Abstract
Cytochrome P450 monooxygenases play a prominent role in the biosynthesis of the diterpenoid anticancer drug Taxol, as they appear to constitute about half of the 19 enzymatic steps of the pathway in yew (Taxus) species. A combination of classical biochemical and molecular methods, including cell-free enzyme studies and differential-display of mRNA-reverse transcription polymerase chain reaction (RT-PCR) combined with a homology-based searching and random sequencing of a cDNA library from induced T. cuspidata cells, led to the discovery of six novel cytochrome P450 taxoid (taxane diterpenoid) hydroxylases. These genes show unusually high sequence similarity with each other (>70%) but low similarity (<30%) to, and significant evolutionary distance from, other plant P450s. Despite their high similarity, functional analysis of these hydroxylases demonstrated distinctive substrate specificities responsible for an early bifurcation in the biosynthetic pathway after the initial hydroxylation of the taxane core at C5, leading into a biosynthetic network of competing, but interconnected, branches. The use of surrogate substrates, in cases where the predicted taxoid precursors were not available, led to the discovery of two core oxygenases, the 2α- and the 7β-hydroxylase. This general approach could accelerate the functional analysis of candidate cDNAs from the extant family of P450 genes to identify the remaining oxygenation steps of this complex pathway.
Keywords: Taxoid hydroxylases, Taxane diterpenoid, Taxadiene, Taxusin, Taxus
Introduction
The highly functionalized diterpenoid Taxol is one of about 360 known taxoid (taxane diterpenoid) secondary metabolites produced by Taxus species (Baloglu and Kingston 1999; Itokawa 2003). After the discovery of the antineoplastic potential of Taxol and the elucidation of its complex structure (Wall 1998; Brown 2003), this natural product developed into one of the most powerful drugs now available for the treatment of refractory ovarian and metastatic breast cancers (Suffness and Wall 1995) when used alone or in combination therapies with other antineoplastic agents such as platinum compounds (Brown 2003). Taxol also represents a platform for the development of the next generation of taxoid drugs and prodrugs (Wang et al. 2003), for which the total market is expected to expand three-fold in the next several years (McCoy 2004). For the production of Taxol, elegant total syntheses have been devised [see (Kington 1995) for review] but they are too costly to be commercially viable. The supply of Taxol must therefore rely on the isolation of the drug and its immediate precursors from Taxus plants (Croom 1995; Kikuchi and Yatagai 2003) or cell cultures (Gibson et al. 1995; Takeya 2003). For the improvement of these production processes, an understanding of Taxol biosynthesis, the pathway enzymes, and the corresponding structural genes is essential.
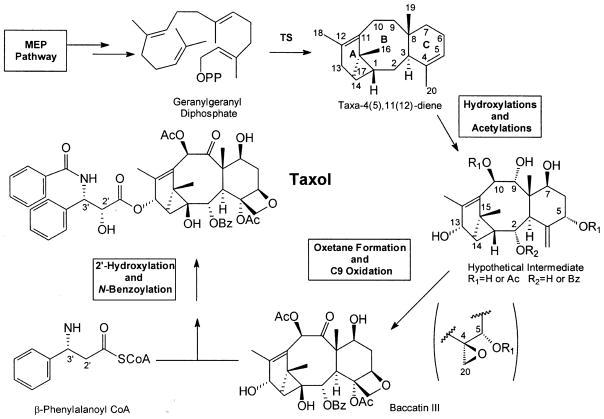
Typical of plant secondary metabolism (Schuler 1996), cytochrome P450-mediated oxygenations play a mayor role in Taxol biosynthesis. Approximately one-half of the proposed 19 distinct enzymatic steps of the pathway are considered to be catalyzed by cytochrome P450 oxygenases (Croteau et al. 2006). The committed step of the Taxol biosynthetic pathway (Koepp et al. 1995; Hezari et al. 1995) is the cyclization of the universal diterpenoid precursor geranylgeranyl diphosphate to taxa-4(5),11(12)-diene, possessing the unique taxane core structure, which is then decorated with eight oxygen functional groups (six alcohol functions, one carbonyl and one cyclic ether). Two subsequent acetylations, a benzoylation, oxetane ring formation, and oxidation at the C9 lead to the late intermediate baccatin III, to which the C13-side chain is attached (derived by transfer from β-phenylalanoyl CoA, followed by 2′-hydroxylation and N-benzoylation) to complete the pathway (Fig. 1). The order of the oxygenations on the parental taxane core can be roughly predicted based on the relative abundances of the several hundred defined taxoids bearing oxy-functional groups at the various positions (Floss and Mocek 1995; Ketchum and Croteau 2006). The sequence of reactions begins with C5 then C10, followed by C13 and C9, and later C7 and C2. The final reactions involve the epoxidation of the C4,C20-double bond (ultimately leading to the oxetane ring) and C1 oxygenation; the precise timing of the acetylations at C5 and C10, and the benzoylation at C2, is uncertain. In this review, we provide a more detailed description of the enzymes and genes of this novel cytochrome P450 taxane oxygenase subfamily.
Fig. 1.
Outline of Taxol biosynthesis (TS: taxadiene synthase)
Approaching taxane cytochrome P450 oxygenases
The first investigations on the hydroxylations of the taxane core were performed with cell-free preparations from T. brevifolia and T. cuspidata (Hefner et al. 1996; Lovy Wheeler et al. 2001) and demonstrated that most if not all of these reactions were conducted by microsomal cytochrome P450 oxygenases, a finding that led to several cloning strategies to acquire the corresponding cDNAs. Initially, a differential display of mRNA-reverse transcription-polymerase chain reaction (DD-RT-PCR) method was employed (Schoendorf et al. 2001) using transcripts isolated from methyl jasmonate-induced T. cuspidata cells versus uninduced cells (Ketchum et al. 1999). This was followed by an additional, homology-based screen and by random sequencing of the same cDNA library from Taxus cuspidata from which over two dozen closely related cytochrome P450 candidate clones were obtained (Jennewein et al. 2004a; Jennewein et al. 2004b). For functional assessment with taxoid precursors, these clones were expressed in Saccharomyces cerevisiae WAT11 cells that coexpress an Arabidopsis NADPH:cytochrome P450 reductase (Pompon et al. 1996), and were screened by in vivo feeding of the transformed yeast cultures, or were expressed using the baculovirus-Spodoptera system (Kutchan et al. 1994) from which microsomes were isolated and assayed.
Functional evaluation of this group of clones led to the acquisition of the taxoid 2α-, 5α-, 7β-, 10β-, 13α-, and 14β-hydroxylases, representing a family of cytochrome P450 sequences with high deduced similarity to each other (>70%) but with limited resemblance (< 30% similarity) to other genes of this type (Jennewein et al. 2004b). The typical conserved structural elements of cytochrome P450 oxygenases (von Wachtenfeldt and Johnson 1995; Kahn and Durst 2000) are present in these Taxus P450s (encoding proteins from 54 to 57 kDa), including the heme-binding motif with PFG element (around position aa420), the PERF motif (~aa410 as the ‘PSRF motif’), the oxygen and reductase binding motifs, the essential cysteine position (~aa440), and an EXXR salt bridge (~aa360). All of these hydroxylases encode a putative N-terminal membrane (endoplasmic reticulum) insertion sequence which is somewhat variable between acquisitions (Jennewein et al. 2003).
Because of its essential role in electron transfer in cytochrome P450-mediated oxygenations, the NADPH:cytochrome P450 reductase gene from T. cuspicata was also isolated (Jennewein et al. 2004b), and has been useful in functional screening of oxygenase clones. This reductase has been co-expressed in yeast and in Spodoptera cells to improve the catalytic efficiency of expressed Taxus P450 oxygenases (Jennewein et al. 2005), and as a preliminary step to reconstructing taxoid biosynthesis in yeast (De Jong et al. 2006).
Gene function and enzyme characteristics
Early oxygenation steps (C5-, C10-, C13-, C9- and C14-hydroxylases)
The first hydroxylation of the Taxol pathway for the conversion of taxa-4(5),11(12)-diene to taxa-4(20),11(12)-dien-5α-ol (Hefner et al. 1996), and as many as five additional oxygenation steps (Lovy Wheeler et al. 2001), were initially observed with T. cuspidata microsomes using standard cytochrome P450 assay conditions. The demonstration of this reaction sequence leading to a taxadien-hexaol suggested that most, if not all, of the relevant taxane core hydroxylations were cytochrome P450-mediated and localized to the endoplasmic reticulum.
The cDNA for the taxa-4(5),11(12)-diene 5α-hydroxylase was obtained in the homology-based screen and encodes a 57 kDa enzyme that utilizes taxa-4(5),11(12)-diene (with Ks of 5–8 μM and Vrel of 100) but also accepts taxa-4(20),11(12)-diene (with Ks of 3–5 μM and Vrel of 135) as substrate (Jennewein et al. 2004a). The mechanism of the hydroxylation reaction involves an unusual allylic transposition of the 4,5-double bond, presumably via a common delocalized radical intermediate derived from either olefin substrate (Fig. 2). The taxadiene-5α-hydroxylase transcript shows a modest abundance (0.8‰) in the induced Taxus cell EST library and the encoded enzyme possesses a low kcat, suggesting that 5α-hydroxylation is a slow step of the Taxol pathway. Nevertheless, most of the known taxoids are functionalized at C5 (Itokawa 2003) indicating that the 5α-hydroxylase is catalytically efficient in vivo. Because taxa-4(20),11(12)-dien-5α-ol is found only in trace amounts in Taxus (Ketchum and Croteau 2006; Hefner et al. 1996), this intermediate is likely rapidly consumed in subsequent acylation and/or hydroxylation reactions.
Fig. 2.
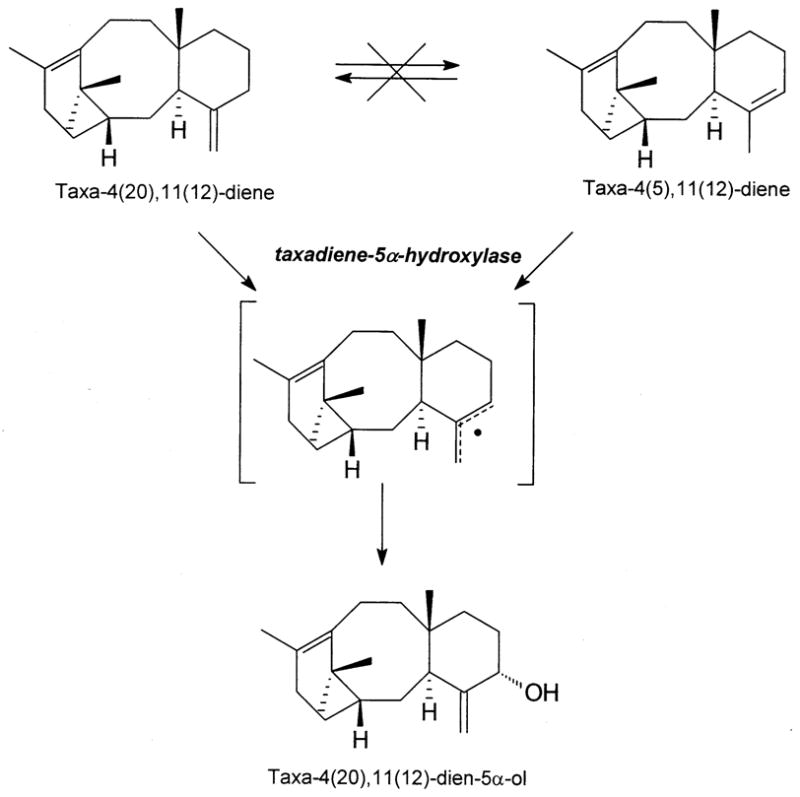
Proposed mechanism for cytochrome P450-mediated conversion of taxa-4(5),11(12)-diene and taxa-4(20),11(12)-diene to taxa-4(20),11(12)-dien-5α-ol involving hydrogen abstraction at C5 or C20 to provide the common allylic radical, followed by oxygen insertion at the C5α-face. Taxa-4(5),11(12)-diene and taxa-4(20),11(12)-diene isomerization was not observed (Jennewein et al. 2004)
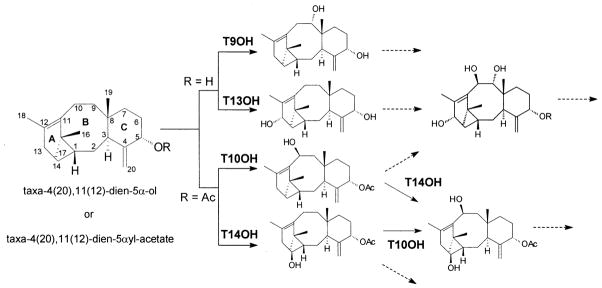
The taxoid 13α-hydroxylase (Jennewein et al. 2001) and the 10β-hydroxylase (Schoendorf et al. 2001) were acquired by DD-RT-PCR, and the recombinant enzymes possess properties typical of the taxane oxygenases. Both enzymes show complementary but opposite substrate selectivities; the 13α-hydroxylase favors taxadien-5α-ol as substrate whereas the 10β-hydroxylase prefers taxadien-5α-acetate but both enzymes accept the other substrate with lower efficiency. These subtly distinct selectivities and the regiospecifities, as well as feeding studies of Taxus cells with the relevant intermediates (Ketchum and Croteau 2006), suggest a bifurcation in the early stages of Taxol biosynthesis (Fig. 3). This early pathway divergence via taxadien-5α,13α-diol or taxadien-5α-acetoxy-10β-ol could in part explain the large number of taxoid intermediates involved in multiple anastomosing routes to Taxol, a process further complicated by competing side routes (e.g. to the 14β-hydroxy taxanes, see below). The oxygenation at C9 presumably also occurs early in the pathway (Floss and Mocek 1995). A cDNA clone for the taxoid 9α-hydroxylase has been tentatively identified by in vivo feeding of yeast transformed with candidate P450 sequences using taxadien-5α-ol as substrate, but product identity has not yet been confirmed by NMR due to insufficient amounts of this material (Croteau et al. 2006).
Fig. 3.
Early bifurcation of the Taxol biosynthetic pathway following the 5α-hydroxylation step, and involving the taxoid 13α- (T13OH), 10β- (T10OH), 14β- (T14OH) and presumptive 9α- (T9OH) hydroxylases. The broken arrows indicate subsequent, but as yet undefined, metabolic steps
A clone for the taxoid 14β-hydroxylase was also discovered by in vivo screening of transformed yeast cells bearing candidate P450s (Jennewein et al. 2003). The encoded enzyme converts only 5α-acetylated derivatives (Fig. 3) to the corresponding 14β-hydroxylated taxadienes, a reaction step which is not part of the Taxol biosynthetic pathway because Taxol is not functionalized at C14. This distinctive substrate recognition feature demonstrates the importance of the 5-acetyl transfer step (Walker et al. 1999) at this early branch point of the pathway (Fig. 3). 14β-Hydroxylase transcripts are of relatively high abundance (1.5‰) in the induced Taxus cell cDNA library (Jennewein et al. 2004b), and this enzyme is clearly responsible for an early, and presumably major, diversion of the pathway towards 14β-hydroxy taxoids (e.g. taxuyunnanine C) which occur as prominent metabolites in Taxus cell cultures (Ketchum et al. 2003). The availability of this cDNA suggests a strategy to suppress 14β-hydroxylase gene expression in order to permit significant redirection of pathway flux towards Taxol.
Intermediate oxygenation steps (C2- and C7-hydroxylases)
The intermediate oxygenation steps of Taxol biosynthesis have been more difficult to approach because the reaction order is not yet precisely known, and highly functionalized taxoid substrates required for the assays are not easily accessible. Taxusin (5α,9α,10β,13α-tetraacetoxy taxa-5(20),11(12)-diene), considered a dead end metabolite of yew heartwood (Koepp et al. 1995) and not an intermediate on the pathway to Taxol, was nevertheless successfully used as a surrogate substrate for the identification of two clones responsible for the respective, regioselective taxoid 2α- (Chau and Croteau 2004) and 7β-hydroxylation steps (Chau et al. 2004). By generating the corresponding hydroxy taxusins using Taxus microsomes, and then demonstrating the reciprocal conversions to the common hexaol tetraacetate (2α-,7β-dihydroxy taxusin) in transformed yeast, it was shown that both enzymes can operate sequentially, with 7β-hydroxylation probably followed by 2α-hydroxylation; the 2α-hydroxylase also utilized taxusin with the lower turnover. The 7β-hydroxylase did not convert less functionalized taxadienes (monool, diol and triol derivatives), and substrate binding studies with the recombinant enzyme (expressed in Spodoptera) demonstrated a stronger preference for peracetylated polyols than for partially or fully deacetylated polyols. Acetylation at the 5α-position significantly promoted tight substrate binding, whereas the influence of 13α-acetylation was less important then 9α- and 10β-acetylation (Chau et al. 2004). The preference for highly functionalized taxadiene substrates clearly suggests roles for the 2α- and 7β-hydroxylases in the central portion of the Taxol biosynthetic pathway.
Characteristics and evolutionally origins of the taxane oxygenases
With an amino acid sequence similarity greater than 70% (Jennewein et al. 2004b), the taxane cytochrome P450 monooxygenases from T. cuspidata are more closely related to each other than to any other known plant cytochrome P450 type. By way of illustrative example, within the cytochrome P450 monooxygenases of steroid biosynthesis (Kagawa and Waterman 1995), those hydroxylases with the same catalytic capability are 68% similar but those with different functions belong to different families (Feldmann et al. 2002; Kim and Tsukaya 2002); the taxoid hydroxylases, with their strict regiochemistries and substrate selectivities, thus form an unusually cohesive group. The taxane hydroxylases genes were sorted by the generally accepted nomenclature (Nelson 1999) into the class A-type genes encoding proteins responsible for secondary metabolite generation, and the subfamily CYP725 was created in which the genes for the 10β(A1)-, 13α(A2)-, 14β(A3)-, and the 5α- and 7β-hydroxylases (A-like) are included (http://www.members.shaw.ca/P450sinPlants/P450s_in_Other_Plants.html); the 2α-hydroxylase should also now be added to this subfamily. Phylogenetic considerations demonstrate the tight coherence of the taxoid hydroxylases and the distant relationship to other plant cytochrome P450s (< 35% similarity) by placing them all on the same branch with the catalytically similar diterpenoid hydroxylase abietadienol/abietadienal oxidase from loblolly pine (Pinus taeda) (Fig. 4), whereas the evolutionary distance from other, seemingly similar terpenoid oxygenases, such as the steroid hydroxylases, is remarkably great.
Fig. 4.
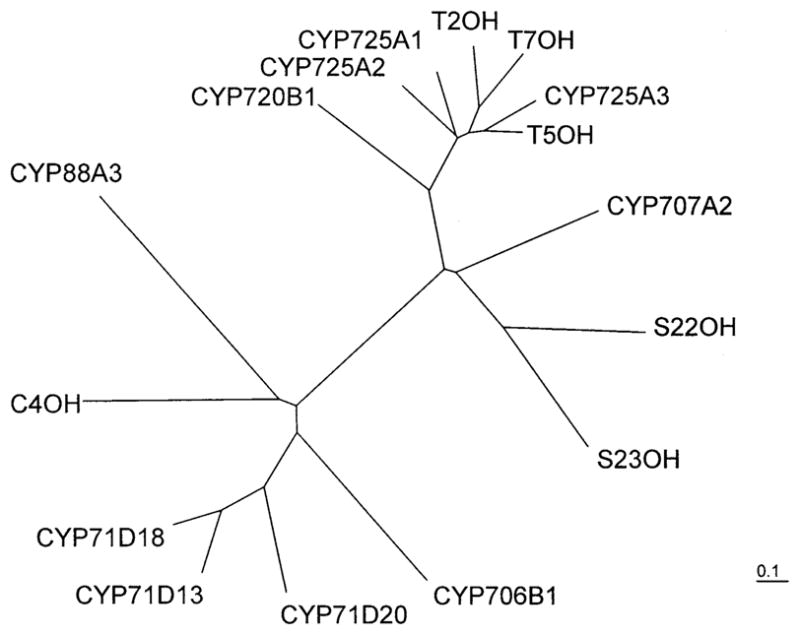
Phylogenetic distance tree of taxoid and related cytochrome P450 monooxygenases generated by sequence alignment with Pileup (Genetics Computing Group Version 11.0), manual editing and conversion by Genedoc (2.6.002), and using ClustalW (neighbor joining, distance corrected), and Treeview 1.6.6 for displaying the tree (CYP725A1: taxoid-10β-hydroxylase [GENBANK accession number: AF318211]; CYP725A2: taxoid-13α-hydroxylase [AY056019]; CYP725A3: taxoid-14β-hydroxylase [AY188177]; T2OH: taxoid-2α-hydroxylase [AY518383]; T5OH: taxadiene-5α-hydroxylase [AY364469]; T7OH: taxoid-7β-hydroxylase [AY307951]; CYP720B1: abietadienol/abietadienal oxidase [AY779537-40]; S23OH: steroid-23α-hydroxylase [Zinnia elegans, AB231154]; S22OH: steroid-22α-hydroxylase [Z. elegans, AB231155]; CYP707A2: abscissic acid-8′-hydroxylase [DQ206631]; CYP88A3: ent-kaurene oxidase [AF047719.1]; C4OH: cinnamic acid-4-hydroxylase [Z32563]; CYP706B1: (+)-δ-cadinene-8-hydroxylase [AF332974]; CYP71D20: 5-epiaristolochene-1,3-dihydroxylase [AF368376]; CYP71D13: limonene-3-hydroxylase [AY281027.1]; CYP71D18: limonene-6-hydroxylase [AY281025.1]
Two full-length cDNAs were obtained recently from Taxus chinensis (Tu et al. 2004) that closely resemble the previously cloned (Schoendorf et al. 2001) 10β-hydroxylase (92% identical) and the previously described (Chau et al. 2004) 7β-hydroxylase from T. cuspidata (97% identical). BLAST database searching (Altschul et al. 1997) revealed several entries for presumptive 2α-, 5α-, and 13α-hydroxylases from T. chinensis and for presumptive 10β- and 13α-hydroxylases from T. x media that were deposited by Kai and coworkers. The 13α-hydroxylase sequences of T. cuspidata and T. x media differ in only one deduced amino acid (99% identity), and both are 96% similar to the 13α-hydroxylase from T. chinensis. The great similarity between homologous Taxol biosynthetic genes from various Taxus species has been previously noted (Jennewein et al. 2004b); for example, the taxadiene synthase sequences from T. brevifolia, T. cuspidata and T. chinensis are virtually identical. The production of Taxol by a fungal symbiont of the Pacific yew (T. brevifolia) was reported over a decade ago (Stierle et al. 1995), and recently, a presumptive taxoid 10β-hydroxylase sequence from a Botrytis sp. symbiont of T. chinensis var mairei was deposited by Guo and coworkers. The presumptive fungal gene is essentially identical to the taxoid 10β-hydroxylase of T. chinensis from which the fungus was isolated. Should the fungal gene prove not to be an artifact of plant origin, the observation would raise some very interesting questions about the timing and mechanism of acquisition of the symbiont gene from the yew host. Our own attempts to isolate taxadiene synthase fungal homologues from various yew symbionts have proved fruitless (Croteau unpublished).
Although it is obvious that these new cytochrome P450 sequences should be placed within the CYP725 subfamily, it has been generally advised (Chapple 1998) that in vivo functions should not be ascribed to such newly identified genes based on sequence alone, and that confirmatory expression studies are required to establish catalytic capabilities.
The structural basis underlying functional diversity of the taxoid hydroxylases is not understood. Differences in regiochemistry and substrate selectivity almost certainly reside in differences in substrate recognition sites (Gotoh 1992) [some of which have been described (Chau and Croteau 2004)], substrate entry channels (Wade et al. 2004), and reductase binding orientations (Ohta and Mizutami 2004) but these structural differences do not readily lend themselves to the prediction (or rationalization) of observed catalytic differences by homology modeling (Poulus and Johnson 2005). Interestingly, the 14β-hydroxylase, which oxygenates the taxane A-ring, displays higher similarity to the 10β-hydroxylase for B-ring functionalization than to the 13β-hydroxylase which is also directed to A-ring functionalization. This unexpected observation indicates the near impossibility of predicting hydroxylation regiochemistry based solely on homology (Jennewein et al. 2003). The taxane oxygenases, such as the 13α-, and 10β-hydroxylases, also lack absolute specificity in substrate utilization (both alcohols and acetate esters are used). This feature is not uncommon for P450 enzymes of plant secondary metabolism (Schuler and Werck-Reichhart 2003), has been observed for other terpene oxygenases, such as the abietadienol/abietadienal oxidase (Ro et al. 2005), limonene hydroxylase (Wüst et al. 2001) and kaurene oxidase (Helliwell et al. 1999), and further complicates the attempt to understand structure-function relationships. Neither mutagenesis nor crystallographic studies have yet been attempted with the taxoid hydroxylases.
A variety of taxoids have antifeedant (Daniewski et al. 1998) or antibiotic activity (Elmer et al. 1994), or are toxic to mammals, and thus are thought to play a role in plant defense (Odgen 1988). Taxoid hydroxylase genes of yew are inducible by treatment with methyl jasmonate (Ketchum et al. 1999) or fungal elicitors (Yuan et al 2002; Chen et al. 1999), consistent with a role in defense and suggestive that this evolutionary pressure has driven taxoid metabolic complexity in this long-lived species. It seems reasonable to assume that the taxoid hydroxylases derived from a common progenitor by gene duplication and differentiation (Pichersky and Gang 2000) to evolve alternative substrate selectivities and stereo-/regio-selectivities of the hydroxylation reactions on the taxadiene core. Because the 5α-hydroxylase represents the initial oxygenation of the committed taxadiene precursor, this gene can be regarded as the parental sequence from which other hydroxylase genes evolved in a pattern reflecting the order of hydroxylations in the Taxol biosynthetic pathway.
To evaluate this possibility, a cladogram of the taxoid oxygenases from T. cuspidata was constructed with the 5α-hydroxylase as root (Jennewein et al. 2005). The pattern, based on sequence relatedness, is consistent with the preliminary hydroxylation steps leading from C5 to C10 hydroxylation (67% sequence identity to the 5α-hydroxylase) or C13 hydroxylation (63% identity), and with early emergence of C14 hydroxylation (70% identity) as a major side-route after C5 hydroxylation. The 2α- and 7β-hydroxylases are more similar to each other (65% identity) than to the early pathway hydroxylases (e.g. 55%: identity to the 10β-hydroxylase). Nevertheless, phylogenetic considerations place the taxoid 7β-hydroxylase and 2α-hydroxylase (which only hydroxylate highly functionalized taxadienes) closer to the 5α-hydroxylase (62–63% identity) than might have been anticipated based on the proposed sequence of hydroxylations deduced from the relative abundances of taxoid metabolites functionalized at the various positions (Floss and Mocek 1995) but in agreement with recent experimental data (see 3.2) for the relative placement of the 7β-hydroxylase (Chau and Croteau 2004; Chau et al. 2004). With at least two pathway oxygenases that operate on the taxane core still unidentified, and with many of the cytochrome P450 genes of still uncertain function, this preliminary phylogeny of the pathway oxygenases is very tentative.
A second taxoid 10β-hydroxylase was subsequently discovered by screening cytochrome P450 acquisitions from the induced Taxus cell EST library (Jennewein et al. 2004b). This second 10β-hydroxylase is too distant from the first (Schoendorf et al. 2001) to be considered an allelic variant (68% identity), thus further supporting an extended phylogeny for this gene family and, more importantly, indicating catalytic redundancy in at least one pathway hydroxylation step. The enzymatic redundancy in at least one case, the above-mentioned multiple routes from taxadienol, and the observed plasticity in substrate utilization by some hydroxylases suggest that the Taxol biosynthetic pathway is not as linear as initially imagined but likely involves multiple routes diverging early in the pathway and converging at a late stage intermediate (Croteau et al. 2006). The cytochrome P450 taxoid hydroxylases can be seen to play a central role in Taxol biosynthesis and a major role in diversification of the pathways to other metabolites.
Remaining oxygenation steps
Functionalization of the taxane core to baccatin III
The C1β-hydroxylation and the C4,C20β-epoxidation leading to oxetane ring formation are two remaining steps on route to the intermediate bacccatin III (Fig. 1). Both reactions occur relatively late in the pathway, prior to C13-side chain transfer, and they presumably are mediated by cytochrome P450 monooxygenases. The responsible genes likely reside in the family of unassigned cytochrome P450 clones and await the determination of function by screening with appropriate substrates or surrogates. The general approach to these genes, and specific assay design, are constrained by uncertainties in the precise timing of these reactions in the pathway. Relatively few diagnostic metabolites (i.e. advanced polyoxygenated taxoids bearing the C4,20-methylene or β-epoxide, and devoid of the 1β-hydroxyl function) accumulate in Taxus cell cultures (Ketchum and Croteau 2006) to assist in predicting the sequence of reactions (or to provide ready access to relevant substrates). Nevertheless, the available evidence suggests that C4,C20-epoxidation occurs at the level of a taxadien-hexaol (of an uncertain degree of acetylation/benzoylation), and is followed by ring expansion to the oxetane (perhaps by a transferase-type mechanism) and finally hydroxylation at C1.
Oxidation of the C9-hydroxyl group to the corresponding carbonyl also likely occurs after oxetane ring formation but the timing with respect to C1-hydroxylation is less clear (Floss and Mocek 1995). This reaction could be mediated by a cytochrome P450 monooxygenase via the transient ketone hydrate, a mechanism similar to that involved in diterpene resin acid formation in gymnosperms (Funk and Croteau 1994; Ro et al. 2005). The reaction might also be catalyzed by a pyridine nucleotide-dependent dehydrogenase, for which the EST acquisitions from T. cuspidata have revealed many possible candidates. Because this step occurs relatively late in the pathway, uncertainties in the precise timing similarly constrain substrate design for the required assay. It is clear that cell-free biochemical studies with suitable substrates must precede a cloning effort for the C9-oxidase and for the epoxidase as well, since it is conceivable that the latter reaction could be conducted not only by a cytochrome P450 but also by a flavin-dependent monooxygenase like squalene epoxidase (Abe and Prestwich 1999).
2′-Hydroxylation of the C13-side chain
The order of the construction steps in C13-side chain assembly, particularly the placement of the 2′-hydroxylation, has been one of the more difficult parts of the Taxol pathway to elucidate. It is clear that the process begins with the aminomutase-mediated conversion of α-phenylalanine to β-phenylalanine (Walker et al. 2004) and that the side chain precursor (as the CoA ester) is transferred to baccatin III (Walker et al. 2002); however, it was not clear whether 2′-hydroxylation occurred before or after transfer of the amino phenylpropanoid side chain. The recent demonstration of the cytochrome P450-mediated conversion of β-phenylalanoyl baccatin III (N-debenzoyl-2′-deoxy Taxol), previously reported as a minor metabolite of Taxus (Baloglu and Kingston 1999; Itokawa 2003), to 3-phenylisoserinoyl baccatin III (N-debenzoyl taxol) in Taxus cell microsomes (Long and Croteau 2005) now strongly implies the involvement of a cytochrome P450 monooxygenase in 2′-hydroxylation following the transfer of the amino phenylpropanoid side chain (i.e., as β-phenylalanoyl CoA rather than as isoserinoyl CoA). This observation is consistent with the fact that all other oxygenations of the Taxol pathway are seemingly mediated by cytochrome P450 enzymes (Croteau et al. 2006) and suggests that the 2′-hydroxylase gene will be found within the family of Taxus cytochrome P450 cDNAs now available (Croteau et al. 2006; Jennewein et al. 2004b). Side chain assembly can now be most reasonably formulated as involving the formation of β-phenylalanoyl CoA and acyl transfer to baccatin III, followed by 2′-hydroxylation and N-benzoylation of the appended side chain to complete the pathway to Taxol.
Conclusion
The taxoid cytochrome P450 oxygenases represent a particularly interesting subfamily of plant terpenoid hydroxylases because of their remarkable structural similarity, yet distinct catalytic capabilities, and their probable sequential evolution during the development of the Taxol biosynthetic pathway. The complexity and length of the pathway, and the difficulty in accessing appropriate substrates for use in biochemical studies and cDNA functional screening, have presented major challenges in searching for missing oxygenation steps. The use of surrogate substrates, such as taxusin, in the functional screening of cytochrome P450 clones has proved to be productive in the elucidation of the intermediate 2α- and 7β-hydroxylation pathway steps. Other surrogate substrates, such as baccatin I, 1-dehydroxy baccatin VI and acetylated taxadien polyols prepared from Taxus cell extracts, although not likely to reside on the pathway to Taxol, could be useful in identifying P450 clones responsible for the remaining taxane core oxygenations. Taxoid hydroxylase genes have been used in a preliminary attempt to reconstruct the early section of the Taxol pathway in yeast (DeJong et al. 2006), and completion of cloning of the remaining genes of this extended pathway should allow transgenic manipulation of Taxus cell cultures to increase production yields and, potentially, may provide the biotechnical means to generate tailor-made taxoids.
The demonstration that recombinant taxoid hydroxylases are capable of using more than one substrate suggests similar plasticity in substrate utilization in planta, which implies highly complex relationships between these heme proteins and their potential substrates. It now seems reasonable to assume that the pathway to Taxol is not linear but rather a metabolic grid of permissible reactions, with perhaps several common nodes on the way to the final product. Because of the very large number of defined taxoids, and the fact that only 19 enzymatic steps are required for the conversion of geranylgeranyl diphosphate to Taxol, several side routes, and diversions to taxines, taxinines, taxayunnanines and other dead-end metabolites, must exist (Chau and Croteau 2004). Thus, the substrate selectivities of these taxoid hydroxylases, and of the intervening transferases, must play a central role in structural diversification of the taxoid family of natural products.
Acknowledgments
We thank Christopher J.D. Mau and Raymond E.B. Ketchum for critical reading of the manuscript, and Robert M. Long for helpful discussions. The work from the authors’ laboratory described in this paper was supported by National Institutes of Health Grant CA-55254 and by Project 0967 from The Agricultural Research Center, Washington State University.
References
- Abe I, Prestwich GD. Squalene epoxidase and oxidosqualene: lanosterol cyclase-key enzymes in cholesterol biosynthesis. In: Cane DE, editor. Comprehensive natural product chemistry. Elsevier; Amsterdam, NL: 1999. pp. 267–298. [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucl Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baloglu E, Kingston DGI. The taxane diterpenoids. J Nat Prod. 1999;62:1448–1472. doi: 10.1021/np990176i. [DOI] [PubMed] [Google Scholar]
- Brown DT. Preclinical and clinical studies of the taxanes. In: Itokawa H, Lee KH, editors. Taxus – The Genus Taxus. Taylor & Francis; London. UK: 2003. pp. 387–435. [Google Scholar]
- Chapple C. Molecular-genetic analysis of plant cytochrome P450-dependent monooxygenases. Ann Rev Plant Physiol Plant Mol Biol. 1998;49:311–343. doi: 10.1146/annurev.arplant.49.1.311. [DOI] [PubMed] [Google Scholar]
- Chau M, Croteau R. Molecular cloning and characterization of a cytochrome P450 taxoid 2α-hydroxylase involved in Taxol biosynthesis. Arch Biochem Biophys. 2004;427:48–57. doi: 10.1016/j.abb.2004.04.016. [DOI] [PubMed] [Google Scholar]
- Chau M, Jennewein S, Walker K, Croteau R. Taxol biosynthesis: molecular cloning and characterization of a cytochrome P450 taxoid 7β-hydroxylase. Chem Biol. 2004;11:663–672. doi: 10.1016/j.chembiol.2004.02.025. [DOI] [PubMed] [Google Scholar]
- Chen Y, Zhu W, Wu Y, Hu Q. Effects of fungus elicitors on taxol production in suspension cells of Taxus yunnanensis. Shengwu Gongcheng Xuebao. 1999;15:522–524. [Google Scholar]
- Croom EM., Jr . Taxus for taxol and taxoids. In: Suffness M, editor. Taxol – science and applications. CRC Press; Boca Raton, USA: 1995. pp. 37–70. [Google Scholar]
- Croteau R, Ketchum REB, Long RM, Kaspera R, Wildung MR. Taxol biosynthesis and molecular genetics. Phytochem Rev. 2006 doi: 10.1007/s11101-005-3748-2. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniewski WM, Gumulka M, Anczemski M, Masnyk M, Bioszyk E, Gupta KK. Why the yew tree (Taxus baccata) is not attacked by insects. Phytochemistry. 1998;49:1279–1282. [Google Scholar]
- De Jong JM, Liu Y, Bollon AP, Long RM, Jennewein S, Williams D, Croteau R. Genetic engineering of Taxol biosynthetic genes in Saccharomyces cerevisiae. Biotechnol Bioeng. 2006;93:212–224. doi: 10.1002/bit.20694. [DOI] [PubMed] [Google Scholar]
- Elmer WH, Mattine MJI, MacEachern GI. Sensitivity of plant pathogenic fungi to taxane extracts from ornamental yews. Phytopathology. 1994;84:1179–1185. [Google Scholar]
- Feldmann KA, Choe S, Kim H, Park J-H. Functional genomics of cytochromes P450 in plants. Rec Adv Phytochem. 2002;36:125–143. [Google Scholar]
- Floss HG, Mocek U. Biosynthesis of taxol. In: Suffness M, editor. Taxol – science and applications. CRC Press; Boca Raton, USA: 1995. pp. 191–208. [Google Scholar]
- Funk C, Croteau R. Diterpenoid resin acid biosynthesis in conifers – characterization of two cytochrome P450-dependent monooxygenases and an aldehyde dehydrogenase involved in abietic acid biosynthesis. Arch Biochem Biophys. 1994;308:258–266. doi: 10.1006/abbi.1994.1036. [DOI] [PubMed] [Google Scholar]
- Gibson DM, Ketchum REB, Hirasuna TJ, Shuler ML. Potential of plant cell cultures for taxane production. In: Suffness M, editor. Taxol – Science and Applications. CRC Press; Boca Raton, USA: 1995. pp. 71–95. [Google Scholar]
- Gotoh O. Substrate recognition sites in cytochrome P450 Family 2 (CYP2) proteins inferred from comparative analyses of amino acid and coding nucleotide sequences. J Biol Chem. 1992;267:83–90. [PubMed] [Google Scholar]
- Hefner J, Rubenstein SM, Ketchum REB, Gibson DM, Williams RM, Croteau R. Cytochrome P450-catalyzed hydroxylation of taxa-4(5),11(12)-diene to taxa-4(20),11(12)-dien-5α-ol: the first oxygenation step in taxol biosynthesis. Chem Biol. 1996;3:479–489. doi: 10.1016/s1074-5521(96)90096-4. [DOI] [PubMed] [Google Scholar]
- Helliwell CA, Poole A, Peacock J, Dennis ES. Arabidopsis ent-kaurene oxidase catalyzes three steps of gibberellin biosynthesis. Plant Physiol. 1999;119:507–510. doi: 10.1104/pp.119.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hezari M, Lewis NG, Croteau R. Purification and characterization of taxa-4(5),11(12)-diene synthase from Pacific yew (Taxus brevifolia) that catalyzes the first commited step of Taxol biosynthesis. Arch Biochem Biophys. 1995;322:437–444. doi: 10.1006/abbi.1995.1486. [DOI] [PubMed] [Google Scholar]
- Itokawa H. Taxoids occuring in the genus Taxus. In: Itokawa H, Lee KH, editors. Taxus – The Genus Taxus. Taylor & Francis; London, UK: 2003. pp. 35–78. [Google Scholar]
- Jennewein S, Long R, Williams RM, Croteau R. Cytochrome P450 taxadien 5α-hydroxylase, a mechanistically unusual monooxygenase catalyzing the first oxygenation step of Taxol biosynthesis. Chem Biol. 2004a;11:379–387. doi: 10.1016/j.chembiol.2004.02.022. [DOI] [PubMed] [Google Scholar]
- Jennewein S, Park H, DeJong JM, Long RM, Bollon AP, Croteau RB. Coexpression in yeast of Taxus cytochrome P450 reductase with cytochrome P450 oxygenases involved in Taxol biosynthesis. Biotechnol Bioeng. 2005;89:588–598. doi: 10.1002/bit.20390. [DOI] [PubMed] [Google Scholar]
- Jennewein S, Rithner CD, Williams RM, Croteau R. Taxol biosynthesis: taxane 13α-hydroxylase is a cytochrome P450-dependent monooxygenase. Proc Natl Acad Sci USA. 2001;98:13595–13600. doi: 10.1073/pnas.251539398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennewein S, Rither CD, Williams RM, Croteau R. Taxoid metabolism: taxoid 14β-hydroxylase is a cytochrome P450-dependent monooxygenase. Arch Biochem Biophys. 2003;413:262–270. doi: 10.1016/s0003-9861(03)00090-0. [DOI] [PubMed] [Google Scholar]
- Jennewein S, Wildung MR, Chau M, Walker K, Croteau R. Random sequencing of an induced Taxus cell cDNA library for identification of clones involved in Taxol biosynthesis. Proc Natl Acad Sci USA. 2004b;101:9149–9154. doi: 10.1073/pnas.0403009101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagawa N, Waterman MR. Regulation of steroidogenic and related P450s. In: Ortiz de Montellano PR, editor. Cytochrome P450: structure, mechanism, and biochemistry. Plenum Press; New York, USA: 1995. pp. 419–442. [Google Scholar]
- Kahn RA, Durst F. Function and evolution of plant cytochrome P450. Rec Adv Phytochem. 2000;34:151–189. [Google Scholar]
- Ketchum REB, Croteau R. The Taxus metabolome and the elucidation of the Taxol® biosynthetic pathway in cell suspension cultures. In: Saito K, Dixon R, Willmetzer L, editors. Plant metabolomics (biotechnology in agriculture and forestry) Vol. 57. Springer; Heidelberg, D: 2006. pp. 291–309. [Google Scholar]
- Ketchum REB, Rithner CD, Qiu D, Kim YS, Williams RM, Croteau RB. Taxus metabolomics: methyl jasmonate preferentially induces production of taxoids oxygenated at C-13 in Taxus × media cell cultures. Phytochemistry. 2003;62:901–909. doi: 10.1016/s0031-9422(02)00711-2. [DOI] [PubMed] [Google Scholar]
- Ketchum REB, Tandon M, Gibson DM, Begley T, Shuler ML. Isolation of labeled 9-dihydrobaccatin III and related taxoids from cell cultures of Taxus canadensis elicited with methyl jasmonate. J Nat Prod. 1999;62:1395–1398. doi: 10.1021/np990201k. [DOI] [PubMed] [Google Scholar]
- Kikuchi Y, Yatagai M. The commercial cultivation of Taxus species and production of taxoids. In: Itokawa H, Lee KH, editors. Taxus – The genus Taxus. Taylor & Francis; London, UK: 2003. pp. 151–178. [Google Scholar]
- Kim G-T, Tsukaya H. Regulation of the biosynthesis of plant hormones by cytochrome P450s. J Plant Res. 2002;115:169–177. doi: 10.1007/s102650200022. [DOI] [PubMed] [Google Scholar]
- Kingston DGI. Recent advances in the chemistry and structure-activity relationships of paclitaxel. Am Chem Soc Symp Ser. 1995;583:203–216. [Google Scholar]
- Koepp AE, Hezari M, Zajicek J, Vogel BS, Lafever RE, Lewis NG, Croteau R. Cyclization of geranylgeranyl diphosphate to taxa-4(5),11(12)-diene is the committed step of Taxol biosynthesis in Pacific yew. J Biol Chem. 1995;270:8686–8690. doi: 10.1074/jbc.270.15.8686. [DOI] [PubMed] [Google Scholar]
- Kutchan TM, Bock A, Dittrich H. Heterologous expression of the plant proteins strictosidine synthase and berberine bridge enzyme in insect cell culture. Phytochemistry. 1994;35:353–360. doi: 10.1016/s0031-9422(00)94763-0. [DOI] [PubMed] [Google Scholar]
- Long RM, Croteau R. Preliminary assessment of the C13-side chain 2′-hydroxylase involved in Taxol biosynthesis. Biochem Biophys Res Comm. 2005;338:410–417. doi: 10.1016/j.bbrc.2005.08.119. [DOI] [PubMed] [Google Scholar]
- Lovy Wheeler A, Long RM, Ketchum REB, Rither CD, Williams RM, Croteau R. Taxol biosynthesis: differential transformations of taxadien-5α-ol and its acetate ester by cytochrome P450 hydroxylases from Taxus suspension cultures. Arch Biochem Biophys. 2001;390:265–278. doi: 10.1006/abbi.2001.2377. [DOI] [PubMed] [Google Scholar]
- McCoy M. Lining up to make a cancer drug. Chem Eng News. 2004;82:12–14. [Google Scholar]
- Nelson DR. Cytochrome P450 and the individuality of species. Arch Biochem Biophys. 1999;369:1–10. doi: 10.1006/abbi.1999.1352. [DOI] [PubMed] [Google Scholar]
- Odgen L. Taxus (Yews) - A highly toxic plant. Vet Hum Toxicol. 1998;30:563–564. [PubMed] [Google Scholar]
- Ohta D, Mizutani M. Redundancy or flexibility: molecular diversity of the electron transfer components for P450 monooxygenases in higher plants. Front Biosci. 2004;9:1587–1597. doi: 10.2741/1356. [DOI] [PubMed] [Google Scholar]
- Pichersky E, Gang DR. Genetics and biochemistry of secondary metabolites in plants: an evolutionary perspective. Trends Plant Sci. 2000;5:439–445. doi: 10.1016/s1360-1385(00)01741-6. [DOI] [PubMed] [Google Scholar]
- Pompon D, Louerat B, Bronine A, Urban P. Yeast expression of animal and plant P450s in optimized redox environments. Methods Enzymol. 1996;272:51–64. doi: 10.1016/s0076-6879(96)72008-6. [DOI] [PubMed] [Google Scholar]
- Poulos TL, Johnson EF. Structures of cytochrome P450 enzymes. In: Ortiz de Montellano PR, editor. Cytochrome P450: structure, mechanism, and biochemistry. Kluwer; New York, USA: 2005. pp. 87–114. [Google Scholar]
- Ro DK, Arimura GL, Lau SYW, Piers E, Bohlmann J. Loblolly pine abietadienol/abietadienal oxidase PtAO (CYP720B1) is a multifunctional, multi-substrate cytochrome P450 monooxygenase. Proc Natl Acad Sci USA. 2005;102:8060–8065. doi: 10.1073/pnas.0500825102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoendorf A, Rither CD, Williams RM, Croteau RB. Molecular cloning of a cytochrome P450 taxane 10β-hydroxylase cDNA from Taxus and functional expression in yeast. Proc Natl Acad Sci USA. 2001;98:1501–1506. doi: 10.1073/pnas.98.4.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler MA. Plant cytochrome P450 monooxygenases. Crit Rev Plant Sci. 1996;15:235–284. [Google Scholar]
- Schuler MA, Werck-Reichhart D. Functional genomics of P450s. Ann Rev Plant Biol. 2003;54:629–667. doi: 10.1146/annurev.arplant.54.031902.134840. [DOI] [PubMed] [Google Scholar]
- Stierle A, Strobel G, Stierle D, Grothaus P, Bignami G. The search for a Taxol-producing microorganism among the endophytic fungi of the Pacific yew, Taxus brevifolia. J Nat Prod. 1995;58:1315–1324. doi: 10.1021/np50123a002. [DOI] [PubMed] [Google Scholar]
- Suffness M, Wall ME. Discovery and development of taxol. In: Suffness M, editor. Taxol – science and applications. CRC Press; Boca Raton, USA: 1995. pp. 3–25. [Google Scholar]
- Takeya K. Plant tissue culture of taxoids. In: Itokawa H, Lee KH, editors. Taxus – The genus Taxus. Taylor & Francis; London, UK: 2003. pp. 134–150. [Google Scholar]
- Tu J, Zhu P, Cheng KD, Meng C. Cloning and sequencing of hydroxylase genes involved in taxol biosynthesis. Z Naturforsch. 2004;59c:561–564. doi: 10.1515/znc-2004-7-820. [DOI] [PubMed] [Google Scholar]
- von Wachenfeldt CA, Johnson EF. Structures of eucaryotic cytochrome P450 enzymes. In: Ortiz de Montellano PR, editor. Cytochrome P450: structure, metabolism, and biochemistry. Plenum Press; New York USA: 1995. pp. 183–223. [Google Scholar]
- Wade RC, Winn PJ, Schlichting I, Sudarko A survey of active site access channels in cytochrome P450. J Inorgan Biochem. 2004;98:1175–1182. doi: 10.1016/j.jinorgbio.2004.02.007. [DOI] [PubMed] [Google Scholar]
- Walker K, Ketchum REB, Hezari M, Gatfield D, Goleniowski M, Barthol A, Croteau R. Partial purification and characterization of acetyl coenzyme A: taxa-4(20),11(12)-dien-5α-ol O-acetyl transferase that catalyzes the first acylation step of Taxol biosynthesis. Arch Biochem Biophys. 1999;364:273–279. doi: 10.1006/abbi.1999.1125. [DOI] [PubMed] [Google Scholar]
- Walker K, Fujisaki S, Long R, Croteau R. Molecular cloning and heterologous expression of the C-13 phenylpropanoid side chain-CoA acyltransferase that functions in Taxol biosynthesis. Proc Nat Acad Sci USA. 2002;99:12715–12720. doi: 10.1073/pnas.192463699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker KD, Klettke K, Akiyama T, Croteau R. Cloning, heterologous expression, and characterization of a phenylalanine aminomutase involved in taxol biosynthesis. J Biol Chem. 2004;279:53947–53954. doi: 10.1074/jbc.M411215200. [DOI] [PubMed] [Google Scholar]
- Wall ME. Camptothecin and taxol: discovery to clinic. Med Res Rev. 1998;18:299–314. doi: 10.1002/(sici)1098-1128(199809)18:5<299::aid-med2>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- Wang X, Itokawa H, Lee K-H. Structure-activity relationships of taxoids. In: Itokawa H, Lee KH, editors. Taxus – The Genus Taxus. Taylor & Francis; London, UK: 2003. pp. 298–386. [Google Scholar]
- Wüst M, Little DB, Schalk M, Croteau R. Hydroxylation of limonene enantiomers and analogs by recombinant (−)-limonene 3- and 6-hydroxylases from mint (Mentha) species: evidence for catalysis within sterically constrained active sites. Arch Biochem Biophys. 2001;387:125–136. doi: 10.1006/abbi.2000.2248. [DOI] [PubMed] [Google Scholar]
- Yuan Y-J, Li C, Hu Z-D, Wu J-C. A double oxidative burst for taxol production in suspension cultures of Taxus chinensis var. mairei induced by oligosaccharide from Fusarium oxysporum. Enz Microbial Technol. 2002;30:774–778. [Google Scholar]