 htSNPs reported in a study by the MEC. The MEC authors did not find a significant association between PCa risk and rs4764695 (P > 0.30) but they had limited power among whites (23%) for this marker (61). They reported a marginally significant association between PCa and heterozygotes for an IGF1 htSNP (rs7965399) located in the 5′ region (ORhet = 1.25; 95% CI 1.09–1.43), whereas we observed a null association between rs7965399 and PCa with (P = 0.334) and without (P = 0.689) the MEC Caucasian participants. The majority of the MEC-nested PCa case–control study, however, is non-Caucasian (African American = 28%, Latino = 28%, Japanese = 20%, native Hawaiian = 3%) and the different risk associations among overlapping IGF1 htSNPs suggest either racial/ethnic differences within this region, the causative SNP remains unknown or false-positive findings. The IGF1 htSNP results for African Americans presented by Cheng et al. remained unchanged when we included an additional 105 African American PCa cases from PLCO (data not shown).
The Swedish CAPS case–control study (N = 2863 PCa cases, all Caucasian) also evaluated IGF1 genetic variation with PCa by genotyping SNPs identified in the HapMap Phase I data (45). A marginally significant association was reported for carriers of one copy of a haplotype spanning from intron 2 to the 3′ UTR of IGF1 with PCa risk (P = 0.02 adjusting for multiple testing) corresponding to a similar region covered by our IGF1 haplotype block 3. Although the three SNPs in the CAPS haplotype (rs2033178, rs7136446 and rs6220) were not included in our IGF1 htSNP panel, our IGF1 htSNP rs2373722 is in perfect LD (r2 = 1.00 in HapMap CEPH) with the CAPS SNP rs2033178 (45). Our block 3 haplotype association was mainly driven by the htSNP rs2373722 which was null after correcting for multiple testing (Padj = 0.14). Neither the SNP rs474695 identified in our study nor any equivalent proxy was included in the CAPS study.
Although IGF1 SNP rs4764695 was significantly associated with prostate cancer risk and the main effect was slightly greater for high tumor grade (high-grade OR = 1.43; low-grade OR = 1.20; Phet = 0.065), the association with IGF-I blood levels was null. Among the six BPC3 studies with IGF blood levels in this report, elevated IGF-I levels were significantly associated with higher PCa risk (Ptrend = 0.02); the comparison between the highest IGF-I quartile with the lowest quartile yielded a 21% increased risk of PCa (P = 0.02). In contrast, the association between IGFBP-3 blood levels and PCa was null (Ptrend = 0.89). The IGF-I finding in the BPC3 is consistent with results of a recent meta-analysis of 42 studies (OR = 1.21; 95% CI 1.07–1.36) (64). In the same meta-analysis, data from 29 studies showed a significant inverse association for IGFBP-3 (OR 0.88; 95% CI 0.79–0.98) with substantial heterogeneity; the inverse association of IGFBP-3 with PCa risk was seen in retrospective studies, but not prospective studies (64).
Several reasons may explain the lack of association between rs4764695 and IGF-I blood levels. First, the IGF-I measurements reflect systemic levels measured at a single time point prior to PCa diagnosis rather than tissue-specific levels. Free IGF-I blood levels, unavailable across these studies, may be a more biologically relevant measure and impacted by rs4764695. Furthermore, the complexity of the IGF pathway is likely not entirely captured in these simple associations, and a more complete pathway analysis is warranted. As the rs4764695 marker lies ∼34 kb downstream of IGF1, this marker may be involved in another pathway entirely. In addition, measurement error for both the genotypes and IGF-I blood levels would lead to non-differential misclassification and a bias towards the null. Lastly, the result for rs4764695 may be spurious, although we have taken steps by setting a stringent alpha level (0.01) and correcting for multiple comparisons.
We identified several IGFBP3 SNPs strongly associated with IGFBP-3 blood levels among Caucasians but not associated with PCa risk. Given a considerable amount of LD exists within the 5′ region (Fig. 2) of IGFBP3, we tested the independent effect of six correlated SNPs using a multi-marker model and determined that the most significant SNP was rs2854746, a non-synonymous polymorphism in exon 1 (Gly32Ala). This is in contrast to the promoter polymorphism rs2854744 (A-202C) that has been extensively reported to be associated with IGFBP-3 levels (39,41,43,47–54). This observation has been alluded to in two previous reports (50,53), but could not be substantiated due to the limited sample sizes and strong LD between these two markers among Caucasians (r2 = 0.85 among Caucasian PLCO controls). Although each minor allele was associated with 6.3% higher IGFBP-3 blood levels on average, rs2854746 explains only 3.6% of the variation. The lack of an association between rs2854746 and PCa risk, whereas a strong influence on IGFBP3 blood levels exists, supports a Mendelian randomization argument for no etiological effect of IGFBP-3 on incident risk of PCa (65,66). However, caution is needed because confounding or pleiotropic effects would negate this argument (67).
The major strength of this study is the utilization of a large cohort consortium and a comprehensive approach to examine the genetic variation across three genes in the IGF pathway, a strong candidate in prostate carcinogenesis. Specifically, we have the ability to look at the effects of SNPs on pre-diagnostic blood levels as well as risk in the same set of subjects. Although our data in Caucasians limits the generalizability to other ethnic/racial groups, our large sample size allows us to present the overall risk estimates using a 99% CI, reducing the chance of both false-positive and -negative results. In addition, we reduced the probability of a spurious association due to multiple hypothesis testing by applying a correction across all models, traits and genetic markers. The IGF pathway is a complex system and we have limited our study to three primary genes. Additional IGF pathway genes need to be investigated and a more comprehensive pathway analysis would be necessary.
While several genome-wide association scans (GWASs) have recently identified multiple susceptibility loci for PCa risk (68–76), only recently has an IGF variant (IGF2-rs7127900) been among them. The Cancer Genetic Markers of Susceptibility (CGEMS) GWAS is the only publicly available database to compare our IGF1 rs4764695 finding. Although rs4764695 was not present on the CGEMS platform, a proxy rs1980236 showed a similar effect, although not statistically significant (P = 0.57). However, the PLCO study was the first stage of the CGEMS GWAS and more than 70% of this sample is included in this analysis where the observed effect for rs4764695 was 1.11 (P = 0.15). The initial stages of the GWAS studies include less than 2000 PCa cases and have limited power, 67%, to detect an OR of 1.20 for an MAF of 0.50. In contrast, our large consortium study allows for focused and comprehensive evaluation of candidate genes among more than 6000 PCa cases, having the necessary power, 99%, to identify an OR of 1.20 for an MAF of 0.50. This is evidenced by mulitple GWAS studies identifying common as well as different risk loci. Currently, only 6% of PCa genetic variation is explained by the known loci identified in GWAS studies (77). The missing heritability, aka ‘dark matter’, may reside undetected in the ‘lower Manhattan’ plots and represented by multiple variants (78,79). For example, CGEMS replicated several variants, JAZF1 (GWAS rank 24 407; P = 0.04) and MSMB (GWAS rank 24 223; P = 0.04), with little evidence from the first stage. Furthermore, both in vivo (1–3) and in vitro (4–6) studies have demonstrated that the IGF pathway plays a role in tumorigenesis, thus making it a strong biological candidate.
In conclusion, a significant association between prostate cancer risk and an IGF1 SNP, rs4764695, was identified among Caucasians. Although this is a novel finding, the evidence is still preliminary and further confirmation is needed. The estimated population-attributable risk for homozygotic variant carriers is ∼5% due to the high frequency of the minor allele (G = 49%). This variant could be of greater importance due to the potential for a stronger association with high-tumor-grade PCa. The association between rs4764695 and cancer is exclusive with prostate cancer as the association with breast cancer risk and this marker has been reported as null in the NCI BPC3 study (54). Furthermore, we provide strong evidence for a novel association between IGFBP-3 blood levels and a non-synonymous IGFBP3 marker in contrast to the previously reported IGFBP3 promoter polymorphism. Additional studies, such as fine mapping to determine the causal variant in IGF1 and the examination of additional genes in the IGF axis, are needed. In summary, preliminary evidence implicates common genetic variation in the IGF1 locus with PCa risk.
MATERIALS AND METHODS
Study population
The BPC3 and member cohorts have been described in detail elsewhere (80). In brief, the consortium combines resources from seven well-established cohort studies: the American Cancer Society Cancer Prevention Study II (CPS-II) (81), the Alpha-Tocopherol, Beta-Carotene Cancer Prevention (ATBC) Study (82), the European Prospective Investigation into Cancer and Nutrition Cohort (EPIC—comprised of cohorts from Denmark, Great Britain, Germany, Greece, Italy, the Netherlands, Spain, and Sweden) (83), the Health Professionals Follow-up Study (HPFS) (84), the Hawaii/Los Angeles MEC (85), the Physicians' Health Study (PHS) (16), and the Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial (PLCO) (86). These cohorts collectively include over 248 000 men with a blood sample.
The current study was restricted to individuals who self-reported as Caucasian and consists of 6012 prospective PCa cases and 6641 controls. Cases from other ethnic groups were contributed mostly from the MEC and had been reported on previously (53,61,62); we analyzed the data for Caucasians with and without the contribution from the MEC (457 cases and 452 controls) to assess the impact on the overall results. Prospective PCa cases were identified through population-based cancer registries or self-reports confirmed by medical records, including pathology reports. The BPC3 data for PCa consists of a series of matched nested case–control studies within each cohort; controls were matched to cases on a number of potential confounding factors, including age, country and region of recruitment. For the current analysis, PCa cases were matched to available controls by age in 5-year intervals, cohort and country of residence for EPIC. A written informed consent was obtained from all subjects, and each study was approved by the Institutional Review Boards at their respective institutions.
IGF-I and IGFBP-3 blood levels
Pre-diagnostic measurements of IGF-I and IGFBP-3 were available for six of the seven BPC3 cohort members (ATBC, EPIC, HPFS, MEC, PHS and PLCO; IGF-I: N = 6076; IGFBP-3: N = 6059) (16,19–23,53). Most blood samples in the CPS-II were collected post-diagnosis and therefore were only included in the genotyping analyses. Samples from three of the studies (ATBC, HPFS and PHS) were measured in the Pollak laboratory and the remaining three studies (EPIC, MEC and PLCO) were measured in the laboratory of the Hormones and Cancer Team at IARC; all used enzyme-linked immunosorbent assays (Diagnostic System Laboratories, Webster, TX, USA).
SNP discovery and htSNP selection
As previously described by Cheng et al. (61,62), a multi-stage approach was used to characterize genetic variation across IGF1, IGFBP1 and IGFBP3 loci. Most of the exons across the three genes were resequenced in 95 advanced PCa cases, and a multiethnic panel of 349 controls was utilized to determine the patterns of LD encompassing ∼20 kilobases (kb) upstream and ∼10 kb downstream of each gene. Haplotype-tagging SNPs (htSNPs) for each haplotype block, determined by the confidence interval method of Gabriel et al. (87,88), were chosen based on 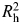 , a measure of the correlation between observed and predicted haplotypes based on the htSNP genotypes (89), to select a minimum set of SNPs that would achieve an
, a measure of the correlation between observed and predicted haplotypes based on the htSNP genotypes (89), to select a minimum set of SNPs that would achieve an 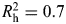 for all common haplotypes with an estimated frequency of ≥5% among Caucasians.
for all common haplotypes with an estimated frequency of ≥5% among Caucasians.
For genetic characterization of IGF1 (chromosome 12q22-q23), 154 SNPs were evaluated over a 156-kb region (one SNP for every 2.4 kb) in a multiethnic panel of subjects with no history of cancer (61). After removing markers that were monomorphic or had poor genotyping results, a panel of 64 SNPs remained, from which 14 htSNPs were selected to predict the common haplotypes among Caucasians (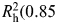 ). Of the 14 htSNPs, 11 SNPs are available in HapMap Phase II CEPH samples and capture 60% of the common genetic variation (MAF > 5%) of IGF1 with an r2 > 0.70.
). Of the 14 htSNPs, 11 SNPs are available in HapMap Phase II CEPH samples and capture 60% of the common genetic variation (MAF > 5%) of IGF1 with an r2 > 0.70.
For genetic characterization of IGFBP1 and IGFBP3 (which are located contiguously in a 35-kb region on chromosome 7p13-p12), 56 SNPs over a 71-kb region were evaluated (one SNP for every 2 kb) in the multiethnic panel (61,62). Twenty markers were removed due to being monomorphic or having poor genotyping results. The final selection included 12 htSNPs to predict the common haplotypes among Caucasians (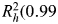 ), 2 genic SNPs in IGFBP3 not part of a haplotype block (rs6670, rs2453839) and 2 additional IGFBP3 SNPs (rs2132570, rs2960436). Ten of these 16 SNPs are available in HapMap Phase II CEPH samples and capture 41% of the common genetic variation (MAF > 5%) of IGFBP1 and IGFBP3 with an r2 > 0.70.
), 2 genic SNPs in IGFBP3 not part of a haplotype block (rs6670, rs2453839) and 2 additional IGFBP3 SNPs (rs2132570, rs2960436). Ten of these 16 SNPs are available in HapMap Phase II CEPH samples and capture 41% of the common genetic variation (MAF > 5%) of IGFBP1 and IGFBP3 with an r2 > 0.70.
Genotyping
Genotyping was conducted by five laboratories (University of Southern California, Los Angeles, CA, USA; University of Hawaii, Honolulu, HI, USA; Harvard School of Public Health, Boston, MA, USA; Core Genotyping Facility, National Cancer Institute, Bethesda, MD, USA; and Cambridge University, Cambridge, UK) using a fluorescent 5′ endonuclease assay and the ABI-PRISM 7900 for sequence detection (TaqMan; Applied Biosystems, Inc.). Assay information is available at the MEC Genetics Web site (http://uscnorris.com/mecgenetics/CohortGCKView.aspx). For each assay, the concordance rate was 100% for 102 samples from the SNP500 Cancer project (http://snp500cancer.nci.nih.gov) (90) and inter-laboratory completion and concordance rates were greater than 99%, based on cross-laboratory assessment of 30 SNPs on 94 samples from the Coriell Biorepository (Camden, NJ, USA). The internal quality of genotype data at each genotyping center was assessed by typing 5–10% blinded samples in duplicate or greater (depending on study).
Statistical analysis
All statistical tests presented are two-sided and were conducted in SAS 9.0 (SAS Institute). Figures and multiple testing corrections were generated in the statistical program R (http://cran.r-project.org/). To account for multiple hypothesis testing, we applied the method implemented in PACT, a flexible and efficient approach that adjusts for correlation between multiple traits, genetic markers and models (91). PACT utilizes less computational time while maintaining the accuracy of permutation or simulation-based tests. As multiple models and traits can be considered, the P-value corrections were computed simultaneously for blood levels and PCa risk across all 30 IGF1 and IGFBP1/IGFBP3 markers. A test of significance was set at the 0.01 level to minimize the chance of both false-positive and -negative results (92,93). We present the uncorrected P-values (Puncorr) for association in the figures and tables. A corrected P-value (Pcorr) is presented for significant SNPs correcting for multiple comparisons across all traits, genetic markers and statistical models among the 30 IGF genetic markers analyzed.
Genetic determinants of blood levels
We identified cohort- and assay batch-specific statistical outliers based on the generalized extreme studentized deviate many-outlier detection approach (94) setting alpha to 0.05 for both IGF-I and IGFBP-3 blood levels. Based on this, we excluded 23 IGF-I samples (n = 14 cases and 9 controls) and 43 IGFBP-3 samples (n = 19 cases and 24 controls). The IGF-I and IGFBP-3 blood levels were log-transformed to provide approximate normal distributions. The geometric mean and 95% CI according to haplotypes or SNPs were calculated using linear regression analysis adjusting for age at blood draw, assay batch, cohort (including country for EPIC) and case–control status. Haplotype frequencies and subject-specific expected haplotype indicators were calculated among cases and controls combined by cohort (and country within EPIC) and then combined for the haplotype analyses. A global haplotype test was performed for each haplotype block by using an F-test to compare the sum of the squared residuals for a full model (all haplotypes within a block) and a nested model (without haplotypes within a block). A multi-SNP model was utilized by including all statistically significant SNPs identified from the univariate analysis to assess independent SNP effects within a gene.
Genetic determinants of prostate cancer
The statistical methods used have been described previously (92,95). In brief, we used conditional logistic regression to estimate ORs and 99% CI for disease associated with genetic markers (SNP or haplotype). The matching factors in the conditional logistic regression were age (in 5-year intervals), cohort and country within EPIC. We estimated the genotypic ORs for disease by using the most common genotype as the referent group for the SNP analyses. We estimated haplotype-specific ORs using an expectation-substitution approach to account for haplotype uncertainty given unphased genotype data (96,97). To test the global null hypothesis of no association between IGF genetic variation and risk of PCa, we used a likelihood ratio test (LRT) comparing a model with additive effects for each common haplotype (treating the most common haplotype as the referent) to the intercept-only model. We considered haplotypes with greater than 5% frequency in at least one cohort to be ‘common’. All other haplotypes were excluded.
When assessing genetic effects (SNPs or haplotype), we tested for heterogeneity across cohort and several potential effect modifiers. We used an LRT by including an interaction term between the genetic effect (SNP or haplotype) and variable of interest in comparison to the null model. The tests for heterogeneity by cohort were not statistically significant (P > 0.01), and therefore we present the pooled results. We assessed for the presence of effect modification by family history (at least one first-degree relative or two second-degree relatives diagnosed with PCa), age at diagnosis (<65, ≥65), BMI at baseline (<25, 25–<30, >30) and height (tertiles and quartiles, cohort-specific cut-points among controls were used). Finally, we evaluated the association of gene variants with advanced stage PCa (stage C or D) and high-grade PCa (Gleason score ≥8 or poorly differentiated).
SUPPLEMENTARY MATERIAL
FUNDING
This work was funded by National Cancer Institute (NCI) grants U01 CA098216 (EPIC), U01 CA098233 (Harvard), U01 CA098758 (MEC) and U01 CA098710 (ACS). F.R.S. was supported by the National Research Service Award Training Program in Cancer Epidemiology (T32 CA009001-32). The Physicians' Health Study was supported by the NCI (CA-34944, CA-40360 and CA-097193) and the NHLBI (HL-26490 and HL-34595). The Multiethnic Cohort was supported by CA54281 and CA63464. The ATBC Study was supported by US Public Health Service contracts N01-CN-45165, N01-RC-45035 and N01-RC-37004 from the Department of Health and Human Services, National Cancer Institute.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Hardeep Ranu and Patrice Soule of the Dana-Farber/Harvard Cancer Center High Throughput Polymorphism Detection Core.
Conflict of Interest statement. None declared.
REFERENCES
- 1.DiGiovanni J., Kiguchi K., Frijhoff A., Wilker E., Bol D.K., Beltran L., Moats S., Ramirez A., Jorcano J., Conti C. Deregulated expression of insulin-like growth factor 1 in prostate epithelium leads to neoplasia in transgenic mice. Proc. Natl Acad. Sci. USA. 2000;97:3455–3460. doi: 10.1073/pnas.97.7.3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ruan W., Powell-Braxton L., Kopchick J.J., Kleinberg D.L. Evidence that insulin-like growth factor I and growth hormone are required for prostate gland development. Endocrinology. 1999;140:1984–1989. doi: 10.1210/endo.140.5.6721. [DOI] [PubMed] [Google Scholar]
- 3.Yu H., Rohan T. Role of the insulin-like growth factor family in cancer development and progression. J. Natl Cancer Inst. 2000;92:1472–1489. doi: 10.1093/jnci/92.18.1472. [DOI] [PubMed] [Google Scholar]
- 4.Boudon C., Rodier G., Lechevallier E., Mottet N., Barenton B., Sultan C. Secretion of insulin-like growth factors and their binding proteins by human normal and hyperplastic prostatic cells in primary culture. J. Clin. Endocrinol. Metab. 1996;81:612–617. doi: 10.1210/jcem.81.2.8636277. [DOI] [PubMed] [Google Scholar]
- 5.Cohen P., Peehl D.M., Lamson G., Rosenfeld R.G. Insulin-like growth factors (IGFs), IGF receptors, and IGF-binding proteins in primary cultures of prostate epithelial cells. J. Clin. Endocrinol. Metab. 1991;73:401–407. doi: 10.1210/jcem-73-2-401. [DOI] [PubMed] [Google Scholar]
- 6.Pietrzkowski Z., Mulholland G., Gomella L., Jameson B.A., Wernicke D., Baserga R. Inhibition of growth of prostatic cancer cell lines by peptide analogues of insulin-like growth factor 1. Cancer Res. 1993;53:1102–1106. [PubMed] [Google Scholar]
- 7.Parrizas M., LeRoith D. Insulin-like growth factor-1 inhibition of apoptosis is associated with increased expression of the bcl-xL gene product. Endocrinology. 1997;138:1355–1358. doi: 10.1210/endo.138.3.5103. [DOI] [PubMed] [Google Scholar]
- 8.Pollak M. Insulin-like growth factor physiology and neoplasia. Growth Horm. IGF Res. 2000;10(Suppl A):S6–S7. doi: 10.1016/s1096-6374(00)90002-9. [DOI] [PubMed] [Google Scholar]
- 9.Pollak M. Insulin-like growth factor physiology and cancer risk. Eur. J. Cancer. 2000;36:1224–1228. doi: 10.1016/s0959-8049(00)00102-7. [DOI] [PubMed] [Google Scholar]
- 10.Pollak M.N., Schernhammer E.S., Hankinson S.E. Insulin-like growth factors and neoplasia. Nat. Rev. Cancer. 2004;4:505–518. doi: 10.1038/nrc1387. [DOI] [PubMed] [Google Scholar]
- 11.Renehan A.G., Zwahlen M., Minder C., O'Dwyer S.T., Shalet S.M., Egger M. Insulin-like growth factor (IGF)-I, IGF binding protein-3, and cancer risk: systematic review and meta-regression analysis. Lancet. 2004;363:1346–1353. doi: 10.1016/S0140-6736(04)16044-3. [DOI] [PubMed] [Google Scholar]
- 12.Baxter R.C. Characterization of the acid-labile subunit of the growth hormone-dependent insulin-like growth factor binding protein complex. J. Clin. Endocrinol. Metab. 1988;67:265–272. doi: 10.1210/jcem-67-2-265. [DOI] [PubMed] [Google Scholar]
- 13.Lee P.D., Giudice L.C., Conover C.A., Powell D.R. Insulin-like growth factor binding protein-1: recent findings and new directions. Proc. Soc. Exp. Biol. Med. 1997;216:319–357. doi: 10.3181/00379727-216-44182. [DOI] [PubMed] [Google Scholar]
- 14.Baxter R.C. Signalling pathways involved in antiproliferative effects of IGFBP-3: a review. Mol. Pathol. 2001;54:145–148. doi: 10.1136/mp.54.3.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Firth S.M., Baxter R.C. Cellular actions of the insulin-like growth factor binding proteins. Endocr. Rev. 2002;23:824–854. doi: 10.1210/er.2001-0033. [DOI] [PubMed] [Google Scholar]
- 16.Chan J.M., Stampfer M.J., Ma J., Gann P., Gaziano J.M., Pollak M., Giovannucci E. Insulin-like growth factor-I (IGF-I) and IGF binding protein-3 as predictors of advanced-stage prostate cancer. J. Natl Cancer Inst. 2002;94:1099–1106. doi: 10.1093/jnci/94.14.1099. [DOI] [PubMed] [Google Scholar]
- 17.Shi R., Berkel H.J., Yu H. Insulin-like growth factor-I and prostate cancer: a meta-analysis. Br. J. Cancer. 2001;85:991–996. doi: 10.1054/bjoc.2001.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stattin P., Rinaldi S., Biessy C., Stenman U.H., Hallmans G., Kaaks R. High levels of circulating insulin-like growth factor-I increase prostate cancer risk: a prospective study in a population-based nonscreened cohort. J. Clin. Oncol. 2004;22:3104–3112. doi: 10.1200/JCO.2004.10.105. [DOI] [PubMed] [Google Scholar]
- 19.Allen N.E., Key T.J., Appleby P.N., Travis R.C., Roddam A.W., Rinaldi S., Egevad L., Rohrmann S., Linseisen J., Pischon T., et al. Serum insulin-like growth factor (IGF)-I and IGF-binding protein-3 concentrations and prostate cancer risk: results from the European Prospective Investigation into Cancer and Nutrition. Cancer Epidemiol. Biomarkers. Prev. 2007;16:1121–1127. doi: 10.1158/1055-9965.EPI-06-1062. [DOI] [PubMed] [Google Scholar]
- 20.Platz E.A., Pollak M.N., Leitzmann M.F., Stampfer M.J., Willett W.C., Giovannucci E. Plasma insulin-like growth factor-1 and binding protein-3 and subsequent risk of prostate cancer in the PSA era. Cancer Causes Control. 2005;16:255–262. doi: 10.1007/s10552-004-3484-8. [DOI] [PubMed] [Google Scholar]
- 21.Weiss J.M., Huang W.Y., Rinaldi S., Fears T.R., Chatterjee N., Chia D., Crawford E.D., Kaaks R., Hayes R.B. IGF-1 and IGFBP-3: risk of prostate cancer among men in the Prostate, Lung, Colorectal and Ovarian Cancer Screening Trial. Int. J. Cancer. 2007;121:2267–2273. doi: 10.1002/ijc.22921. [DOI] [PubMed] [Google Scholar]
- 22.Woodson K., Tangrea J.A., Pollak M., Copeland T.D., Taylor P.R., Virtamo J., Albanes D. Serum insulin-like growth factor I: tumor marker or etiologic factor? A prospective study of prostate cancer among Finnish men. Cancer Res. 2003;63:3991–3994. [PubMed] [Google Scholar]
- 23.Chan J.M., Stampfer M.J., Giovannucci E., Gann P.H., Ma J., Wilkinson P., Hennekens C.H., Pollak M. Plasma insulin-like growth factor-I and prostate cancer risk: a prospective study. Science. 1998;279:563–566. doi: 10.1126/science.279.5350.563. [DOI] [PubMed] [Google Scholar]
- 24.Roddam A.W., Allen N.E., Appleby P., Key T.J., Ferrucci L., Carter H.B., Metter E.J., Chen C., Weiss N.S., Fitzpatrick A., et al. Insulin-like growth factors, their binding proteins, and prostate cancer risk: analysis of individual patient data from 12 prospective studies. Ann. Intern. Med. 2008;149:461–471,. doi: 10.7326/0003-4819-149-7-200810070-00006. W483–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harman S.M., Metter E.J., Blackman M.R., Landis P.K., Carter H.B. Serum levels of insulin-like growth factor I (IGF-I), IGF-II, IGF-binding protein-3, and prostate-specific antigen as predictors of clinical prostate cancer. J. Clin. Endocrinol. Metab. 2000;85:4258–4265. doi: 10.1210/jcem.85.11.6990. [DOI] [PubMed] [Google Scholar]
- 26.Stattin P., Bylund A., Rinaldi S., Biessy C., Dechaud H., Stenman U.H., Egevad L., Riboli E., Hallmans G., Kaaks R. Plasma insulin-like growth factor-I, insulin-like growth factor-binding proteins, and prostate cancer risk: a prospective study. J. Natl Cancer Inst. 2000;92:1910–1917. doi: 10.1093/jnci/92.23.1910. [DOI] [PubMed] [Google Scholar]
- 27.Morris J.K., George L.M., Wu T., Wald N.J. Insulin-like growth factors and cancer: no role in screening. Evidence from the BUPA study and meta-analysis of prospective epidemiological studies. Br. J. Cancer. 2006;95:112–117. doi: 10.1038/sj.bjc.6603200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaaks R., Lukanova A. Energy balance and cancer: the role of insulin and insulin-like growth factor-I. Proc. Nutr. Soc. 2001;60:91–106. doi: 10.1079/pns200070. [DOI] [PubMed] [Google Scholar]
- 29.Thissen J.P., Ketelslegers J.M., Underwood L.E. Nutritional regulation of the insulin-like growth factors. Endocr. Rev. 1994;15:80–101. doi: 10.1210/edrv-15-1-80. [DOI] [PubMed] [Google Scholar]
- 30.Hall K., Hilding A., Thoren M. Determinants of circulating insulin-like growth factor-I. J. Endocrinol. Invest. 1999;22:48–57. [PubMed] [Google Scholar]
- 31.Harrela M., Koistinen H., Kaprio J., Lehtovirta M., Tuomilehto J., Eriksson J., Toivanen L., Koskenvuo M., Leinonen P., Koistinen R., et al. Genetic and environmental components of interindividual variation in circulating levels of IGF-I, IGF-II, IGFBP-1 and IGFBP-3. J. Clin. Invest. 1996;98:2612–2615. doi: 10.1172/JCI119081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hong Y., Brismar K., Hall K., Pedersen N.L., de Faire U. Associations between insulin-like growth factor-I (IGF-I), IGF-binding protein-1, insulin and other metabolic measures after controlling for genetic influences: results from middle-aged and elderly monozygotic twins. J. Endocrinol. 1997;153:251–257. doi: 10.1677/joe.0.1530251. [DOI] [PubMed] [Google Scholar]
- 33.Kao P.C., Matheny A.P., Jr, Lang C.A. Insulin-like growth factor-I comparisons in healthy twin children. J. Clin. Endocrinol. Metab. 1994;78:310–312. doi: 10.1210/jcem.78.2.8106617. [DOI] [PubMed] [Google Scholar]
- 34.Verhaeghe J., Loos R., Vlietinck R., Herck E.V., van Bree R., Schutter A.M. C-peptide, insulin-like growth factors I and II, and insulin-like growth factor binding protein-1 in cord serum of twins: genetic versus environmental regulation. Am. J. Obstet. Gynecol. 1996;175:1180–1188. doi: 10.1016/s0002-9378(96)70025-x. [DOI] [PubMed] [Google Scholar]
- 35.Frayling T.M., Hattersley A.T., McCarthy A., Holly J., Mitchell S.M., Gloyn A.L., Owen K., Davies D., Smith G.D., Ben-Shlomo Y. A putative functional polymorphism in the IGF-I gene: association studies with type 2 diabetes, adult height, glucose tolerance, and fetal growth in UK populations. Diabetes. 2002;51:2313–2316. doi: 10.2337/diabetes.51.7.2313. [DOI] [PubMed] [Google Scholar]
- 36.Rietveld I., Janssen J.A., Hofman A., Pols H.A., van Duijn C.M., Lamberts S.W. A polymorphism in the IGF-I gene influences the age-related decline in circulating total IGF-I levels. Eur. J. Endocrinol. 2003;148:171–175. doi: 10.1530/eje.0.1480171. [DOI] [PubMed] [Google Scholar]
- 37.Rietveld I., Janssen J.A., van Rossum E.F., Houwing-Duistermaat J.J., Rivadeneira F., Hofman A., Pols H.A., van Duijn C.M., Lamberts S.W. A polymorphic CA repeat in the IGF-I gene is associated with gender-specific differences in body height, but has no effect on the secular trend in body height. Clin. Endocrinol. (Oxf) 2004;61:195–203. doi: 10.1111/j.1365-2265.2004.02078.x. [DOI] [PubMed] [Google Scholar]
- 38.Vaessen N., Heutink P., Janssen J.A., Witteman J.C., Testers L., Hofman A., Lamberts S.W., Oostra B.A., Pols H.A., van Duijn C.M. A polymorphism in the gene for IGF-I: functional properties and risk for type 2 diabetes and myocardial infarction. Diabetes. 2001;50:637–642. doi: 10.2337/diabetes.50.3.637. [DOI] [PubMed] [Google Scholar]
- 39.Fletcher O., Gibson L., Johnson N., Altmann D.R., Holly J.M., Ashworth A., Peto J., Silva Idos S. Polymorphisms and circulating levels in the insulin-like growth factor system and risk of breast cancer: a systematic review. Cancer Epidemiol. Biomarkers Prev. 2005;14:2–19. [PubMed] [Google Scholar]
- 40.Allen N.E., Davey G.K., Key T.J., Zhang S., Narod S.A. Serum insulin-like growth factor I (IGF-I) concentration in men is not associated with the cytosine–adenosine repeat polymorphism of the IGF-I gene. Cancer Epidemiol. Biomarkers Prev. 2002;11:319–320. [PubMed] [Google Scholar]
- 41.Chen C., Freeman R., Voigt L.F., Fitzpatrick A., Plymate S.R., Weiss N.S. Prostate cancer risk in relation to selected genetic polymorphisms in insulin-like growth factor-I, insulin-like growth factor binding protein-3, and insulin-like growth factor-I receptor. Cancer Epidemiol. Biomarkers Prev. 2006;15:2461–2466. doi: 10.1158/1055-9965.EPI-06-0541. [DOI] [PubMed] [Google Scholar]
- 42.DeLellis K., Ingles S., Kolonel L., McKean-Cowdin R., Henderson B., Stanczyk F., Probst-Hensch N.M. IGF1 genotype, mean plasma level and breast cancer risk in the Hawaii/Los Angeles multiethnic cohort. Br. J. Cancer. 2003;88:277–282. doi: 10.1038/sj.bjc.6600728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lai J.H., Vesprini D., Zhang W., Yaffe M.J., Pollak M., Narod S.A. A polymorphic locus in the promoter region of the IGFBP3 gene is related to mammographic breast density. Cancer Epidemiol. Biomarkers Prev. 2004;13:573–582. [PubMed] [Google Scholar]
- 44.Rosen C.J., Kurland E.S., Vereault D., Adler R.A., Rackoff P.J., Craig W.Y., Witte S., Rogers J., Bilezikian J.P. Association between serum insulin growth factor-I (IGF-I) and a simple sequence repeat in IGF-I gene: implications for genetic studies of bone mineral density. J. Clin. Endocrinol. Metab. 1998;83:2286–2290. doi: 10.1210/jcem.83.7.4964. [DOI] [PubMed] [Google Scholar]
- 45.Johansson M., McKay J.D., Stattin P., Canzian F., Boillot C., Wiklund F., Adami H.O., Balter K., Gronberg H., Kaaks R. Comprehensive evaluation of genetic variation in the IGF1 gene and risk of prostate cancer. Int. J. Cancer. 2007;120:539–542. doi: 10.1002/ijc.22344. [DOI] [PubMed] [Google Scholar]
- 46.Johansson M., McKay J.D., Wiklund F., Rinaldi S., Verheus M., van Gils C.H., Hallmans G., Balter K., Adami H.O., Gronberg H., et al. Implications for prostate cancer of insulin-like growth factor-I (IGF-I) genetic variation and circulating IGF-I levels. J. Clin. Endocrinol. Metab. 2007;92:4820–4826. doi: 10.1210/jc.2007-0887. [DOI] [PubMed] [Google Scholar]
- 47.Al-Zahrani A., Sandhu M.S., Luben R.N., Thompson D., Baynes C., Pooley K.A., Luccarini C., Munday H., Perkins B., Smith P., et al. IGF1 and IGFBP3 tagging polymorphisms are associated with circulating levels of IGF1, IGFBP3 and risk of breast cancer. Hum. Mol. Genet. 2006;15:1–10. doi: 10.1093/hmg/ddi398. [DOI] [PubMed] [Google Scholar]
- 48.Canzian F., McKay J.D., Cleveland R.J., Dossus L., Biessy C., Rinaldi S., Landi S., Boillot C., Monnier S., Chajes V., et al. Polymorphisms of genes coding for insulin-like growth factor 1 and its major binding proteins, circulating levels of IGF-I and IGFBP-3 and breast cancer risk: results from the EPIC study. Br. J. Cancer. 2006;94:299–307. doi: 10.1038/sj.bjc.6602936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jernstrom H., Deal C., Wilkin F., Chu W., Tao Y., Majeed N., Hudson T., Narod S.A., Pollak M. Genetic and nongenetic factors associated with variation of plasma levels of insulin-like growth factor-I and insulin-like growth factor-binding protein-3 in healthy premenopausal women. Cancer Epidemiol. Biomarkers Prev. 2001;10:377–384. [PubMed] [Google Scholar]
- 50.Le Marchand L., Kolonel L.N., Henderson B.E., Wilkens L.R. Association of an exon 1 polymorphism in the IGFBP3 gene with circulating IGFBP-3 levels and colorectal cancer risk: the multiethnic cohort study. Cancer Epidemiol. Biomarkers Prev. 2005;14:1319–1321. doi: 10.1158/1055-9965.EPI-04-0847. [DOI] [PubMed] [Google Scholar]
- 51.Ren Z., Cai Q., Shu X.O., Cai H., Li C., Yu H., Gao Y.T., Zheng W. Genetic polymorphisms in the IGFBP3 gene: association with breast cancer risk and blood IGFBP-3 protein levels among Chinese women. Cancer Epidemiol. Biomarkers Prev. 2004;13:1290–1295. [PubMed] [Google Scholar]
- 52.Schernhammer E.S., Hankinson S.E., Hunter D.J., Blouin M.J., Pollak M.N. Polymorphic variation at the −202 locus in IGFBP3: influence on serum levels of insulin-like growth factors, interaction with plasma retinol and vitamin D and breast cancer risk. Int. J. Cancer. 2003;107:60–64. doi: 10.1002/ijc.11358. [DOI] [PubMed] [Google Scholar]
- 53.Cheng I., Delellis Henderson K., Haiman C.A., Kolonel L.N., Henderson B.E., Freedman M.L., Le Marchand L. Genetic determinants of circulating insulin-like growth factor (IGF)-I, IGF binding protein (BP)-1, and IGFBP-3 levels in a multiethnic population. J. Clin. Endocrinol. Metab. 2007;92:3660–3666. doi: 10.1210/jc.2007-0790. [DOI] [PubMed] [Google Scholar]
- 54.Patel A.V., Cheng I., Canzian F., Le Marchand L., Thun M.J., Berg C.D., Buring J., Calle E.E., Chanock S., Clavel-Chapelon F., et al. IGF-1, IGFBP-1 and IGFBP-3 polymorphisms predict circulating IGF levels but not breast cancer risk: findings from the Breast and Prostate Cancer Cohort Consortium (BPC3) PLoS One. 2008;3:e2578. doi: 10.1371/journal.pone.0002578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Deal C., Ma J., Wilkin F., Paquette J., Rozen F., Ge B., Hudson T., Stampfer M., Pollak M. Novel promoter polymorphism in insulin-like growth factor-binding protein-3: correlation with serum levels and interaction with known regulators. J. Clin. Endocrinol. Metab. 2001;86:1274–1280. doi: 10.1210/jcem.86.3.7280. [DOI] [PubMed] [Google Scholar]
- 56.Li L., Cicek M.S., Casey G., Witte J.S. No association between genetic polymorphisms in IGF-I and IGFBP-3 and prostate cancer. Cancer Epidemiol. Biomarkers Prev. 2004;13:497–498. [PubMed] [Google Scholar]
- 57.Nam R.K., Zhang W.W., Trachtenberg J., Jewett M.A., Emami M., Vesprini D., Chu W., Ho M., Sweet J., Evans A., et al. Comprehensive assessment of candidate genes and serological markers for the detection of prostate cancer. Cancer Epidemiol. Biomarkers Prev. 2003;12:1429–1437. [PubMed] [Google Scholar]
- 58.Neuhausen S.L., Slattery M.L., Garner C.P., Ding Y.C., Hoffman M., Brothman A.R. Prostate cancer risk and IRS1, IRS2, IGF1, and INS polymorphisms: strong association of IRS1 G972R variant and cancer risk. Prostate. 2005;64:168–174. doi: 10.1002/pros.20216. [DOI] [PubMed] [Google Scholar]
- 59.Schildkraut J.M., Demark-Wahnefried W., Wenham R.M., Grubber J., Jeffreys A.S., Grambow S.C., Marks J.R., Moorman P.G., Hoyo C., Ali S., et al. IGF1 (CA)19 repeat and IGFBP3 -202 A/C genotypes and the risk of prostate cancer in Black and White men. Cancer Epidemiol. Biomarkers Prev. 2005;14:403–408. doi: 10.1158/1055-9965.EPI-04-0426. [DOI] [PubMed] [Google Scholar]
- 60.Tsuchiya N., Wang L., Horikawa Y., Inoue T., Kakinuma H., Matsuura S., Sato K., Ogawa O., Kato T., Habuchi T. CA repeat polymorphism in the insulin-like growth factor-I gene is associated with increased risk of prostate cancer and benign prostatic hyperplasia. Int. J. Oncol. 2005;26:225–231. [PubMed] [Google Scholar]
- 61.Cheng I., Stram D.O., Penney K.L., Pike M., Le Marchand L., Kolonel L.N., Hirschhorn J., Altshuler D., Henderson B.E., Freedman M.L. Common genetic variation in IGF1 and prostate cancer risk in the Multiethnic Cohort. J. Natl Cancer Inst. 2006;98:123–134. doi: 10.1093/jnci/djj013. [DOI] [PubMed] [Google Scholar]
- 62.Cheng I., Penney K.L., Stram D.O., Le Marchand L., Giorgi E., Haiman C.A., Kolonel L.N., Pike M., Hirschhorn J., Henderson B.E., et al. Haplotype-based association studies of IGFBP1 and IGFBP3 with prostate and breast cancer risk: the multiethnic cohort. Cancer Epidemiol. Biomarkers Prev. 2006;15:1993–1997. doi: 10.1158/1055-9965.EPI-06-0361. [DOI] [PubMed] [Google Scholar]
- 63.Hunter D.J., Riboli E., Haiman C.A., Albanes D., Altshuler D., Chanock S.J., Haynes R.B., Henderson B.E., Kaaks R., Stram D.O., et al. A candidate gene approach to searching for low-penetrance breast and prostate cancer genes. Nat. Rev. Cancer. 2005;5:977–985. doi: 10.1038/nrc1754. [DOI] [PubMed] [Google Scholar]
- 64.Rowlands M.A., Gunnell D., Harris R., Vatten L.J., Holly J.M., Martin R.M. Circulating insulin-like growth factor peptides and prostate cancer risk: a systematic review and meta-analysis. Int. J. Cancer. 2009;124:2416–2429. doi: 10.1002/ijc.24202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ebrahim S., Davey Smith G. Mendelian randomization: can genetic epidemiology help redress the failures of observational epidemiology? Hum. Genet. 2008;123:15–33. doi: 10.1007/s00439-007-0448-6. [DOI] [PubMed] [Google Scholar]
- 66.Smith G.D., Ebrahim S. Mendelian randomization: prospects, potentials, and limitations. Int. J. Epidemiol. 2004;33:30–42. doi: 10.1093/ije/dyh132. [DOI] [PubMed] [Google Scholar]
- 67.Davey Smith G., Ebrahim S. ‘Mendelian randomization’: can genetic epidemiology contribute to understanding environmental determinants of disease? Int. J. Epidemiol. 2003;32:1–22. doi: 10.1093/ije/dyg070. [DOI] [PubMed] [Google Scholar]
- 68.Amundadottir L.T., Sulem P., Gudmundsson J., Helgason A., Baker A., Agnarsson B.A., Sigurdsson A., Benediktsdottir K.R., Cazier J.B., Sainz J., et al. A common variant associated with prostate cancer in European and African populations. Nat. Genet. 2006;38:652–658. doi: 10.1038/ng1808. [DOI] [PubMed] [Google Scholar]
- 69.Eeles R.A., Kote-Jarai Z., Al Olama A.A., Giles G.G., Guy M., Severi G., Muir K., Hopper J.L., Henderson B.E., Haiman C.A., et al. Identification of seven new prostate cancer susceptibility loci through a genome-wide association study. Nat. Genet. 2009;41:1116–1121. doi: 10.1038/ng.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Eeles R.A., Kote-Jarai Z., Giles G.G., Olama A.A., Guy M., Jugurnauth S.K., Mulholland S., Leongamornlert D.A., Edwards S.M., Morrison J., et al. Multiple newly identified loci associated with prostate cancer susceptibility. Nat. Genet. 2008;40:316–321. doi: 10.1038/ng.90. [DOI] [PubMed] [Google Scholar]
- 71.Gudmundsson J., Sulem P., Gudbjartsson D.F., Blondal T., Gylfason A., Agnarsson B.A., Benediktsdottir K.R., Magnusdottir D.N., Orlygsdottir G., Jakobsdottir M., et al. Genome-wide association and replication studies identify four variants associated with prostate cancer susceptibility. Nat. Genet. 2009;41:1122–1126. doi: 10.1038/ng.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gudmundsson J., Sulem P., Rafnar T., Bergthorsson J.T., Manolescu A., Gudbjartsson D., Agnarsson B.A., Sigurdsson A., Benediktsdottir K.R., Blondal T., et al. Common sequence variants on 2p15 and Xp11.22 confer susceptibility to prostate cancer. Nat. Genet. 2008;40:281–283. doi: 10.1038/ng.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sun J., Zheng S.L., Wiklund F., Isaacs S.D., Li G., Wiley K.E., Kim S.T., Zhu Y., Zhang Z., Hsu F.C., et al. Sequence variants at 22q13 are associated with prostate cancer risk. Cancer Res. 2009;69:10–15. doi: 10.1158/0008-5472.CAN-08-3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Thomas G., Jacobs K.B., Yeager M., Kraft P., Wacholder S., Orr N., Yu K., Chatterjee N., Welch R., Hutchinson A., et al. Multiple loci identified in a genome-wide association study of prostate cancer. Nat. Genet. 2008;40:310–315. doi: 10.1038/ng.91. [DOI] [PubMed] [Google Scholar]
- 75.Yeager M., Chatterjee N., Ciampa J., Jacobs K.B., Gonzalez-Bosquet J., Hayes R.B., Kraft P., Wacholder S., Orr N., Berndt S., et al. Identification of a new prostate cancer susceptibility locus on chromosome 8q24. Nat. Genet. 2009;41:1055–1057. doi: 10.1038/ng.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yeager M., Orr N., Hayes R.B., Jacobs K.B., Kraft P., Wacholder S., Minichiello M.J., Fearnhead P., Yu K., Chatterjee N., et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat. Genet. 2007;39:645–649. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- 77.Xu J., Sun J., Kader A.K., Lindstrom S., Wiklund F., Hsu F.C., Johansson J.E., Zheng S.L., Thomas G., Hayes R.B., et al. Estimation of absolute risk for prostate cancer using genetic markers and family history. Prostate. 2009;69:1565–1572. doi: 10.1002/pros.21002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Maher B. Personal genomes: the case of the missing heritability. Nature. 2008;456:18–21. doi: 10.1038/456018a. [DOI] [PubMed] [Google Scholar]
- 79.Manolio T.A., Collins F.S., Cox N.J., Goldstein D.B., Hindorff L.A., Hunter D.J., McCarthy M.I., Ramos E.M., Cardon L.R., Chakravarti A., et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hunter D.J. Gene–environment interactions in human diseases. Nat. Rev. Genet. 2005;6:287–298. doi: 10.1038/nrg1578. [DOI] [PubMed] [Google Scholar]
- 81.Calle E.E., Rodriguez C., Jacobs E.J., Almon M.L., Chao A., McCullough M.L., Feigelson H.S., Thun M.J. The American Cancer Society Cancer Prevention Study II Nutrition Cohort: rationale, study design, and baseline characteristics. Cancer. 2002;94:500–511. doi: 10.1002/cncr.10197. [DOI] [PubMed] [Google Scholar]
- 82.The ATBC Cancer Prevention Study Group. The alpha-tocopherol, beta-carotene lung cancer prevention study: design, methods, participant characteristics, and compliance. Ann. Epidemiol. 1994;4:1–10. doi: 10.1016/1047-2797(94)90036-1. [DOI] [PubMed] [Google Scholar]
- 83.Riboli E., Hunt K.J., Slimani N., Ferrari P., Norat T., Fahey M., Charrondiere U.R., Hemon B., Casagrande C., Vignat J., et al. European Prospective Investigation into Cancer and Nutrition (EPIC): study populations and data collection. Public Health Nutr. 2002;5:1113–1124. doi: 10.1079/PHN2002394. [DOI] [PubMed] [Google Scholar]
- 84.Giovannucci E., Pollak M., Liu Y., Platz E.A., Majeed N., Rimm E.B., Willett W.C. Nutritional predictors of insulin-like growth factor I and their relationships to cancer in men. Cancer Epidemiol. Biomarkers Prev. 2003;12:84–89. [PubMed] [Google Scholar]
- 85.Kolonel L.N., Altshuler D., Henderson B.E. The multiethnic cohort study: exploring genes, lifestyle and cancer risk. Nat. Rev. Cancer. 2004;4:519–527. doi: 10.1038/nrc1389. [DOI] [PubMed] [Google Scholar]
- 86.Hayes R.B., Reding D., Kopp W., Subar A.F., Bhat N., Rothman N., Caporaso N., Ziegler R.G., Johnson C.C., Weissfeld J.L., et al. Etiologic and early marker studies in the prostate, lung, colorectal and ovarian (PLCO) cancer screening trial. Control Clin. Trials. 2000;21:349S–355S. doi: 10.1016/s0197-2456(00)00101-x. [DOI] [PubMed] [Google Scholar]
- 87.Barrett J.C., Fry B., Maller J., Daly M.J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 88.Gabriel S.B., Schaffner S.F., Nguyen H., Moore J.M., Roy J., Blumenstiel B., Higgins J., DeFelice M., Lochner A., Faggart M., et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 89.Stram D.O., Haiman C.A., Hirschhorn J.N., Altshuler D., Kolonel L.N., Henderson B.E., Pike M.C. Choosing haplotype-tagging SNPs based on unphased genotype data using a preliminary sample of unrelated subjects with an example from the Multiethnic Cohort Study. Hum. Hered. 2003;55:27–36. doi: 10.1159/000071807. [DOI] [PubMed] [Google Scholar]
- 90.Packer B.R., Yeager M., Staats B., Welch R., Crenshaw A., Kiley M., Eckert A., Beerman M., Miller E., Bergen A., et al. SNP500Cancer: a public resource for sequence validation and assay development for genetic variation in candidate genes. Nucleic Acids Res. 2004;32:D528–D532. doi: 10.1093/nar/gkh005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Conneely K.N., Boehnke M. So many correlated tests, so little time! Rapid adjustment of P-values for multiple correlated tests. Am. J. Hum. Genet. 2007;81 doi: 10.1086/522036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kraft P., Pharoah P., Chanock S.J., Albanes D., Kolonel L.N., Hayes R.B., Altshuler D., Andriole G., Berg C., Boeing H., et al. Genetic variation in the HSD17B1 gene and risk of prostate cancer. PLoS Genet. 2005;1:e68. doi: 10.1371/journal.pgen.0010068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wacholder S., Chanock S., Garcia-Closas M., El Ghormli L., Rothman N. Assessing the probability that a positive report is false: an approach for molecular epidemiology studies. J. Natl Cancer Inst. 2004;96:434–442. doi: 10.1093/jnci/djh075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rosner B. Percentage points for a generalized ESD many-outlier procedure. Technometrics. 1983;25:165–172. [Google Scholar]
- 95.Feigelson H.S., Cox D.G., Cann H.M., Wacholder S., Kaaks R., Henderson B.E., Albanes D., Altshuler D., Berglund G., Berrino F., et al. Haplotype analysis of the HSD17B1 gene and risk of breast cancer: a comprehensive approach to multicenter analyses of prospective cohort studies. Cancer Res. 2006;66:2468–2475. doi: 10.1158/0008-5472.CAN-05-3574. [DOI] [PubMed] [Google Scholar]
- 96.Kraft P., Cox D.G., Paynter R.A., Hunter D., De Vivo I. Accounting for haplotype uncertainty in matched association studies: a comparison of simple and flexible techniques. Genet. Epidemiol. 2005;28:261–272. doi: 10.1002/gepi.20061. [DOI] [PubMed] [Google Scholar]
- 97.Zaykin D.V., Westfall P.H., Young S.S., Karnoub M.A., Wagner M.J., Ehm M.G. Testing association of statistically inferred haplotypes with discrete and continuous traits in samples of unrelated individuals. Hum. Hered. 2002;53:79–91. doi: 10.1159/000057986. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.