Abstract
Fragile X syndrome (FXS) is the most common form of inherited mental retardation and is caused by the loss of function for Fragile X protein (FMRP), an RNA-binding protein thought to regulate synaptic plasticity by controlling the localization and translation of specific mRNAs. We have recently shown that FMRP is required to control the proliferation of the germline in Drosophila. To determine whether FMRP is also required for proliferation during brain development, we examined the distribution of cell cycle markers in dFmr1 brains compared with wild-type throughout larval development. Our results indicate that the loss of dFmr1 leads to a significant increase in the number of mitotic neuroblasts (NB) and BrdU incorporation in the brain, consistent with the notion that FMRP controls proliferation during neurogenesis. Developmental studies suggest that FMRP also inhibits neuroblast exit from quiescence in early larval brains, as indicated by misexpression of Cyclin E. Live imaging experiments indicate that by the third instar larval stage, the length of the cell cycle is unaffected, although more cells are found in S and G2/M in dFmr1 brains compared with wild-type. To determine the role of FMRP in neuroblast division and differentiation, we used Mosaic Analysis with a Repressible Marker (MARCM) approaches in the developing larval brain and found that single dFmr1 NB generate significantly more neurons than controls. Our results demonstrate that FMRP is required during brain development to control the exit from quiescence and proliferative capacity of NB as well as neuron production, which may provide insights into the autistic component of FXS.
INTRODUCTION
Fragile X syndrome (FXS) is a devastating neurological disorder which affects the cognitive abilities of 1/4000 males and 1/8000 females worldwide (1). The disease is caused by the loss of function for the RNA-binding protein, Fragile X protein (FMRP), which is thought to regulate synaptic plasticity by controlling the localized translation of specific mRNAs, including futsch, profilin, Rac1 and PSD-95 mRNAs, all of which are involved in various aspects of synaptic development and function (2–6). In addition to its synaptic function, FMRP was shown to play a role in germline proliferation and differentiation (7–9). Previous studies using neurospheres, an in vitro model for neural stem cell development, have suggested that FMRP controls the balance between glia and neuron production in the brain with more neurons being produced at the expense of glia (10). Similar experiments using human neural progenitor cells identified several genes misexpressed in FMR1 mutant cells but found no neurogenesis defects (11). Recent in vivo studies have identified defects within the glutamatergic lineage in the FMR1 KO mouse brain (12), and have implicated FMRP in adult neurogenesis (13). Taken together, these data implicate FMRP in neural stem cell proliferation and differentiation and indicate that its role may be developmentally regulated, although this aspect of FMRP biology remains poorly understood.
Drosophila larval brain neuroblasts (NB) have emerged as a neural stem cell model for elucidating the mechanisms controlling proliferation and differentiation in the developing nervous system. The powerful genetic tools available in Drosophila have led to the identification of several genes required for neural stem cell renewal and differentiation, including brain tumor (brat), prospero (pros) and miranda (mir) (reviewed in 14). To determine whether FMRP is required during neurogenesis, we performed loss of function and clonal analyses using the larval neuroblast model. Here we report that loss of dFmr1 leads to cell cycle defects: in late third instar larval brains, there are more mutant cells found in mitosis and S phase, as indicated by PhosphoHistone H3 and BrdU incorporation. Clonal analyses indicate that dFmr1 mutant NB have an increased proliferative capacity and generate more neurons in the developing brain, in which they persist through adulthood. Developmental studies show that dFmr1 mutant NB exit quiescence prematurely and begin their proliferative activities sooner than their wild-type counterparts. Taken together, these data demonstrate that FMRP controls the developmental timing and proliferative capacity of larval brain NB and suggest new mechanisms for FXS.
RESULTS
Loss of dFmr1 alters the cell cycle profile of the larval brain
We have previously shown that FMRP controls the proliferation of the germline during Drosophila oogenesis (7). To test whether FMRP plays a similar role in the developing brain, we compared the distribution of cell cycle markers in control brains (w1118 and dFmr1 genomic rescue, P[dFmr1+]; dFmr13, see Materials and Methods) versus dFmr1 mutant brains [dFmr13, dFmr13/50M and dFmr13/Df 6265, see Zhang et al. (2) and Wan et al. (15)]. We began by examining the distribution of the mitotic marker PhosphoHistone H3 (PH3) in dFmr1 mutant brains at the late third instar larval stage. As seen in Figure 1, we found more PH3 positive cells in mutant brains compared with wild-type (Fig. 1A–F). Indeed, the mitotic index of dFmr1 mutant brains measured in squashed brain preparations was significantly higher than wild-type. We found an average of 540 ± 65 mitotic cells in wild-type (n = 3) and 831 ± 11 mitotic cells in dFmr13/Df 6265 (n = 3, Pvalue = 0.01, data not shown). To determine whether FMRP controls NB specifically, we used the neural stem cell marker Miranda in conjunction with PH3 stainings, and identified significantly more dFmr1 mutant NB in mitosis [24.8 ± 2.4 for dFmr13 (n = 6, Pvalue = 0.04) and 26.1 ± 1.1 for dFmr13/50M brains (n = 8, Pvalue < 0.001) compared with 20.4 ± 0.9 for genomic rescue brains, P[dFmr1+]; dFmr13 (n = 14), see Fig. 1K]. There was no significant difference in PH3 positive NB between the two controls (dFmr1 genomic rescue and w1118, see Fig. 1K). Interestingly, some mutant NB contain chromosomes that appear ‘spread out’ (compare Fig. 1E and F), suggesting potential chromatin organization, or mitotic checkpoint defects, which remains to be investigated. These results are consistent with a role for FMRP in regulating neural stem cell mitosis.
Figure 1.
Loss of dFmr1 alters cell cycle progression in the larval brain. (A–F) Staining for the M-phase marker PH3 in wild-type (A) and dFmr13 mutant (B) larval brains. Dashed lines delineate the CB areas. The stem cell-specific protein Miranda marks NBs (C–F). Higher magnification indicates that the dFmr1 mutant brains contain more PH3 positive neuroblasts (F, arrowheads) compared with wild-type (E, arrowheads). (K) Total number of mitotic neuroblasts (NBs) are significantly increased in both dFmr13 homozygotes (P = 0.04) and dFmr13/50M (P < 0.001) brains compared with the genetic rescue, P[dFmr1];dFmr13. There is no statistical significance (n.s.) between the w1118 and genomic rescue controls. Student's t-test was used to calculate statistical significance. (G–J) Staining for the S-phase marker BrdU in wild-type (G and I) and dFmr1 mutant (H and J) larval brains. The cortical protein Lgl was used to delineate the CB (see also dashed areas) region, where the neuroblasts and associated cells display higher levels of BrdU incorporation in dFmr1 mutants (H and J) compared with wild-type (G and I). Stainings and genotypes as indicated. Brains were dissected from age matched late third instar larvae. Scale bar in (A) 120 µm, in (E) 30 µm. All images are projections of three confocal slices, slice size = 2 µm.
To further test the role of FMRP throughout the cell cycle, we used BrdU incorporation as an S-phase marker and found that dFmr1 mutant brains contain more BrdU positive cells (Fig. 1H and J) compared with wild-type (Fig. 1G and I). These data indicate that more cells in the central brain (CB) region are actively synthesizing DNA due to the loss of dFmr1. Flow cytometry experiments using whole brains support these results by indicating that more mutant cells are in S phase (15.4%) as well as in G2/M (12%) compared with controls (9.6% in S and 4.6% in G2/M, respectively). Consistent with this abnormal distribution, there are fewer dFmr1 mutant cells in G1 (72.6%) compared with wild-type (85.8%) (data not shown). Taken together, our data indicate that, in the absence of FMRP, cell cycle progression is altered during brain development.
FMRP controls the size of neuroblast lineages in the central larval brain
Our findings of more mutant cells in S and G2/M in the whole brain analyses can be explained by either an increase in proliferation or a cell cycle delay. To distinguish between these two scenarios, we set out to determine the number of cells produced by individual NB in the developing brain. To this end, we performed clonal analyses using the MARCM (Mosaic Analysis with a Repressible Cell Marker) technique (16) using two independently derived null alleles of dFmr1, namely dFmr13 and dFmr150M (2,15). In brief, using this technique, we can generate dFmr1 mutant NB in an otherwise heterozygous background. The mutant NB are concomitantly marked with GFP, thus allowing us to trace all the daughter cells produced by a single neuroblast (16). Individual NB exhibit neural stem cell-like behaviors: they divide asymmetrically to renew themselves and to generate a smaller daughter, called a Ganglion Mother Cell (GMC) (17). For type I NB, which comprise the majority of NB in the larval CB, each GMC divides symmetrically a single time to produce two Ganglion Cells (GC), which differentiate into neurons. Type II NB, which have been shown to reside mostly on the dorsal-medial side of the brain, generate GMCs that divide asymmetrically, generate multiple neurons and are referred to as intermediate neural progenitor cells (INPs) (18–20). Individual NB in the larval brain can be distinguished by Asense, which is expressed in type I but not type II NB (20).
Using the MARCM technique, we induced wild-type and dFmr1 mutant neuroblast clones at 4–10 h after larval hatching (ALH) (Fig. 2A–H) and then allowed the larvae to develop to the late third instar stage. This early clone induction regime allowed us to quantify the proliferative potential of individual NB throughout larval development. Brains of carefully staged larvae were then dissected and imaged using confocal microscopy. Immunohistochemistry experiments confirmed that FMRP, which is ubiquitously expressed in the brain, was below detectable levels in mutant clones (data not shown). Individual neuroblast lineage sizes were determined by counting the number of cells in each clone, based on the number of GFP positive cells in conjunction with DAPI stainings. The loss of dFmr1 results in significantly larger mutant clones in both type I (Fig. 2A–D and M) and type II (Fig. 2E–H and N) NB. Using the type I neuroblast-specific marker Asense (Fig. 2A–D), we found that type I wild-type clones induced at 4–10 h ALH had an average of 34.9 cells per clone (n = 21 clones). In contrast, dFmr1 mutant NB generated significantly more cells, with an average of 47.0 cells per dFmr13 clone (n = 18 clones, P = 0.001), and an average of 50.5 cells per dFmr150M clone (n = 31 clones, P < 0.001) (Fig. 2M). Type II NB, identified by the presence of Miranda and lack of Asense (Fig. 2E–H), had an average of 96 cells per clone in the controls (n = 3 clones), while dFmr1 mutants had significantly higher numbers, with an average of 148 cells per clone for dFmr13 clones (n = 5, P = 0.02) and 141 cells per clone in dFmr150M clones (n = 4, P = 0.04) (Fig. 2N). The lower sample size for type II clones is attributed to the fact that there are only eight type II NB per brain lobe, compared with about 90 type I NB, which reduces considerably the frequency of such clones compared with type I neuroblast clones. Despite the smaller sample size for type II clones, loss of dFmr1 led to significantly larger clones, similar to our results obtained for type I NB. Importantly, no detectable changes were found in the dimensions of individual cells due to loss of dFmr1, indicating that FMRP controls cellular proliferation and not cell size in the developing larval brain (data not shown).
Figure 2.
FMRP regulates neural stem cell proliferation in the larval brain. (A–H) Single CB neuroblast clones (marked with GFP) in late third instar larval brains. Control clones [wild-type (A, B, E, F)] have fewer cells than dFmr1 mutant clones (C, D, G, H) after induction at first instar stage (4–10 h ALH). The type I neuroblast-specific marker, Asense, was used to differentiate type I (A–D) from type II (E–H) clones. Individual clone sizes were quantified by counting cells within each clone, grouped by genotype for type I (M) and type II (N) clones. Statistical significance (see text for P-values) was calculated using Student's t-test. Error bars indicate standard error of the mean. (I–L) Late clone induction (48–54 h ALH) results in single cell clones in both wild-type (I, J) and mutant (K, L) backgrounds in the ventral brain. Stainings, genotypes and clone induction regimes as indicated. Scale bar in (A) 25 µm, (I) 60 µm. Panels shown represent projections of two to three individual confocal slices, as needed, to include the entire clone.
Since both NB and GMCs are the actively proliferating cell types with the lineage, we next investigated whether the loss of dFmr1 leads to overproliferation of the GMCs as well. To test this, we induced GMC clones at 48–54 h ALH, when wild-type clones contain either one or two cells, depending on which cell underwent the Flp-mediated recombination event. As shown in Figure 2 (compare Fig. 2I and J with K and L), all mutant clones contained either one or two cells just like their wild-type counterparts (n = 5 wild-type, 11 dFmr13 and 6 dFmr150M clones). The inability to produce clones containing several cells from a single, dFmr1 mutant GMC supports the notion that FMRP controls cellular proliferation in NB only and has no effect on the proliferation potential of individual GMCs.
Loss of dFmr1 leads to supranumerary neurons
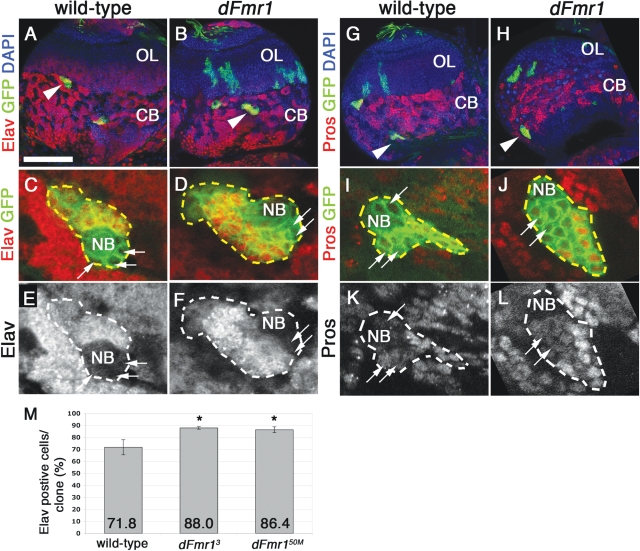
Our clonal analyses demonstrate that the loss of dFmr1 leads to increased numbers of cells within neuroblast lineages in the larval brain. An important question remaining is whether these supranumerary cells differentiate into neurons, retain their GMC fate or apoptose. To address this important issue, we examined the distribution of the GMC/neuronal marker Prospero, as well as the neuronal marker Elav. Prospero protein is inherited from the neuroblast and expressed in the GMC cytoplasm (21–23). Prospero then translocates into the nucleus and causes the GMC to exit the cell cycle, divide symmetrically one time and generate two daughter ganglion cells. As they acquire their neuronal fate, these two daughters begin expressing the neuronal marker Elav (24). Differentiated neurons should therefore express nuclear Prospero and Elav. Since type II NB generate complex lineages containing both neurons and glia (25), we focused our analyses on type I lineages, as marked by the presence of Asense in the neuroblast (data not shown). Immunohistochemistry experiments show that both wild-type and mutant type I clones contain a comparable number of GMCs surrounding the neuroblast as indicated by the presence of cytoplasmic Prospero (compare arrows in Fig. 3I and K with J and L) and the lack of Elav (compare arrows, Fig. 3C and E with D and F). The remainder of the cells within all clones examined expresses nuclear Prospero (Fig. 3K and L) as well as the neuronal marker Elav (Fig. 3E and F). Quantification of Elav positive cells per clone indicated that dFmr1 mutant clones contain significantly more neurons [88.0 ± 1.0% in dFmr13 (n = 4 clones, Pvalue = 0.02) and 86.4 ± 2.5% in dFmr150M (n = 7 clones, Pvalue = 0.05) compared with 71.8 ± 6.2% in wild-type clones (n = 6), see Fig. 3M]. Taken together, our data suggest that the supranumerary cells produced by type I dFmr1 mutant NB take on a neuronal fate, and predict that Fragile X brains may contain up to 16% more neurons, which could lead to wiring defects that precede the known synaptic dysfunction in FXS.
Figure 3.
Loss of dFmr1 causes an increase in the number of Elav positive neurons with no effect on Prospero expression. (A, B, G, H) Low magnification images of larval brains containing control (A and B) and dFmr1 (G and H) clones (see arrowheads). (C–F) High magnification views of representative clones show that Elav is expressed in all cells within the clone but the neuroblast (NB) and a few surrounding cells, which are presumably GMCs (see arrows). Quantification of the Elav positive cells/per clone results in a significant increase in both dFmr13 and dFmr150M clones compared with control (M). Student's t-test was used to calculate statistical significance. (G–L) Prospero stainings in control and dFmr1 clones show that a comparable number of cells surrounding the NB exhibit cytoplasmic Prospero corresponding to GMCs (arrows) with the remainder showing nuclear localization characteristic to differentiated neurons. Genotypes and stainings as indicated. Scale bar in (A) 100 µm. Panels shown represent projections of two to three individual confocal slices.
Supranumerary dFmr1 neurons survive into adulthood
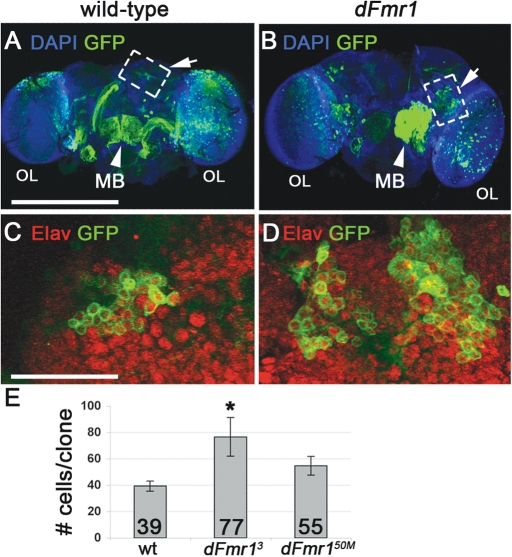
The fly brain undergoes a significant amount of neuronal remodeling during morphogenesis. To test whether the overproliferation of dFmr1 NB during larval development may impact neuronal circuitry in the adult brain, we set out to determine whether the supranumerary neurons found in dFmr1 mutant clones persist into adulthood. To address this issue, we induced clones at 4–10 h ALH as previously described, allowed the flies to develop into adults and quantified the size of NB clones in the CB. These experiments indicate that dFmr1 mutant clones not only persist into adulthood but they are larger than wild-type clones and contain supranumerary Elav positive neurons (Fig. 4). The average number of persisting GFP positive cells that also expressed the neuronal marker Elav in newly enclosed adults was higher in both dFmr13 clones (Fig. 4D and E, average number of neurons per clone = 76.7 ± 14, n = 8, Pvalue = 0.03) and dFmr150M clones (Fig. 4E, average number of neurons per clone = 54.8 ± 7, n = 10, Pvalue = 0.08) compared with controls (Fig. 4C and E, average number of neurons per clone = 39.4 ± 3.8, n = 8). Mushroom body (mb) clones as well as optic lobe (OL) clones are also visible (Fig. 4A and B), but were not quantified. Taken together, our data show that the loss of dFmr1 in neural stem cells early in development results in long-term defects in the size of neuronal populations, and suggest that wiring defects may precede the well-established synaptic plasticity defects found in Fragile X brains (26).
Figure 4.
Increased neurons in mutant clones survive to adulthood. (A and B) Low magnification images of adult brains, clones induced at 4–10 h ALH. (C and D) High magnification images indicate GFP positive cells also stain positive for neuronal marker Elav. (E) Quantification of adult clone numbers. Increased number of cells per clone is statistically significant in dFmr13 mutants compared with control clones. Scale bar in (A) 275 µm, (C) 35 µm. (A) and (B) are projections of the entire brain. (C) and (D) are projections of two to three individual confocal slices to capture entire clone.
Loss of dFmr1 does not affect the length of the cell cycle in third instar larval NB
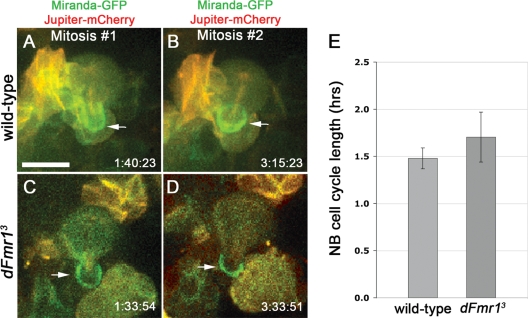
A possible explanation for the ability of dFmr1 NB to generate more daughters is that they divide faster. To address this possibility, we performed live imaging analyses of early third instar wild-type and dFmr1 mutant brains, which express the stem cell marker Miranda fused to GFP (Miranda-GFP) and the microtubule associated protein Jupiter fused to mCherry (Jupiter-mCherry) (27). We then measured the time between neuroblast mitoses in wild-type and dFmr1 mutant brains. As seen in Fig. 5, our live imaging experiments show that the loss of dFmr1 has no significant effect on the timing of the cell cycle in these explanted early third instar brains (n = 18 mitoses in wild-type and 27 mitoses in dFmr13, Pvalue = 0.36). Furthermore, no obvious defects in asymmetric cell division were noted in the mutant brains, at least in regard to Miranda distribution. These results, together with our clonal analyses, suggest that the defects seen in dFmr1 mutant brains must occur earlier during development.
Figure 5.
Live imaging of dividing neuroblasts indicates no change in the duration of the cell cycle in third instar dFmr1 mutant brains compared to wild-type. (A–D) Miranda-GFP (shown in green) and Jupiter-mCherry (shown in red) label neuroblasts, GMC daughters and mitotic spindles, respectively. Several mitoses were imaged live in cultured explanted brains. Two mitoses are shown at the time of GMC pinching off (arrows). Genotypes and division times as indicated. Note: dFmr1 neuroblasts shown were located deep within the brain, hence the lower quality image. (E) Quantification of cell cycle time shows no significant difference between wild-type (1.48 ± 0.11 h) and mutant brains (1.70 ± 0.26 h). Imaging was performed using early third instar brains. Student's t-test was used to calculate statistical significance (see text for P-value). Scale bar in (A) 10 µm.
Cyclin E is misexpressed and NB exit quiescence prematurely in dFmr1 brains
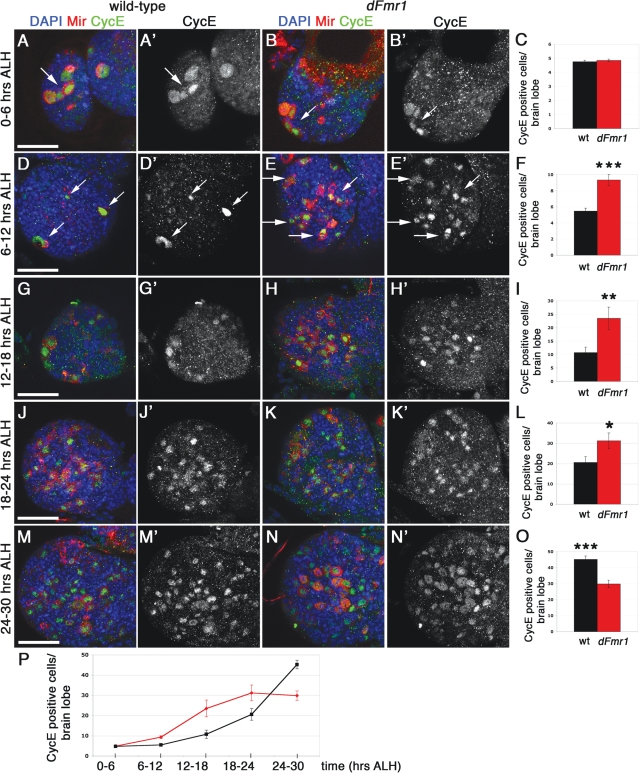
To test the possibility that FMRP controls proliferation earlier than the third instar stage, we set out to perform a careful developmental analysis of dFmr1 mutant brains compared with wild-type. It has been established that each brain lobe contains about 100 NB, which are generated during embryogenesis (28). At hatching, most NB are in a state of quiescence, with only four mb NB and one lateral neuroblast actively proliferating (29), and expressing the G1/S transition cyclin, Cyclin E (CycE) (Fig. 6A and B). As the larvae develop, more NB exit the state of quiescence and reenter the cell cycle to divide and generate multiple GMCs and neurons (28). Their proliferative capacity increases exponentially during first instar larval stages, then it reaches a saturation point during the third instar stage. The only NB known to continue their proliferation and differentiation program beyond third instar are the four mb NB that were active at larval hatching. In wild-type brains, NB normally begin to reenter the cell cycle by 8 h ALH (29) gradually increasing the number of proliferative NB. To determine whether FMRP regulates neuroblast exit from quiescence, we examined the localization of Miranda in conjunction with the cell cycle marker CycE in five consecutive 6 h windows during early larval brain development (0–30 h ALH). We discovered that the loss of FMRP impacts the timing of NB exiting quiescence after 6–12 h ALH. As seen in Fig. 6, at 0–6 h ALH, no significant differences were found between wild-type and mutant brains with regard to the number of Miranda/CycE positive NB (Fig. 6A–C, n = 24 wild-type and 32 dFmr13/50M brain lobes), with only the four mb NB and one lateral neuroblast (not shown) actively dividing. At 6–12 h ALH, however (Fig. 6D–F), there is a highly significant increase in the number of Miranda/CycE positive NB in the dFmr1 mutant brains (9.3 ± 0.7, n = 21, Pvalue < 0.001) compared with wild-type (5.5 ± 0.4, n = 15). At 12–18 h ALH (Fig. 6G–I), the dFmr1 mutant brains exhibit approximately 2-fold more Miranda/CycE positive NB (24.5 ± 4.2, n = 6, Pvalue = 0.007) compared with wild-type (10.8 ± 2.0, n = 8). By 18–24 h ALH (Fig. 6J–L), wild-type brains (20.0 ± 2.9, n = 7) begin to catch up to the mutants (31.2 ± 3.9, n = 10), although the difference is still significant at this time point (Pvalue = 0.05). Interestingly, 24–30 h ALH (Fig. 6M–O), there is a switch, with more wild-type brain NB (45.2 ± 2.6, n = 5) actively dividing compared with dFmr1 mutants (29.9 ± 2.3, n = 7). Compared with the profile of wild-type neuroblast exit from quiescence throughout the first 30 h ALH (Fig. 6P), dFmr1 mutant NB exhibit a precocious increase in the rate of exit from quiescence, followed by a decrease, possibly due to a homeostatic mechanism not yet understood.
Figure 6.
Loss of dFmr1 leads to premature exit from quiescence in the developing larval brain. Wild-type (w1118) and dFmr13/50M larval brains analyzed at different time points ALH, as shown. Brains were immunostained for Miranda and CycE (A–N′). DAPI was used to label nuclear DNA. (A–B′) At 0–6 h ALH, both wild-type (A, B, see arrows) and dFmr13/50M mutant brains (A′, B′, see arrows) display large, Miranda/CycE positive mb neuroblasts (NBs) only. (C) Quantification of Miranda/CycE double positive cells shows no significant change between genotypes at this timepoint. (D–E′) At 6–12 h ALH, wild-type brains contain four large mb NBs (D, D′, arrows). Note: only three out of the four mb NBs clearly visible in the single confocal slice shown. Mutant brains show an increased number of smaller, Miranda/CycE positive NBs (E, E′, arrows) compared with controls. (F) The total number of Miranda/CycE positive NBs at this time point is significantly increased in the dFmr13/50M mutant brains. (G–H′) At 12–18 h ALH, the number of Miranda/CycE positive cells continues to be significantly increased in dFmr13/50M mutant brains (H, H′), quantified in (I). (J–K′) At 18–24 h ALH, the number of Miranda/CycE positive cells remains significantly increased in dFmr13/50M mutant brains (K, K′), quantified in (L). (M–N′) At 24–30 h ALH, the number of Miranda/CycE positive cells is now significantly increased in w1118 brains (M, M′), compared with the mutant brains (N, N′), quantified in (O). (P) Line plot of average CycE/Miranda positive neuroblasts at each 6 h interval ALH. Student's t-test was used to calculate statistical significance (see text for P-values). Scale bars: (A) 30 µm, (D) 30 µm, (G) 30 µm, (J) 40 µm, (M) 40 µm. Panels (A–B′) represent projections of two to three individual confocal slices to capture all mb neuroblasts. All remaining images represent single confocal slices (2 µm thick).
Our results indicate that dFmr1 brains exhibit a misregulation of CycE expression, which in turn controls the G1/S transition during the cell cycle (30) and are consistent with our previous findings of CycE misexpression due to the loss of dFmr1 in the ovaries (7). Taken together, our data suggest that FMRP controls proliferation by regulating the timing of CycE expression in NB. Furthermore, these results suggest that FMRP controls the dynamic behavior of proliferating NB during brain development and provide further support to the notion that FMRP controls neurogenesis in the developing brain (Fig. 7 and Discussion below for model).
Figure 7.
Proposed model for FMRP function in the developing brain. FMRP controls the dynamic proliferative activity of neuroblasts during development. See text for details.
DISCUSSION
FMRP is an RNA-binding protein with an established role in the transport and translation of specific mRNAs in neurons (31–33). In addition to its established role in differentiated neuronal cells, FMRP has been recently implicated in proliferation and differentiation in neural progenitors, albeit with somewhat conflicting results (10–13). To determine the role of FMRP during early neurogenesis in vivo, we used loss of function and clonal approaches in the Drosophila larval brain. Here we show that whole dFmr1 mutant brains from late third instar larvae exhibit altered cell cycle profiles, with more cells found in S and G2/M at the expense of G1. These cell cycle defects indicate that FMRP is necessary for correct cell cycle progression in neural stem cells. Our developmental studies coupled with live imaging experiments indicate that FMRP controls the exit from quiescence and proliferative capacity of larval brain NB. To our knowledge, this is the first evidence that FMRP controls the exit from quiescence of neural progenitors in the developing brain. Such developmental defects could lead to significant problems in neural connections that are dependent on precise timing for proper function.
Our clonal analyses provide further support to these findings, by showing that dFmr1 mutant NB produce an increased number of neurons, which persist in the adult brain. In the future, it will be interesting to determine whether some neuronal populations are more sensitive to loss of FMRP than others and what are the functional consequences of having supranumerary neurons. Taken together, our results show that FMRP is required cell-autonomously for neurogenesis in vivo and are consistent with recent reports indicating that neural stem cell proliferation is increased (13) and that the density of intermediate progenitors and pyramidal cells is increased in the early postnatal cortex of FMR1 KO mice (12). In addition, our work suggest a mechanism involving the control of the G1/S transition point possibly through the regulation of CycE expression in NB during early larval brain development.
A model for FMRP function in the developing brain
Our data reveal a novel and surprising role for FMRP during brain development. In the young first instars, there are comparable numbers of Miranda/CycE expressing NB in the mutant compared with wild-type but only 6 h later (6–12 h ALH), there is a shift, with more dFmr1 mutant cells expressing Miranda/CycE compared with wild-type. The coexpression of the neuroblast marker Miranda and the G1/S transition marker CycE indicates that these additional cells correspond to NB exiting the G0 quiescence phase and entering the cell cycle (29). Based on these results, we propose that FMRP controls the timing of neuroblast exit from quiescence. It remains to be seen whether CycE is a direct target or whether FMRP acts through other factors such as E2F, Rb or the SCF/cullin complex [for review of CycE regulation, see Lee and Orr-Weaver (30)]. Interestingly, recent work in mice has shown that FMRP plays a role in adult neurogenesis by regulating the expression of cell cycle regulators such as Cyclin D and CDK4 (13).
While we found no significant change in the number of mitotic cells in early larval stages, at the onset of the third instar larval stage, we observed a slight increase in PH3 positive cells in dFmr1 mutant brains compared with wild-type (data not shown). In late third instar larval brains, however, the difference in PH3 positive NB became statistically significant, which could be accounted for by seemingly opposite scenarios whereby the mutant NB either divide faster, or are progressing through mitosis at a slower pace. It remains to be determined what mRNA targets mediate FMRP's role in the timing of mitotic events. Given that dFmr1 mutant NB generate significantly larger lineages containing more neurons and that the live imaging experiments showed no change in the total length of the cell cycle of third instars, we propose a model whereby FMRP controls neuroblast proliferation during early brain development by regulating the timing of reentry into the cell cycle. This is supported by our developmental studies showing misexpression of CycE, which suggests a premature exit from quiescence due to the loss of dFmr1. Our clonal analyses indicate that dFmr1 mutant NB produce on average 15–16 more neurons than wild-type by the end of third instar. This correlates with the mutant neuroblast completing approximately eight more divisions than its wild-type counterparts during larval development (every two neurons are the result of a single neuroblast division). By the end of the larval stage, however, the proliferative activity of dFmr1 NB is comparable to that of their wild-type counterparts, which supports the notion that these additional cell cycles are restricted to the early stages of brain development (see Fig. 7 for proposed model).
Lessons from the Drosophila brain
Neural stem cell proliferation and differentiation are the basis for generating the correct number of neurons and support cells in the developing nervous system. The Drosophila larval brain NB have emerged as a premiere model for neural stem cells that have been successfully used to elucidate the molecular mechanisms underlying stem cell renewal and differentiation in the brain (34). The fly larval type I NB give rise to only neurons as opposed to both neurons and glia, while type II NB give rise to both (25), as is the case with mammalian neural stem cells. Despite some differences, it is clear that Drosophila NB follow general stem cell principles and utilize conserved pathways. Given the powerful genetic tools available in the fly, including the ability to study individual neuroblast lineages, the clues obtained from the fly are likely to provide useful insights into mammalian brain development and the mechanisms for RNA regulation in neural stem cells.
Our clonal analyses demonstrate that dFmr1 mutant NB generate more neurons than their wild-type counterparts. These findings are consistent with previous in vitro studies using neurospheres (10) as well as a recent study using the FMR1 KO mouse (12), both of which reported the presence of supranumerary neurons in the absence of FMRP. Recently, the loss of FMRP from adult neural progenitors was shown to generate more glial cells at the expense of neurons (13). The differences in neuronal numbers between our study and that of Luo et al. (13) may reflect inherent differences between the fly and mouse models. Alternatively, FMRP has a developmental component and exerts distinct modes of regulation on neural stem cell proliferation and differentiation during the various stages of brain development. Supporting this notion is the study by Castren et al. (10), which found more neurons at the expense of glia in embryonically derived neurospheres lacking FMRP.
Quantification of the Elav positive cells within clones suggests that dFmr1 NB generate an estimated 16% more neurons. With approximately 15 000 neurons present in the larval brain (35) and considering that each brain lobe is shaped as a sphere, a 16% increase in neurons is predicted to amount to a 1.13 change in the area occupied by the brain when mounted on a microscope slide (1.13 = ratio between mutant and wild-type surface area of two spheres that differ by 16% in volume). While such a small increase in brain size may go unnoticed by the experimentalist, inter-neuronal connections and circuit formation are likely to be more sensitive. The presence of wiring defects would suggest that the loss of FMRP affects brain development earlier than previously thought and may account for the autistic component of FXS.
Finally, our finding that FMRP regulates the timing of neuroblast reentry into the cell cycle is particularly interesting, as this critical aspect of brain development remains poorly understood. Our whole brain analyses suggest that FMRP may control the exit from quiescence by regulating the expression of CycE. These results are consistent with our previous findings that the loss of dFmr1 leads to CycE misexpression in the fly ovary (7). Another interesting parallel to our oogenesis study is that loss of FMRP leads to both an increase and a delay in proliferation. In the larval brain, we detected an increase in proliferation during early larval stages, followed by a slowdown towards the end of the larval life. This suggests the presence of compensatory mechanisms that alleviate the consequences due to loss of FMRP in the brain and provide an explanation for the lack of an obvious effect on brain size in dFmr1 mutants.
Previous studies have shown that neuroblast exit from quiescence is controlled in part by the glycoprotein Anachronism (Ana), which is secreted by the surrounding glial cells (36). Ana's role in neuroblast proliferation suggests that stem cell proliferation in the brain is controlled by its microenvironment. Whether FMRP is required in glial cells remains to be elucidated. In further support of this notion, a recent study has implicated both Branchless and Hedgehog signaling pathways in NB' exit from quiescence (37). It will be interesting to see whether FMRP cooperates with these genes or utilizes distinct mechanisms to regulate neural stem cell quiescence during brain development.
MATERIALS AND METHODS
Drosophila stocks
Fly stocks were maintained at 25°C on standard cornmeal agar medium. All larvae and adults were obtained from 6 h egg collections to minimize developmental effects. The dFmr13/Df 6265 mutant was produced by crossing w1118;; dFmr13/TM6C Tb Sb with the deficiency Df 6265/TM6B Tb Hu, which removes the dFmr1 locus. The dFmr1 genomic rescue line (w1118; P[dFmr1+; dFmr13/TM6C Sb Tb) was kindly provided by Dr Tom Jongens, University of Pennsylvania. MARCM clones were generated using the following stocks (kindly provided by C.-Y. Lee, University of Michigan): elav Gal4 UAS mCD8-GFP hsflp;; FRT82B Tub Gal80/TM6B Tb Hu crossed with w1118;;[P neoFRT]82B P[Ubi-GFP.D]83] (for control clones) or w1118;; FRT82B dFmr13/TM6B-GFP and w1118;;FRT82B dFmr150M/TM6B-GFP (for dFmr1 clones). The FRT82B dFmr1 stocks were generously provided by K. Broadie (Vanderbilt University). For developmental studies, transheterozygote larvae were produced by crossing w1118;;FRT82B dFmr13/TM6B-GFP with w1118;;FRT82B dFmr150M/TM6B-GFP. Timed larvae were selected against GFP, then dissected and processed as described below.
For live imaging experiments, yw; wor-Gal4, UAS-cherry::Jupiter, UAS-GFP::Mira was used as a wild-type control. For mutant studies, the control stock was introduced into a dFmr13 background by standard genetic techniques. Larval brains were imaged live for several hours as previously described (27).
Brain squashes
Whole brain squashes were prepared as described in Pimpinelli et al. (38) (protocol 1.9, method 3 w/o steps necessary for immunodetection). Briefly, DAPI stained late third instar larval brains were squashed between the slide and the coverslip to visualize the chromosome morphology. Mitotic figures were counted manually using a Nikon E800 (Nikon Instruments Inc.) with standard filters for DAPI, FITC and Cy3.
Mosaic analysis with a repressible cell marker (MARCM) clones
MARCM clones were generated as described (16). In brief, clones were induced by placing the larvae at 37°C for 1 h at 4–10 or 48–54 h ALH. Brains were dissected from larvae of the appropriate genotype at late third instar stage or from newly enclosed adults. Brains were immunolabeled and imaged as described below.
Flow cytometry
To detect the DNA content of larval brain cells, 10 third instar larvae brains were dissected in Grace's insect cell culture medium (GIBCO). After removal of Grace's, 700 µl of filtered ice-cold Partec Buffer (200 mm Tris–HCl, pH 7.5, 4 mm MgCl2, 0.1% Triton X-100) was added to the brains. The samples (three biological replicates) were processed as previously described (7). After adding DAPI (100 µg/ml), samples were kept on ice for 60 min before analysis on a BD FACS Aria high-speed cell sorter (BD BioSciences). As controls, we used either dFmr1 heterozygous larvae processed separately or a Histone2A-GFP control line processed simultaneously with the mutant samples.
Antibodies
Antibodies used in this study were: Primary antibodies: rabbit anti-Asense (1:400, a gift from Cheng-Yu Lee, University of Michigan), rat anti-Miranda (1:100), rat anti-Prospero (1:100, Developmental Studies Hybridoma Bank), rabbit anti-PH3 (1:500, Millipore, Temecula, CA, USA), mouse anti-Elav (1:500, Developmental Studies Hybridoma Bank), rabbit anti-Lgl [1:500, see Zarnescu et al. (39)], mouse anti-BrdU [1:20, BD Bioscience, San Jose, CA, see Gratzner (40)], guinea pig anti-CycE (1:500, a gift from Giovanni Bosco, University of Arizona and Terry Orr-Weaver, MIT) and mouse anti-FMRP (6A15 at 1:500, Abcam). DNA was visualized using DAPI (Sigma-Aldrich, St Louis, MO, USA). Secondary antibodies were goat anti-mouse, rabbit or guinea pig IgG, coupled to the FITC, Alexa 568, 594 or 633 fluorophores and used in conjunction with the appropriate primary antibodies at 1:1000 (1:250 for FITC goat anti-guinea pig).
Immunohistochemistry
Staged larvae were dissected in PBS, and the central nervous system was immediately fixed in 4% formaldehyde (Ted Pella, Inc.) in PBS for 20 min. All incubations were performed at room temperature unless otherwise noted. After three washes in PBS (10 min each), the brains were permeabilized in 0.3% Triton X-100 in PBS (PBT) with 3 × 10 min washes. Non-specific binding was blocked with 10% normal goat serum (in PBT) for 30 min. Primary antibody incubation was performed at 4°C, overnight. After three washes (10 min each), the brains were blocked again for 30 min. Secondary antibody incubations were performed for 3 h. DAPI was added to the secondary antibody solution for the final 30 min. Following three washes in PBS (10 min each), the samples were mounted in Vectashield (Vector Labs, Orton Southgate, UK) and imaged as described below.
For BrdU incorporation, brains were dissected in Grace's insect Media, and incubated for 30 min in 5 mg/ml BrdU/Grace's. Following three washes in Grace's, the brains were fixed in 4% formaldehyde (in PBS). Immunostaining was performed as described above, with the addition of a 30 min 2 N HCl treatment before blocking and primary antibody incubation.
Imaging
Images were acquired using a Zeiss Axiovert 200 510 LSM META confocal microscope (Carl Zeiss Inc., Thornwood, NY, USA), using 40× and 63× objectives. Confocal slices were acquired every 2 µm when using the 40× objective and every 1 µm when using the 63× objective. Images were processed using LSM software and Adobe Photoshop (Adobe).
Live imaging
Live imaging was performed as previously described (27). In brief, early third instar larvae (48–72 ALH) were dissected and mounted in Schneider's insect media (Sigma) supplemented with 1% bovine growth serum (BGS; HyClone), 0.5 mm ascorbic acid and the fat bodies of 10 wild-type larvae. Movies were acquired on a McBain spinning disc confocal microscope equipped with a Hamamatsu EM-CCD camera, using a 63× 1.4NA oil-immersion lens.
FUNDING
This work was supported by the Arizona Biomedical Research Commission (ABRC, contract no. 820, to D.C.Z.), by the Howard Hughes Medical Institute (to C.Q.D.), by the National Institutes of Health (training grant no. GM08569 to M.A.C.), by the American Heart Association (postdoctoral fellowship no. 09POST2250423 to C.C.) and in part by the Undergraduate Biology Research Program funded by the Howard Hughes Medical Institute (no. 52003740 to J.H.).
ACKNOWLEDGEMENTS
We are grateful to the following colleagues for sharing reagents and/or providing advice: Tom Jongens (University of Pennsylvania), Kendal Broadie (Vanderbilt University), Cheng-Yu Lee (University of Michigan), Giovanni Bosco (University of Arizona), Terry Orr-Weaver (MIT), Barb Carolus (Flow Cytometry Facility, University of Arizona), Carl Boswell (Imaging Facility, MCB UA) and JMST discussion group (University of Arizona). We acknowledge the Developmental Studies Hybridoma Bank (University of Iowa) for Pros and Elav monoclonal antibodies. We would like to thank Brad Davidson and Jeanne Louderbough for helpful comments on the manuscript.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Penagarikano O., Mulle J.G., Warren S.T. The pathophysiology of fragile X syndrome. Annu. Rev. Genomics Hum. Genet. 2007;8:109–129. doi: 10.1146/annurev.genom.8.080706.092249. doi:10.1146/annurev.genom.8.080706.092249. [DOI] [PubMed] [Google Scholar]
- 2.Zhang Y.Q., Bailey A.M., Matthies H.J., Renden R.B., Smith M.A., Speese S.D., Rubin G.M., Broadie K. Drosophila fragile X-related gene regulates the MAP1B homolog Futsch to control synaptic structure and function. Cell. 2001;107:591–603. doi: 10.1016/s0092-8674(01)00589-x. doi:10.1016/S0092-8674(01)00589-X. [DOI] [PubMed] [Google Scholar]
- 3.Reeve S.P., Bassetto L., Genova G.K., Kleyner Y., Leyssen M., Jackson F.R., Hassan B.A. The Drosophila fragile X mental retardation protein controls actin dynamics by directly regulating profilin in the brain. Curr. Biol. 2005;15:1156–1163. doi: 10.1016/j.cub.2005.05.050. doi:10.1016/j.cub.2005.05.050. [DOI] [PubMed] [Google Scholar]
- 4.Schenck A., Bardoni B., Langmann C., Harden N., Mandel J.L., Giangrande A. CYFIP/Sra-1 controls neuronal connectivity in Drosophila and links the Rac1 GTPase pathway to the fragile X protein. Neuron. 2003;38:887–898. doi: 10.1016/s0896-6273(03)00354-4. doi:10.1016/S0896-6273(03)00354-4. [DOI] [PubMed] [Google Scholar]
- 5.Zalfa F., Eleuteri B., Dickson K.S., Mercaldo V., De Rubeis S., di Penta A., Tabolacci E., Chiurazzi P., Neri G., Grant S.G., et al. A new function for the fragile X mental retardation protein in regulation of PSD-95 mRNA stability. Nat. Neurosci. 2007;10:578–587. doi: 10.1038/nn1893. doi:10.1038/nn1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Muddashetty R.S., Kelic S., Gross C., Xu M., Bassell G.J. Dysregulated metabotropic glutamate receptor-dependent translation of AMPA receptor and postsynaptic density-95 mRNAs at synapses in a mouse model of fragile X syndrome. J. Neurosci. 2007;27:5338–5348. doi: 10.1523/JNEUROSCI.0937-07.2007. doi:10.1523/JNEUROSCI.0937-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Epstein A.M., Bauer C., Ho A., Bosco G., Zarnescu D.C. Fragile X protein controls cellular proliferation by regulating cbl levels in the ovary. Dev. Biol. 2009;330:83–92. doi: 10.1016/j.ydbio.2009.03.011. doi:10.1016/j.ydbio.2009.03.011. [DOI] [PubMed] [Google Scholar]
- 8.Costa A., Wang Y., Dockendorff T.C., Erdjument-Bromage H., Tempst P., Schedl P., Jongens T.A. The Drosophila fragile X protein functions as a negative regulator in the orb autoregulatory pathway. Dev. Cell. 2005;8:331–342. doi: 10.1016/j.devcel.2005.01.011. doi:10.1016/j.devcel.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 9.Yang L., Duan R., Chen D., Wang J., Chen D., Jin P. Fragile X mental retardation protein modulates the fate of germline stem cells in Drosophila. Hum. Mol. Genet. 2007;15:1814–1820. doi: 10.1093/hmg/ddm129. [DOI] [PubMed] [Google Scholar]
- 10.Castren M., Tervonen T., Karkkainen V., Heinonen S., Castren E., Larsson K., Bakker C.E., Oostra B.A., Akerman K. Altered differentiation of neural stem cells in fragile X syndrome. Proc. Natl Acad. Sci. USA. 2005;102:17834–17839. doi: 10.1073/pnas.0508995102. doi:10.1073/pnas.0508995102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bhattacharyya A., McMillan E., Wallace K., Tubon T.C., Jr, Capowski E.E., Svendsen C.N. Normal neurogenesis but abnormal gene expression in human fragile X cortical progenitor cells. Stem Cells Dev. 2008;17:107–117. doi: 10.1089/scd.2007.0073. doi:10.1089/scd.2007.0073. [DOI] [PubMed] [Google Scholar]
- 12.Tervonen T.A., Louhivuori V., Sun X., Hokkanen M.E., Kratochwil C.F., Zebryk P., Castren E., Castren M.L. Aberrant differentiation of glutamatergic cells in neocortex of mouse model for fragile X syndrome. Neurobiol. Dis. 2009;33:250–259. doi: 10.1016/j.nbd.2008.10.010. doi:10.1016/j.nbd.2008.10.010. [DOI] [PubMed] [Google Scholar]
- 13.Luo Y., Shan G., Guo W., Smrt R.D., Johnson E.B., Li X., Pfeiffer R.L., Szulwach K.E., Duan R., Barkho B.Z., et al. Fragile x mental retardation protein regulates proliferation and differentiation of adult neural stem/progenitor cells. PLoS Genet. 2010;6:e1000898. doi: 10.1371/journal.pgen.1000898. doi:10.1371/journal.pgen.1000898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chia W., Somers W.G., Wang H. Drosophila neuroblast asymmetric divisions: cell cycle regulators, asymmetric protein localization, and tumorigenesis. J. Cell. Biol. 2008;180:267–272. doi: 10.1083/jcb.200708159. doi:10.1083/jcb.200708159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wan L., Dockendorff T.C., Jongens T.A., Dreyfuss G. Characterization of dFMR1, a Drosophila melanogaster homolog of the fragile X mental retardation protein. Mol. Cell. Biol. 2000;20:8536–8547. doi: 10.1128/mcb.20.22.8536-8547.2000. doi:10.1128/MCB.20.22.8536-8547.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee T., Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. doi:10.1016/S0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- 17.Doe C.Q. Molecular markers for identified neuroblasts and ganglion mother cells in the Drosophila central nervous system. Development. 1992;116:855–863. doi: 10.1242/dev.116.4.855. [DOI] [PubMed] [Google Scholar]
- 18.Boone J.Q., Doe C.Q. Identification of Drosophila type II neuroblast lineages containing transit amplifying ganglion mother cells. Dev. Neurobiol. 2008;68:1185–1195. doi: 10.1002/dneu.20648. doi:10.1002/dneu.20648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bello B.C., Izergina N., Caussinus E., Reichert H. Amplification of neural stem cell proliferation by intermediate progenitor cells in Drosophila brain development. Neural. Dev. 2008;3:5. doi: 10.1186/1749-8104-3-5. doi:10.1186/1749-8104-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bowman S.K., Rolland V., Betschinger J., Kinsey K.A., Emery G., Knoblich J.A. The tumor suppressors Brat and Numb regulate transit-amplifying neuroblast lineages in Drosophila. Dev. Cell. 2008;14:535–546. doi: 10.1016/j.devcel.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Broadus J., Fuerstenberg S., Doe C.Q. Staufen-dependent localization of prospero mRNA contributes to neuroblast daughter-cell fate. Nature. 1998;391:792–795. doi: 10.1038/35861. [DOI] [PubMed] [Google Scholar]
- 22.Ikeshima-Kataoka H., Skeath J.B., Nabeshima Y., Doe C.Q., Matsuzaki F. Miranda directs Prospero to a daughter cell during Drosophila asymmetric divisions. Nature. 1997;390:625–629. doi: 10.1038/37641. [DOI] [PubMed] [Google Scholar]
- 23.Spana E.P., Doe C.Q. The prospero transcription factor is asymmetrically localized to the cell cortex during neuroblast mitosis in Drosophila. Development. 1995;121:3187–3195. doi: 10.1242/dev.121.10.3187. [DOI] [PubMed] [Google Scholar]
- 24.Robinow S., White K. Characterization and spatial distribution of the ELAV protein during Drosophila melanogaster development. J. Neurobiol. 1991;22:443–461. doi: 10.1002/neu.480220503. doi:10.1002/neu.480220503. [DOI] [PubMed] [Google Scholar]
- 25.Izergina N., Balmer J., Bello B., Reichert H. Postembryonic development of transit amplifying neuroblast lineages in the Drosophila brain. Neural. Dev. 2009;4:44. doi: 10.1186/1749-8104-4-44. doi:10.1186/1749-8104-4-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Comery T.A., Harris J.B., Willems P.J., Oostra B.A., Irwin S.A., Weiler I.J., Greenough W.T. Abnormal dendritic spines in fragile X knockout mice: maturation and pruning deficits. Proc. Natl Acad. Sci. USA. 1997;94:5401–5404. doi: 10.1073/pnas.94.10.5401. doi:10.1073/pnas.94.10.5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cabernard C., Doe C.Q. Apical/basal spindle orientation is required for neuroblast homeostasis and neuronal differentiation in Drosophila. Dev. Cell. 2009;17:134–141. doi: 10.1016/j.devcel.2009.06.009. doi:10.1016/j.devcel.2009.06.009. [DOI] [PubMed] [Google Scholar]
- 28.Younossi-Hartenstein A., Nassif C., Green P., Hartenstein V. Early neurogenesis of the Drosophila brain. J. Comp. Neurol. 1996;370:313–329. doi: 10.1002/(SICI)1096-9861(19960701)370:3<313::AID-CNE3>3.0.CO;2-7. doi:10.1002/(SICI)1096-9861(19960701)370:3<313::AID-CNE3>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 29.Ito K., Hotta Y. Proliferation pattern of postembryonic neuroblasts in the brain of Drosophila melanogaster. Dev. Biol. 1992;149:134–148. doi: 10.1016/0012-1606(92)90270-q. doi:10.1016/0012-1606(92)90270-Q. [DOI] [PubMed] [Google Scholar]
- 30.Lee L.A., Orr-Weaver T.L. Regulation of cell cycles in Drosophila development: intrinsic and extrinsic cues. Annu. Rev. Genet. 2003;37:545–578. doi: 10.1146/annurev.genet.37.110801.143149. doi:10.1146/annurev.genet.37.110801.143149. [DOI] [PubMed] [Google Scholar]
- 31.Estes P.S., O'Shea M., Clasen S., Zarnescu D.C. Fragile X Protein controls the efficacy of mRNA transport in Drosophila neurons. Mol. Cell. Neurosci. 2008;39:170–179. doi: 10.1016/j.mcn.2008.06.012. doi:10.1016/j.mcn.2008.06.012. [DOI] [PubMed] [Google Scholar]
- 32.Dictenberg J.B., Swanger S.A., Antar L.N., Singer R.H., Bassell G.J. A direct role for FMRP in activity-dependent dendritic mRNA transport links filopodial-spine morphogenesis to fragile X syndrome. Dev. Cell. 2008;14:926–939. doi: 10.1016/j.devcel.2008.04.003. doi:10.1016/j.devcel.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zalfa F., Bagni C. Another view of the role of FMRP in translational regulation. Cell. Mol. Life Sci. 2005;62:251–252. doi: 10.1007/s00018-004-4521-1. doi:10.1007/s00018-004-4521-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu F., Kuo C.T., Jan Y.N. Drosophila neuroblast asymmetric cell division: recent advances and implications for stem cell biology. Neuron. 2006;51:13–20. doi: 10.1016/j.neuron.2006.06.016. doi:10.1016/j.neuron.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 35.Truman J.W., Taylor B.J., Awad T.A. Bate M., Arias A.M. The Development of Drosophila melanogaster. 1993. Formation of the adult nervous system; pp. 1245–1275. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, Vol. II. [Google Scholar]
- 36.Ebens A.J., Garren H., Cheyette B.N., Zipursky S.L. The Drosophila anachronism locus: a glycoprotein secreted by glia inhibits neuroblast proliferation. Cell. 1993;74:15–27. doi: 10.1016/0092-8674(93)90291-w. doi:10.1016/0092-8674(93)90291-W. [DOI] [PubMed] [Google Scholar]
- 37.Barrett A.L., Krueger S., Datta S. Branchless and Hedgehog operate in a positive feedback loop to regulate the initiation of neuroblast division in the Drosophila larval brain. Dev. Biol. 2008;317:234–245. doi: 10.1016/j.ydbio.2008.02.025. doi:10.1016/j.ydbio.2008.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pimpinelli S., Bonaccorsi S., Fanti L., Maurizio G. Preparation and analysis of Drosophila mitotic chromosomes. In: Sullivan W., Ashburner M., Hawley R.S., editors. Drosophila Protocols. Cold Spring Harbor (New York): Cold Spring Harbor Laboratory Press.; 2000. pp. 3–23. [Google Scholar]
- 39.Zarnescu D.C., Jin P., Betschinger J., Nakamoto M., Wang Y., Dockendorff T.C., Feng Y., Jongens T.A., Sisson J.C., Knoblich J.A., et al. Fragile X protein functions with lgl and the par complex in flies and mice. Dev. Cell. 2005;8:43–52. doi: 10.1016/j.devcel.2004.10.020. doi:10.1016/j.devcel.2004.10.020. [DOI] [PubMed] [Google Scholar]
- 40.Gratzner H.G. Monoclonal antibody to 5-bromo- and 5-iododeoxyuridine: a new reagent for detection of DNA replication. Science. 1982;218:474–475. doi: 10.1126/science.7123245. doi:10.1126/science.7123245. [DOI] [PubMed] [Google Scholar]